Abstract
This study monitored changes in bacterial and fungal structure in a mine water in a monthly basis over 4 months. Over the 4-month study period, mine water samples contained more bacteria (91.06 %) compared to fungi (8.94 %). For bacteria, mine water samples were dominated by Proteobacteria (39.14 to 65.06 %) followed by Firmicutes (26.34 to 28.9 %) in summer, and Cyanobacteria (27.05 %) in winter. In the collected samples, 18 % of bacteria could not be assigned to a phylum and remained unclassified suggesting hitherto vast untapped microbial diversity especially during winter. The fungal domain was the sole eukaryotic microorganism found in the mine water samples with unclassified fungi (68.2 to 91 %) as the predominant group, followed by Basidiomycota (6.9 to 27.8 %). The time of collection, which was linked to the weather, had higher impact on bacterial community than fungal community. The bacterial operational taxonomic units (OTUs) ranged from 865 to 4052 over the 4-month sampling period, while fungal OTUs varied from 73 to 249. The diversity indices suggested that the bacterial community inhabiting the mine water samples were more diverse than the fungal community. The canonical correspondence analysis (CCA) results highlighted that the bacterial community variance had the strongest relationship with water temperature, conductivity, pH, and dissolved oxygen (DO) content, as compared to fungi and water characteristics, had the greatest contribution to both bacterial and fungal community variance. The results provided the relationships between microbial community and environmental variables in the studied mining sites.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Water is the most important of all elements sustaining life and without which nothing will live. However, the quality of this precious resource is function of both natural and human influences (Kamika and Momba 2014). The decline of the quality of water has become a global concern due to fast population growth, drastic industrial and agricultural activities, as well as climate change (Kamika et al. 2014). In South Africa, as elsewhere in the world, human activities such as mining have disrupted natural environment by releasing large amount of pollutants into surface and ground water (Xiao et al. 2009). This became alarming as over 80 % of polluted wastewater from both industrial and domestic sources is discharged untreated into water bodies (Corcoran et al. 2010).
Mining activities have long been reported to cause dramatic impacts on water quality (Sekar et al. 2014). This mine water effluent at its acidic form has dramatically increased the level of toxic metals to at least 100 times more than natural sources for lead, cadmium, and zinc (Roane and Peper 2000). Public complaints have often been heard about mining activities causing pollution and public health concerns to both humans and animals (Kamika and Momba 2014). However, the severity of this pollution is highly variable and dependent on many factors such as the nature of the ore body and associated geological strata, climate, and mining engineering constraints (Johnson and Hallberg 2003).
While this acidic mine water tend to be toxic to most living beings, a number of microorganisms have been found able to grow in such environments as they have developed several adaptive mechanisms to such environment (efflux, complexation, etc.) (Nies and Silver 1995). These adaptive mechanisms have enabled the microorganisms to colonize a large range of environments by being directly or indirectly involved in many chemical alterations and mineral decompositions (Sánchez-Andrea et al. 2014). However, their diversity is influenced by the prevailing physicochemical conditions of each mine water restricting them to a fewer prokaryotic species (Johnson and Hallberg 2005). Of these microorganisms, some have been reported to increase the rate of the formation of acid mine water (Silverman and Ehrlich 1964), while others are indirectly or directly involved in the deposition/precipitation of minerals from acid mine drainage (Bai et al. 2013). According to Johnson and Hallberg (2003), mine waters are mostly populated with bacteria and Archaea having Acidophiles as the most dominant microorganism. Acidophilic microorganisms have been reported to play an important role in processes such as bioleaching (He et al. 2007). Even though extensive investigations have been carried out on the microbial community of various mines around the world (Goebel and Stackebrandt 1994; Baker and Banfield 2003; Johnson and Hallberg 2003; Amaral-Zettler et al. 2011; Kuang et al. 2013), to the best of our knowledge, no study has been carried on bacterial and fungal community of gold mine water in South Africa.
It should also be mentioned that regardless of the fact that fungi have long been recognized as active participants in heavy metal removal from sulfide-rich environments, their diversity from acidic mine water has received less attention when compared to other eukaryotic organisms and prokaryotic organisms (Gross and Robbins 2000; Das et al. 2009). In addition, more studies are still needed as microorganisms in such environment are known to show nonrandom distribution patterns across different habitats at different geographical locations (Kuang et al. 2013). It is widely accepted that approximately 95 % of microorganisms with high physiological and metabolic importance are yet to be discovered and explored in such environment (Hennecke et al. 2013). Recent molecular advances have shown more accuracy in estimating microbial diversity from any environment including acid mine water and this has significantly promoted our understanding of the ecological distribution of multiple genes of several taxonomic groups (Kembel et al. 2011). Therefore, the present study assessed the bacterial as well as fungal composition of acid mine water from a gold mine in South Africa using high throughput pyrosequencing. This was carried out in order to understand the microbial compositions and structures in the East Rand Goldfield environment, South Africa.
Materials and methods
Chemicals
Bacterial DNA isolation kits were supplied by Epigenetics, USA. DreamTaq Green Master Mix for PCR and DNA markers were purchased from Fermentas (Thermo Fisher Scientific, Pittsburgh, PA, USA). TAE and molecular grade agarose were taken from Sigma-Aldrich (Steinheim, Germany).
Sample collection and physicochemical characteristics analysis
Acidic mine water samples (1 L each) were collected in sterile plastic sampling bottles at the month of March, April, June, and July 2014 from the mining effluent of East Rand Goldfield environment, South Africa, and immediately placed in a cooler box (4 °C) for transportation to the laboratory for physicochemical analyses and microbial diversity studies. No specific permit was needed for the collection of the acidic mine water samples in the described sample area and this study did not involve endangered or protected species. For some physicochemical parameters such as dissolved oxygen (DO), pH, electrical conductivity (EC), and temperature were determined on site using multimeter probes (HACH, Germany) while chemical oxygen demand (COD), PO4 −3, NO3 −1 and other metal contents were determined in the laboratory. The mine water samples were first homogenously mixed and filtered using No.1 filter papers (Whatman) prior for the analysis of oxygen demand (COD), nitrate, phosphate and metal contents. The closed reflux method was used to determine COD concentration (APHA 2001), while the concentrations of PO4 −3 and NO3 −1 were determined using standard methods such as method 424f and sodium salicylate method, respectively (Monteiro et al. 2003). Metal contents however, were determined using the inductively couple plasma optical emission spectrometer (ICP-OES) (Spectro Ciros CCD, Spectro Analytical Instruments, Kleve, Germany).
Metagenomic DNA extraction, amplification, and pyrosequencing of bacterial 16S/18S rRNA genes
For molecular analysis, the collected acidic mine water samples were pre-filtered on site through a 1.6-μm GF/A filter in order to remove impurities. As bacterial size vary from 0.3 to 5 μm, further filtration of approximately 1-L pre-filtered acid mine water were done through a 0.22-μm polyethersulfone membrane filter using a peristaltic pump as required to harvest microbial cells. Attached cells from the polyethersulfone membrane filter were suspended in 50 mL 1× PBS buffer, vigorously shacked and centrifuged at 10,000×g for 5 min at 4 °C to collect the cell pellets. Cell pellets were re-suspended in 1× TE buffer (pH 8.0), homogenously mixed, and fungal and bacterial DNA was extracted using the ZR Fungal/Bacterial DNA Kit™ (Zymo Research, Pretoria, South Africa) according to the procedures provided by the manufacturer. Finally, the metagenomic DNA was eluted in 50 μL of MilliQ water then after integrity and purity of the metagenomic DNA assessed on the 1.0 % agarose gel and measured using a NanoDrop spectrophotometer (NanoDrop 2000, Thermo Scientific, Japan). The metagenomic DNA samples were amplified in triplicate using the universal primer 27F and 518R amplifying approximately the 500 bp of 16S ribosomal RNA (rRNA) gene sequence targeting variable region V1 to V3 of the 16S rRNA gene of bacteria species and pooled out to avoid PCR bias (Sekar et al. 2014). Each 50-μL PCR reaction system containing 25 μL of 2X Dream Taq green Master Mix (DNA polymerase, dNTPs, and 4 mM MgCl2), 22 μL of sterile Nuclease-free water, 1 μL of forward primer (0.2 μM) and 1 μL of reverse primer (0.2 μM), and 1 μL of metagenomic DNA (50–100 ng/μL). In order to control nuclease contamination, negative control was included at every reaction. The following PCR reaction was performed: an initial denaturation step at 94 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 1 min, annealing at 55 °C for 30 s and extension at 72 °C for 1 min 30 s, and a final extension at 72 °C for 10 min, followed by cooling to 4 °C. For fungal metagenome, DNA was amplified using ITS1f and ITS4 encoding the internal transcribed spacer region 2 as reported by Amend et al. (2010). Briefly, an initial denaturation step was set at 95 °C for 10 min, followed by 34 PCR cycles of denaturation at 95 °C for 1 min, annealing at 51 °C for 1 min, extension at 72 °C for 1 min, and a final extension at 72 °C for 5 min. The PCR product (10 μL) was loaded in 1 % (m/v) agarose gel (Merck, SA) stained with 5 % of 10 mg/mL ethidium bromide (Merck, SA) and visualized under ultra violet Trans illuminatior (InGenius Bio Imaging System, Syngene, Cambridge, UK). The correct PCR amplicons of both bacteria and fungi were pooled together for the respective samples and purified using the DNA Clean & Concentrator Kits (Zymo Research Corporation, USA). The purified DNA samples were then quantified using a NanoDrop spectrophotometer and sequenced at Inqaba Biotechnology Industries, South Africa, on Roche 454 sequencing platform. All experiments were performed in triplicate for each sample.
Pyrosequencing data processing
Prior to be used, raw reads were processed for quality in order to remove artificial replicate reads and low-quality reads using Mothur pipeline (Schloss et al. 2009). Good quality reads were further pre-screened for ribosomal identity (at approximately 70 % identity) using Qiime-uclust. Ribosomal reads were further screened for the presence of chimeric reads using UCHIME according to de novo method (Edgar et al. 2011). The rRNA non-chimeric reads were later used for further analysis using RDP pyrosequencing pipeline at a confidence threshold of 97 % for microbial classification, and genetic distance was also determined. Reads with similarity more than 97 % were clustered within the same operational taxonomic unit (OTU) and rarefaction curves were also determined (Wang et al. 2007; Cole et al. 2014). The Shannon diversity index and richness estimator Chao1 were performed to estimate the microbial diversity and richness from each water samples. The relative abundance (%) of individual taxa within each community was calculated by comparing the number of sequences assigned to a specific taxon against the number of total sequences obtained for that sample. The similarity and dissimilarity in bacterial community structure within both wastewater treatment plants were analyzed using the Jaccard index (Cole et al. 2014) and rarefaction curve (Legendre and Legendre 1998), respectively. Generated data was later made publicly available at the DDBJ Sequence Read Archive (DRA) under the accession number.
PRJDB4552.
Additional statistical analysis
Statistical package Stata computer software (version: STATA V10, STATA Corp. LP, 2009) was used to analyze the generated data. The one-way analysis of variances (ANOVA) was also used to compare microbial population between both seasons as well as the environmental variables. An additional test to determine the relationship between environmental variables was carried out using the Pearson correlation test and the interpretation was performed at a two-sided 95 % confidence limit.
Results
Community species richness and diversity indices
The present study analyzed the microbial community structure and species richness in the 16S rRNA as well as ITS genes of acid mine water collected from summer (March and April) and winter (June and July). The 454 pyrosequencing generated high bacterial reads (1865 to 4052 reads) when compared to fungal reads (105 to 373 reads) (Table 1). However, samples collected from summer contained more reads for both bacterial and fungal communities compared to winter samples. RDP pyrosequencing pipelines were used to determine microbial community structure by assigning the read tags to different operational taxonomic units (OTUs) at 3 % nucleotide cutoff with a threshold of 97 %. A range of 620–1192 OTUs for bacteria and 73–249 OTUs for fungi were recovered from the collected samples. For bacterial community, the March sample appeared to have the highest OTUs (1192) followed by June, April, and July. Regardless of the fact that the April samples had a high number of reads when compared to June samples, its OTU number was, in a contrary, lower than those of June. For fungal community instead, the April sample appeared to have the highest OTU number at 249 followed by March, June, and July. The species richness of bacteria appeared to be slightly higher with no significant difference (p > 0.05) whereas fungi were higher (p < 0.05) for summer samples compared to winter samples (Table 1). Furthermore, Shannon diversity index and evenness (E) were determined in order to assess the complexity of individual microbial population within samples. The Shannon diversity index indicated higher microbial diversity in samples collected in summer over those collected in winter with March (6.22) and April (5.2) having the highest value for bacteria and fungi, respectively. The evenness values of bacterial samples (0.86 to 0.88) appeared to be lower than those of fungal samples (0.92 to 0.98). No statistical difference (p > 0.05) was noted between summer and winter samples in terms of their evenness for both bacterial and fungal population.
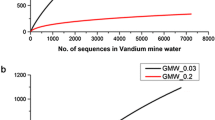
The species richness of the collected samples for both bacterial and fungal communities was also assessed using rarefaction techniques (Fig. 1). The results revealed a short slight plateau on the bacterial samples (Fig. 1a) indicating that a reasonable number of species have been considered from each individual samples. However, for fungal samples (Fig. 1b), the absence of plateau revealed that sequencing depth was still not enough to cover the entire fungal diversity and a large fraction of the different species remains to be discovered. A pairwise community similarity between samples was assessed based on the absence and presence of each OTU using a Jaccard index (Fig. 2, Table S1). The Jaccard index exhibited low or no similarity between all bacterial samples ranging with values from 0.766 to 0.994 with the pair June and July bacterial community showing a slight similarity (0.766). In contrast, for fungal samples, a high and moderately high similarity was exhibited between the pair samples of June/March (0.125) and April/March (0.579).
Microbial community structure in acid mine water
The present study is the first one which comparatively analyzed the bacterial and fungal diversity from acid mine water of South Africa and comprehensively by high throughput sequencing method. A total of six bacterial phyla including unclassified bacteria were observed in the acid mine water samples (Table S3). Winter samples (June and July) were the only ones to contain all six bacterial phyla, while samples collected from summer could contain only three to five phyla for March and April, respectively. Proteobacteria was the most dominant phylum in all collected samples followed by Firmicutes in summer samples (March and April) and unclassified bacteria in winter samples. Actinobacteria was considered as a rare phylum only found in samples collected in winter season. In samples collected in summer time, Cyanobacteria (0.21 %) and Bacteroidetes (0.11 %) were considered as rare phyla as they were found at a very low concentration in April alone. It was also noted that the bacterial community became highly diverse as the sequences were classified into lower taxonomic levels (Tables S3–7). Regardless of their proportion in individual samples, results showed bacterial community up to 15 classes, 31 orders, 54 families, and 99 genera. When considering individual samples, it was observed that winter samples were the most diverse with 24 orders for June and 25 July samples compared to summer samples (March = 12 orders and April = 16 orders). Furthermore, unique orders were also observed in June sample (Lactobacillales and Selenomonadales with 0.04 % each) and July sample (Gammaproteobacteria_incertae_sedis, Nitrosomonadales, Hydrogenophilales, Caulobacterales, Rhodocyclales, Cytophagales with 0.13 % of the total population each except Sphingobacteriales: 0.39 %) (Fig.3). In genus level, there were 38 rare genera that were observed in only one sample, and they accounted for 4.38 % of total classified sequences with most of them found in the July (2.42 %) and June samples (1.82 %).
Taxonomic distribution of different bacterial (a) and fungal (b) orders from Mogalie mine. Analysis of 16S rRNA gene sequences was done in comparison with the RDP II database. The percentages of the phylogenetically classified sequences in each sampling points are the average of each order over the total sequences number of the sampling point
In contrary to bacterial community, only four fungal phyla were noted from all mine water samples with unclassified fungi (68.2 to 91 %) being the predominant followed by Basidiomycota (for April = 26.6 %, June = 27.8 %, and July = 6.9 %) or by Ascomycota (for March = 6.7 %) (Table S8). Chytridiomycota was only found in March (0.56 %) and July (1.98 %) samples. Moreover, fungal community showed a very slight or no change of diversity as the sequences were classified into a lower taxonomic level with a total of 8 classes, 9 orders, 11 families, and 9 genera (Fig. 3, Tables S8–12). When considering individual samples, the March sample showed three phyla, six classes, six orders, seven families, and five genera. Similar to the March sample, the April mine water sample (two phyla, seven classes, six orders, six families, and four genera); the June sample (two phyla, six classes, five orders, five families, and four genera); and the July sample (three phyla, four classes, three orders, three families, and two genera) show a decrease in diversity as the sequences were classified into a lower taxonomic levels. The slight increase or decrease in fungal diversity can be explained by the fact that several sequences were found to be unclassified as the sequences were being classified into a lower taxonomic level.
Physicochemical variables
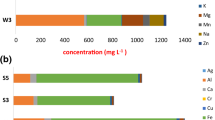
Table 2 summarizes the physicochemical parameters of mine water samples. Through the sampling period, samples appeared to contain several chemical elements with NO3 −, PO4 3−, Zn, Cu, Mn, Hg, As, U, and Fe not exceeding 10 mg/L. Besides sodium (4809.45–7263.48 mg/L) which showed the highest concentration, Ni (19.47–39.01), V (203.19–430.95), Ca (384.74–893.91), Mg (128.87–431.98), K(216.81–346.83), Cr (13.73–28.15), Co (7.46–15.93), Pb (4.91–15.93), and Fe(2.03–43.93) were also found to be the most abundant chemical elements in the mine water samples. The present samples appeared to be highly acidic with pH ranging from 2.46 to 3.11 and samples collected in March having the highest pH value. As the samples were collected during two different seasons (summer and winter), temperature of samples collected in March (21.3 °C) and April (19.55 °C) was statistically different (p < 0.05) to those of June (13.12 °C) and July (7.4 °C). The conductivity and the DO of the samples had values ranging between 525 and 867 μ/Sm, and 3.98 and 5.68 mg/L, respectively, with winter samples having the highest conductivity level. Based on the fact that all chemical contents monitored in these individual samples were considered to be independent, a one-way ANOVA was used to test whether there were any significant differences between the mean values of our individual mine water samples. During the sampling period, no significant difference (p > 0.05) was noted among all parameters except EC that revealed significant difference (p < 0.05) with NO3 −, PO4 3−, temperature, pH, DO, Ni, Zn, Cu, Mn, Hg, Cr, Co, As, Pb, U, and Fe. Furthermore, Na appeared also to be significantly different (p < 0.05) with all parameters investigated. A further statistical analysis revealed no significant difference between specific physicochemical parameters of samples collected in March, April, June, and July with exception of COD (where June had the highest COD levels and March the lowest), temperature (March = highest and July = lowest), conductivity (July = highest and March = lowest), Ca (March = highest and April = lowest), Mg (July = highest and April = lowest), Na (March = highest and April = lowest), and Fe (July = highest and March = lowest).
Due to the fact physicochemical parameters play a major role on microbial diversity of mine water, a canonical correspondence analysis (CCA) was performed to establish the relationship of these variables to the microbial diversity of each individual sample (Fig. 4). In the present study, the CCA diagram displayed 93.83 % of the variance in the bacteria-environmental relationships at the first axis and 6.17 % of the variance at the second axis. It was also noted that the variance of the first axis was more related to pH, temperature, DO, EC, Fe, and Mg whereas Na, Ca, and Ni were related to the variance of second axis. However, the relationship between environmental factors and bacterial community (pyrosequencing data) was significant at both first and second axis (p < 0.05). A strong correlation was noted between pH and Mg, temperature and Fe, and DO and EC. As per CCA results, environmental parameters considered as variables impacted the structure and function of bacterial and fungal communities in acid mine water samples. It should be mentioned that microbial community from summer (March and April) were mostly affected by Mg, Ca, and pH since the latter revealed a strong correlation with both Mg and Ca. In the winter samples (June and July), however, temperature, DO, EC, and Fe appeared to have the most impact on the microbial diversity. Regardless of the statistical difference reported between Na and all other parameters, Na did not show uneven impact on microbial communities from both summer and winter periods. The lack of relationship between samples from individual samples and other environmental parameters suggest that these parameters played a major role in the dissimilarity of microbial community.
Discussion
The tiny microbes dominate the planet and are ubiquitously distributed all over the biosphere tolerating a wide range of physicochemical stresses. Due to their widespread presence and high abundance in almost all ecosystems, it makes them as the most successful organisms on the planet. It is now generally accepted that more than 95 % of microbes living in the environment are yet to be discovered and explored (Johnson 1998). Harsh environment such as acid mine drainage (AMD) have widely been reported to contain a large diversified microbial population (Bond et al. 2000; Johnson and Hallberg 2005; Natarajan 2008; Kuang et al. 2013; Sánchez-Andrea et al. 2014) regardless of the high content of pollutants exceeding largely the maximum limit found in it (Table 2). However, this microbial population is exclusively dominated by unicellular microbes, especially bacteria (Johnson and Hallberg 2005; Sánchez-Andrea et al. 2014). Beside the large abundance of bacteria in AMD, fungal species have also been reported from acid mine drainage (Colmer and Hunkle 1947). But, very few studies have been conducted on fungal community in acid mine drainage especially in South Africa regardless of its historical mine activities. Therefore, the present study is the first one which monitored temporal changes in bacterial and fungal community structures in mine water for a period of 4 months using high throughput sequencing approach. The results revealed that the 454 pyrosequencing generated high bacterial reads (1865 to 4052 reads) when compared to fungal reads (105 to 373 reads) (Table 1). Samples collected during the period considered as summer (March and April) appeared to have the most reads for both bacterial and fungal populations when compared to the winter period (June and July). While assigning the read tags to different OTUs, findings showed a range of 620–1192 OTUs assigned to bacteria community and 73–249 OTUs for fungi (Table 1). In contrary to number of reads, the highest OTUs were noted in March (1192 OTUs) followed by June, April and July for bacterial community, while for fungal community April sample showed the highest OTU number (249) followed by March, June, and July. The high OTUs were also reported by Keshri et al. (2015) when investigating the bacterial profiling of both gold and vanadium mines from South Africa.
The present study revealed a total of six bacterial phyla containing 99 genera throughout the sampling period with the March sample having three phyla, the April sample having five phyla, and the June and July samples having six phyla. Furthermore, only three phyla (Proteobacteria and Firmicutes) including unclassified bacteria were shared in all collected samples with Proteobacteria as the most abundant phylum. This was also true at a lower taxonomic level were most of the genera belonged to Proteobacteria with exception of July samples where genera belonging to Cyanobacteria phylum were the most dominant and suggesting that most of the bacteria belonging to Proteobacteria phylum could not be classified at a lower taxonomic level. Several findings have also reported that Proteobacteria is the dominant phylum in acid mine water (Bond et al. 2000; Bruneel et al. 2006; Keshri et al. 2015). Due to their abilities to widely adapt in any environment, Preteobacteria was found to be well distributed across the surface of the earth (Yergeau et al. 2012; Chao et al. 2012; Kamika et al. 2014). As also revealed by the present study, their abundance and diversity depend on several environmental factors including weather (Fig. 3, Tables S3–7). The present study shows that winter samples were having the highest number of phyla compared to summer samples. It was also noted that the bacterial community become highly diverse as the sequences were classified into lower taxonomic levels (Fig. 3, Tables S3–7) (Keshri et al. 2015). In addition, parametric analyses such as the Shannon index and evenness were also performed to determine an estimated diversity of bacteria indicating no significant shift (p > 0.05) between the summer and winter samples for bacteria (Table 1). This was also confirmed when performing the Chao1 species richness estimator and revealed a very low species richness difference between both the winter and summer samples (Table 1, Fig. 1).
It has been reported that acid mine water is one of the extreme environment that harbors rare or unique bacteria species and this is also seen by the number of unclassified bacteria present in such environment (Keshri et al. 2015). In this study, unique orders were also observed in the June samples (Lactobacillales [Streptococcus] and Selenomonadales [Veillonella] with 0.04 % each) and July sample (Gammaproteobacteria_incertae_sedis [Thiohalobacter], Nitrosomonadales [Nitrosomonas], Hydrogenophilales [Thiobacillus], Caulobacterales [unclassified_Hyphomonadaceae], Rhodocyclales [unclassified_Rhodocyclaceae], Cytophagales with 0.13 % of the total population each and Sphingobacteriales at 0.39 %). Similar to the present finding, Lactobacillales, Hydrogenophilales, and Cytophagales were also found in acid mine drainage at very low concentration (Schrenk et al. 1998; Bond et al. 2000). These rare/unique orders have also been observed in cow rumen (Patil et al. 2012), seawater (Zeng et al. 2014), and Earth-Cave (Zhou et al. 2007). This study disagreed with Baker and Banfield (2003) who stated that Acidithiobacillus, Thiobacillus, Thiomonas, and Leptospirillum were the most dominant genera in acid mine water. The study further disagreed with Kamika and Momba (2014), who revealed that the genus Marinobacter was the most predominant genus in mine water while in this study, Alcaligenes or unclassified_Cyanobacteria were the most dominant genera depending on the samples. Though Acidobacteria has been reported to populate acid mine water with pH ranging between 3.0 and 6.0 (Baker and Banfield 2003), in this study, their presence were not noted.
Unlike bacteria community, fungal species showed a very low population with only four phyla and unclassified fungi occupying a range of approximately 60 to 90 % of the all community followed by Ascomycota, Basidiomycota, and Chytridiomycota (Fig. 3, Table S3). This also corroborates with Ferreira and Goncalves (2010), who stated that streams found in highly contaminated environments such as mine areas usually contain lower fungal diversity and species richness compared to others. Regardless of lower diversity due to the conditions of the environment, Baker et al. (2004) reported higher diversity of fungi than the one reported in the present study. The high abundance of unclassified fungi in the present study suggests that these samples may contain more novel species (Table S3). Furthermore, the diversity of the population appeared to be slightly higher as the sequences were classified in a lower taxonomic level. This was also confirmed by the Shannon diversity index, evenness, and Chao species richness estimator that showed no significant shift (p > 0.05) in diversity and species richness between summer and winter samples (Table 1). However, this diversity decreases from summer (six classes, six orders, seven families, and five genera for March, and seven classes, six orders, six families, and four genera for April) to winter periods (six classes, five orders, five families, and four genera for June, and four classes, three orders, three families, and two genera for July). In addition, rare order was only found in the June sample (unclassified Microbotryomycetes). Rare genera such as Neophaeosphaeria, Epicoccum, and Rhodotorula belonging to the phyla Ascomycota and Basidiomycota, respectively, were noted in all collected samples.
This study further investigated the bacterial and fungal similarity between the collected samples using a pairwise community similarity test such as Jaccard index and revealed a low or no similarity between all bacterial samples ranging with values from 0.766 to 0.994 (Table S2) with the pair June and July bacterial community showing a slight similarity (0.766). In contrast, for fungal samples, a high and moderately high similarity was exhibited between the pair samples of June/March (0.125) and April/March (0.579), respectively. According to both Ranjard et al. (2003) and Somboonna et al. (2012), the microbial abundance as well as diversity in aquatic environment can vary and influence by sampling methods and environmental parameters, including anthropogenic activities. This study showed that the collected samples were highly polluted with metals and metalloids (Table 2). The presence of these chemical elements such as Cd, Cu, and Zn has been reported to play a significant role in similarity and diversity of both bacteria and fungi (Miersch et al. 1997; Jaeckel et al. 2005). This was confirmed while performing the canonical correspondence analysis (CCA) in order to establish the relationship of these variables to the microbial diversity of each individual samples. This study showed that the relationship between environmental factors and bacterial community (pyrosequencing data) was significant at both first and second axis (p < 0.05). Moreover, a strong correlation was also noted between pH and Mg, temperature and Fe, and DO and EC. The lack of relationship between samples from individual samples and other environmental parameters suggested that these parameters played a major role in the dissimilarity of microbial community. Several reports revealed that electrical conductivity (EC) is a significant parameter as it regulates microbial composition and diversity in a particular ecosystem (Lozupone and Knight 2007; Liu et al. 2008; Mohamed and Martiny 2011; Wang et al. 2012). Same as EC, pH, temperature, and DO have also been seen as an important factors as they regulate the overall bacterial diversity and composition in the ecosystem (Fierer and Jackson 2006; Wang et al. 2012). This finding is in agreement with the results of Siggins et al. (2011) who reported that temperature is one of the most important parameter influencing microbial composition in the environment. This study also corroborates to Frey et al. (2013) who reported that soil microorganisms depend largely on both temperature and substrate quality to efficiently use organic matter. Castro et al. (2010) also revealed that factors such as rising CO2, rising temperature (ambient, +3 °C), precipitation considered as climate change drivers and their interactions may cause changes in bacterial and fungal overall abundance.
In conclusion, the present study provided an insight into the bacterial and fungal community structure from acid mine water over summer and winter months period using a high throughput sequencing approach. In summary, bacterial community was mostly dominated with Proteobacteria phylum while fungal community had unclassified fungi as the most abundant. This trend was similar as the reads were classified to lower taxonomic groups expect in July samples were Cyanobacteria became the most abundant phylum. The estimated and observed diversity revealed that bacteria were more diverse than fungi the mine water with winter samples (June and July) more diverse than summer samples (March and April). Further, environmental factors such as temperature, conductivity, dissolved oxygen, pH, and other chemical contents played a major role in species richness, diversity, and similarity between microbial communities from mine water collected in the present study. Although microbial diversity appeared to be high in the winter period, their abundance decreased from the summer to the winter periods. The present study revealed several rare bacterial genera in the mine water samples. The high number of unclassified reads for both bacteria and fungi showed that mine water harbors several novel species. This result can also extend our knowledge about the fungal community in acid mine water which has received less attention compared to their counterparts. Fungal community structure in mine water can also be explored further for their role in bioremediation and environment management. The results provided the relationships between microbial community and environmental variables in mining sites.
References
Amaral-Zettler LA, Zettler ER, Theroux SM, Palacios C, Aguilera A, Amils R (2011) Microbial community structure across the tree of life in the extreme Rio Tinto. ISME J 5:42–50
Amend AS, Seifert KA, Samson R, Bruns TD (2010) Indoor fungal composition is geographically patterned and more diverse in temperate zones than in the tropics. Proc Natl Acad Sci U S A 107:13748–13753
APHA (2001) Standard methods for the examination of Water and Wastewater, 20th edn. American Public Health Association, Washington, DC
Bai H, Kang Y, Quan H, Han Y, Sun J, Feng Y (2013) Treatment of acid mine drainage by sulfate reducing bacteria with iron in bench scale runs. Bioresour Technol 128:818–822
Baker B, Banfield J (2003) Microbial communities in acid mine drainage. FEMS Microbiol Rev 44:139–152
Baker BJ, Lutz MA, Dawson SC, Bond PL, Banfield JF (2004) Metabolically active eukaryotic communities in extremely acidic mine drainage. Appl Environ Microbiol 70(10):6264–6271
Bond PL, Smriga SP, Ban¢eld JF (2000) Phylogeny of microorganisms populating a thick, subaerial, predominantly lithotrophic biofilm at an extreme acid mine drainage site. Appl Environ Microbiol 66:3842–3849
Bruneel O, Duran R, Casiot C, Elbaz-Poulichet F, Personne JC (2006) Diversity of microorganisms in Fe-As-rich acid mine drainage waters of carnoules, France. Appl Environ Microbiol 72(1):551–556
Castro HF, Classen AT, Austin EE, Norby RJ, Schadt CW (2010) Soil microbial community response to multiple experimental climate change drivers. Appl Environ Microbiol 76(4):999–1007
Chao Y, Ma L, Yang Y, Ju F, Zhanh XX, Wu WM, Zhang T (2012) Metagenomic analysis reveals significant changes of microbial compositions and protective functions during drinking water treatment. Sci Report 3:3550. doi:10.1038/srep03550
Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM (2014) Ribosomal Database Project: data and tools for high throughput rRNA analysis nucl. Acids Res 42(Database issue):D633–D642. doi:10.1093/nar/gkt1244
Colmer R, Hunkle ME (1947) The role of microorganisms in acid mine drainage: preliminary report. Science 106:253–256
Corcoran E, Nellemann C, Baker E, Bos R, Osborn D, Savelli H (2010) Sick water? the central role of wastewater management in sustainable development. A Rapid Response Assessment. United Nations Environment Programme, UN-HABITAT, GRID-Arendal
Das BK, Roy A, Koschorreck M, Mandal SM, Wendt-Potthoff K, Bhattacharya J (2009) Occurrence and role of algae and fungi in acid mine drainage environment with special reference to metals and sulfate immobilization. Water Res 43(4):883–894
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27(16):2194–2200
Ferreira V, Goncalves AL (2010) Contamination by uranium mine drainage affects fungal growth and interactions between fungal species and strains. Mycologia 102(5):1004–1011
Fierer N, Jackson RB (2006) The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci 103:626–631
Frey S, Lee J, Melillo JM, Six J (2013) The temperature response of soil microbial efficiency and its feedback to climate. Nat Clim Chang 3:395–398
Goebel BM, Stackebrandt E (1994) Cultural and phylogenetic analysis of mixed microbial populations found in natural and commercial bioleaching environments. Appl Environ Microbiol 60:1614–1621
Gross S, Robbins EI (2000) Acidophilic and acid-tolerant fungi and yeasts. Hydrobiologia 433:91–109
He J, Shen J, Zhang L, Zhu Y, Zheng Y, Xu M, Di H (2007) Quantitative analyses of the abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea of a Chinese upland red soil under long-term fertilization practices. Environ Microbiol 9(9):2364–2374
Hennecke H, Kjelleberg S, Brussaard C (2013) Molecular insights into environmental microbes. FEMS Microbiol Rev 37(3):285
Jaeckel P, Krauss GJ, Krauss G (2005) Cadmium and zinc response of the fungi Heliscus lugdunensis and Verticillium cf. alboatrum isolated from highly polluted water. Sci Total Environ 346:274–279
Johnson DB (1998) Biodiversity and ecology of acidophilic microorganisms. FEMS Microbiol Ecol 27:307–317
Johnson DB, Hallberg KB (2003) The microbiology of acidic mine waters. Res Microbiol 154:466–473
Johnson DB, Hallberg KB (2005) Acid mine drainage remediation options: a review. Sci Total Environ 338:3–14
Kamika I, Momba MNB (2014) Microbial diversity of Emalahleni mine water in South Africa and tolerance ability of the predominant organism to vanadium and nickel. PLoS One 9(1):e86189. doi:10.1371/journal.pone.0086189
Kamika I, Coetzee M, Mamba BB, Msagati T, Momba MNB (2014) The impact of microbial ecology and chemical profile on the enhanced biological phosphorus removal (EBPR) process: a case study of Northern Wastewater treatment works, Johannesburg. Int J Environ Res Publ Health 11:2876–2898
Kembel SW, Eisen JA, Pollard KS, Green JL (2011) The phylogenetic diversity of metagenomes. PLoS One 6(8):e23214. doi:10.1371/journal.pone.0023214
Keshri J, Mankazana BB, Momba MN (2015) Profile of bacterial communities in South African mine-water samples using Illumina next-generation sequencing platform. Appl Microbiol Biotechnol 99(7):3233–3242
Kuang JL, Huang LN, Chen LX, Hua ZS, Li SJ, Hu M, Li JT, Shu WS (2013) Contemporay environmental variation determines microbial diversity patterns in acid mine drainage. ISME J 7:1038–1050
Legendre P, Legendre L (1998) Numerical ecology. 2nd English ed. Elsevier Science BV, Amsterdam
Liu ST, Yang FL, Gong Z, Su ZC (2008) Assessment of the positive effect of salinity on the nitrogen removal performance and microbial composition during the start-up of CANON process. Appl Microbiol Biotechnol 80:339–348
Lozupone C, Knight R (2007) Global patterns in bacterial diversity. Proc Natl Acad Sci U S A 104:11436–11440. doi:10.1073/pnas.0611525104
Miersch J, Barlocher F, Bruns I, Krauss GD (1997) Effects of cadmium, copper, and zinc on growth and thiol content of aquatic hyphomycetes. Hydrobiologia 346:77–84
Mohamed DJ, Martiny JBH (2011) Patterns of fungal diversity and composition along a salinity gradient. ISME J 5:379–388
Monteiro MIC, Ferreira FN, Oliveira NMM, Avila AK (2003) Simplified version of the sodium salicylate method for nitrate analysis in drinking waters. Anal Chim Acta 477(1):125–129
Natarajan KA (2008) Microbial aspects of acid mine drainage and its bioremediation. T Nonferr Metal Soc 18:1352–1360
Nies DH, Silver S (1995) Ion efflux systems involved in bacterial metal resistances. J Ind Microbiol 14(2):186–199
Patil KR, Roune L, McHardy AC (2012) The PhyloPythiaS web server for taxonomic assignment of metagenome sequences. PLoS One 7(6):e38581
Ranjard L, Lejon DP, Mougel C, Schehrer L, Merdinoglu D, Chaussod R (2003) Sampling strategy in molecular microbial ecology: influence of soil sample size on DNA fingerprinting analysis of fungal and bacterial communities. Environ Microbiol 5:1111–1120
Roane TM, Peper IL (2000) Microorganisms and metal pollutants. In: Maier RM, Pepper IL, Gerba CP (eds) Environmental microbiology. Academic Press, San Diego, pp. 403–423
Sánchez-Andrea I, Sanz JL, Bijmans MFM, Stams AJM (2014) Sulfate reduction at low pH to remediate acid mine drainage. J Hazard Mater 269:98–109
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75(23):7537–7541
Schrenk MO, Edwards KJ, Goodman RM, Hamers RJ, Ban¢eld JF (1998) Distribution of thiobacillus ferrooxidans and leptospirillum ferrooxidans: implications for generation of acid mine drainage. Science 279:1519–1522
Sekar S, Zintchem AAEA, Keshri J, Kamika I, Momba MNB (2014) Bacterial profiling in brine samples of the Emalahleni Water Reclamation Plant, South Africa, using 454-pyrosequencing method. FEMS Microbiol Lett 359:55–63
Siggins A, Enright A-M, O’Flaherty V (2011) Temperature dependent (37–15 °C) anaerobic digestion of a trichloroethylene-contaminated wastewater. Bioresour Technol 102(17):7645–7656
Silverman MP, Ehrlich HL (1964) Microbial formation and degradation of minerals. In: Umbreit WW (ed) Advances in applied microbiology, Vol. 6. Academic Press, New York, pp. 153–206
Somboonna N, Assawamakin A, Wilantho A, Tangphatsornruang S, Tongsima S (2012) Metagenomic profiles of free-living archaea, bacteria and small eukaryotes in coastal areas of Sichang island, Thailand. BMC Genomics 13(Suppl 7):S29
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73(16):5261–5267
Wang X, Hu M, Xia Y, Wen X, Ding K (2012) Pyrosequencing analysis of bacterial diversity in 14 wastewater treatment systems in China. Appl Environ Microbiol 78(19):7042–7047
Xiao S, Xie X, Liu J (2009) Microbial communities in acid water environments of two mines. China Environ Pollut 157(3):1045–1050
Yergeau E, Lawrence JR, Sanschagrin S, Waiser MJ, Korber DR, Greer CW (2012) Next-generation sequencing of microbial communities in the Athabasca River and its tributaries in relation to oil sands mining activities. Appl Environ Microbiol 78(21):7626–7637
Zeng YX, Yu Y, Qiao ZY, Jin HY, Li HR (2014) Diversity of bacterioplankton in coastal seawaters of Fildes Peninsula, King George Island. Antarctica Arch Microbiol 196(2):137–147
Zhou JP, Gu YQ, Zou CS, Mo MH (2007) Phylogenetic diversity of bacteria in an eath-cave in Guizhou province, southwest of China. J Microbiol 45(2):105–112
Acknowledgments
The authors are thankful for the University of South Africa (UNISA) research fund.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Funding
This work was supported by a grant from the National Research Fundation (NRF) (Grant number: M590).
Conflicts of interest
The authors declare that they have no competing interests.
Electronic supplementary materials
ESM 1
(PDF 371 kb)
Rights and permissions
About this article
Cite this article
Kamika, I., Azizi, S. & Tekere, M. Microbial profiling of South African acid mine water samples using next generation sequencing platform. Appl Microbiol Biotechnol 100, 6069–6079 (2016). https://doi.org/10.1007/s00253-016-7428-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-016-7428-5