Abstract
Since antibiotics show hormesis effects in cyanobacteria at the nanogram per liter concentration level, the possibility for two commonly used antibiotics (sulfamethoxazole and tetracycline) to increase lipid productivity in Synechocystis sp. PCC 6803 was assessed in the present study. The two target antibiotics significantly promoted (p < 0.05) the biofuel productivity of Synechocystis sp. PCC 6803 through the increase of both biomass and lipid content. Sulfamethoxazole and tetracycline significantly stimulated (p < 0.05) cyanobacterial growth by upregulating proteins related to cell differentiation, cell division, and gene expression; significantly enhanced (p < 0.05) the photosynthetic activity by upregulating photosynthesis-related proteins; and significantly increased (p < 0.05) the lipid content in cyanobacterial cells by downregulating carbohydrate catabolic proteins and carbohydrate transport proteins. Due to the altered expression pattern of biosynthesis-related proteins, the two antibiotics increased the proportion of monounsaturated fatty acids, while tetracycline reduced the proportions of saturated and polyunsaturated fatty acids. The changes in fatty acid composition may improve the combustion performance of biofuel. This study provided insights into the application of antibiotics in cyanobacteria-based biofuel production.

Graphical abstract
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Biofuel is considered to be a compatible alternative to fossil fuels and has various advantages including innocuity, sustainability, and good biodegradability (Aslam et al. 2018). Compared with fossil fuels, biofuel is an environmentally friendly fuel that releases less greenhouse gas and generates less air pollution (Živković et al. 2017). Biofuel comes from oil-rich plants, animals, and photosynthetic microorganisms, among which cyanobacteria, as the third-generation biofuel feedstock, present good application future in biofuel production (Han et al. 2016). Due to the high growth rate, cyanobacterial cultivation requires less land occupation than terrestrial plants and shows good adaptability to extreme culture conditions (Han et al. 2016; Wahlen et al. 2011). Establishing a simple and efficient production process of quality biofuel is the major hurdle for the economization of cyanobacteria-based biofuel (Bharte and Desai 2019).
Cyanobacteria are photosynthetic organisms with the ability to fix CO2 and produce lipids in the form of diacylglycerols and triacylglycerols (Deshmukh et al. 2019a, b), which can be easily transformed to biofuel through transesterification using alcohols (e.g., methanol) and catalysts (base or acid) (Raheem et al. 2018). The reported lipid content in cyanobacteria varies from 4 to 21% (Shuba and Kifle 2018), and increased lipid content is an important issue for the commercial realization of cyanobacteria-based biofuel. The lipid content was reported to be regulated by culture conditions including temperature, light, salinity, and CO2 concentration (Aslam et al. 2018; Chandra et al. 2019; Dineshbabu et al. 2020). Recent studies also observed an increased lipid content in cyanobacteria exposed to exogenous stresses, such as nitrogen starvation, phosphorous deficiency, ultrasonication, ultraviolet radiation, and TiO2 stresses (Casazza et al. 2015; Cordeiro et al. 2017; Ellison et al. 2019). However, the final biofuel production amount is determined by biomass × lipid content. Reports on the simultaneous stimulation of biomass and lipid content during oil-producing cyanobacterial culture are still limited.
Antibiotics are a group of antibacterial chemicals that are broadly used in human disease treatment, veterinary medicine, and aquaculture (Ben et al. 2019). With a bacteria-like structure, cyanobacteria are sensitive to antibiotics (Maul et al. 2006). Various hormetic responses have been observed in cyanobacteria under exposure to low concentrations of antibiotics, including the promotion of biomass growth, photosynthetic activity, and cellular substance synthesis. For instance, the growth of Microcystis flos-aquae was stimulated under exposure to 1–100 ng/L erythromycin (Wan et al. 2015). Exposure to 0.1 mg/L ofloxacin led to increased photosynthetic activity in Microcystis aeruginosa (Deng et al. 2015). Previous studies by our group found that 100–600 ng/L amoxicillin promoted the synthesis of proteins and polysaccharides in M. aeruginosa, and the proteomic responses suggested a stimulatory effect of amoxicillin, ciprofloxacin, and sulfamethoxazole on carbohydrate synthesis (Liu et al. 2016; Liu et al. 2017). If antibiotic stress could increase biomass growth and lipid content at the same time, a significant increase in lipid productivity of oil-producing cyanobacteria could be achieved. However, information on the influence of antibiotics on oil-producing cyanobacteria was still limited.
In this study, two commonly used antibiotics effective against gram-negative bacteria, sulfamethoxazole (SMZ) and tetracycline (TET), were selected as target chemicals (Yang et al. 2018). A commonly used oil-producing cyanobacterial strain with high lipid content, Synechocystis sp. PCC 6803, was selected as the target cyanobacterium (Sivaramakrishnan and Incharoensakdi 2018). The genome of Synechocystis sp. PCC 6803 has been fully recorded (Pattanaik et al. 2020), and it is regarded as an attractive model species for oil-producing cyanobacteria. The regulatory effects of target antibiotics on the biomass, lipid content, and fatty acid composition of Synechocystis sp. PCC 6803 were investigated, based on which the possibility of using antibiotics in cyanobacterial biofuel production was evaluated. In addition, the regulatory mechanisms of antibiotics in Synechocystis sp. PCC 6803 were interpreted according to the responses of photosynthetic activity and the whole proteome.
Materials and methods
Culture of cyanobacterial cells
SMZ and TET with purities of 99.5% and 96.2%, respectively, were purchased from Dr. Ehrenstorfer, Inc. (Germany). The target antibiotics were dissolved in methanol and stored at − 20 °C. Synechocystis sp. PCC 6803 was precultivated using sterile BG11 medium at 25 ± 1 °C under fluorescent white light at an intensity of 2000 lx with a 16:8 light:dark cycle. In the antibiotic exposure test, a certain amount of cyanobacterial cells were inoculated into 200 mL of sterile BG11 medium, and the initial optical density value at 730 nm (OD730) was 0.05 ± 0.002. Next, the cyanobacterial cells were aseptically cultured for 18 days under the same conditions as those used for precultivation. At the beginning of the antibiotic exposure test, each antibiotic was spiked into the culture medium at an exposure dose of 100 ng/L. Before the antibiotic exposure test, a preliminary experiment showed that target antibiotics were degraded by 20–30% every day because of hydrolysis and photolysis. Therefore, TET and SMZ were replenished into the culture medium every 24 h to maintain a stable exposure dose. The final concentration of methanol solvent in each antibiotic-treated group was below 0.005% (v/v). A solvent control group containing Synechocystis sp. PCC 6803 spiked with 0.005% methanol was prepared. Three independent replicate experiments were conducted for two antibiotic-treated groups and solvent control.
Determination of cellular responses
To assess the growth conditions, 3 mL of cyanobacterial culture was aseptically taken from each flask at a regular time interval of 24 h, and the OD730 value was measured through spectrophotometry. The specific growth rate was calculated as Eq. (1):
where ODt0 and ODt1 are the OD730 values at t0 and t1 of the exponential growth phase, respectively.
Three parameters indicating the photosynthetic activity, namely, the chlorophyll a content, the maximum photochemical quantum yield of Photosynthesis system II (Fv/Fm), and the maximum relative electron transport rate (rETRmax), were analyzed every 3 days. The chlorophyll a content was measured according to Bland and Angenent (2016). The Fv/Fm and rETRmax values in the cyanobacterial cells were analyzed using a Dual-PAM-100 fluorometer (Walz GmbH, Germany) after dark-adaption for 10 min. The rapid light curve was determined using a series of 20-s light exposures, and rETRmax was estimated through curve-fitting calculations (Fu et al. 2012). The (Fv/Fm) was calculated as Eq. (2):
where F0 is the minimum fluorescence intensity recorded at a low-frequency light pulse and Fm is the maximum fluorescence intensity recorded at 600-ms blue saturation pulse.
On the last day of exposure, 100 mL of cyanobacterial culture was sampled from each flask and used for the analysis of biofuel production ability. The cyanobacterial cells were collected through centrifugation and freeze-dried under vacuum. The dry cyanobacterial cells were first extracted with a solvent system containing chloroform and methanol (2:1, v/v) and then extracted with 0.9% sodium chloride solution. The final extract was dried under a gentle flow of nitrogen and weighed to assess the lipid content and the lipid productivity. The fatty acids in Synechocystis sp. PCC 6803 were methyl-esterified into fatty acid methyl esters (FAMEs) according to the method developed by Lepage and Roy (1984) and determined through an Agilent (7890A-5975C) GC/MS system according to Anahas and Muralitharan (2015). The extraction of lipid, the calculation of lipid content and lipid productivity, and the transesterification process were described in detail in the Supplementary Material. Student’s t test was used to determine the differences in the biomass, photosynthetic parameters, total lipid content, and fatty acid composition between each antibiotic-treated group and the solvent control using the SPSS software (version 23.0).
Determination of proteomic responses
Through tandem mass tag (TMT)–based quantitative proteomic analysis, the proteomic profiles of Synechocystis sp. PCC 6803 cells were compared between each antibiotic-treated group and the solvent control on the 9th day of antibiotic exposure. Twenty milliliters of culture medium containing cyanobacterial cells was taken from each replicate of each test group. The proteins in the cyanobacterial cells were extracted with SDT lysis buffer, digested through the filter-aided sample preparation (FASP) method (Wiśniewski et al. 2009), and labeled with the TMT 10-plex isobaric label reagent set (Thermo Scientific Corp., USA). Next, the labeled peptides were fractionated and quantitatively analyzed through a UHPLC/MS/MS system. Proteins were identified according to the database of Synechocystis sp. PCC 6803 in UniProt (updated on 30/11/2019). Differentially expressed proteins presented a fold change of higher than 1.5 or lower than 0.67 at p < 0.05 according to Student’s t test. The procedures of protein extraction, digestion, labeling, fractionation, quantitative analysis, and protein identification are described in detail in the Supplementary Material. Each differentially expressed protein was annotated according to the biological process defined in Gene Ontology database. The protein-protein interaction (PPI) network of differentially expressed proteins was constructed through the software Cytoscape (version 3.8.0), according to the interaction information obtained from the STRING web-based tool (https://string-db.org). Functional modules composed of highly interconnected proteins (p < 0.05) within the PPI network were constructed through the ClusterONE plugin of Cytoscape.
Results and discussion
Influence of antibiotics on cyanobacterial growth
Figure 1 shows the variation in biomass in different test groups during the antibiotic exposure test. There was no significant difference (p > 0.05) in the OD730 values between the solvent control and each antibiotic-treated group during the first 3 days of exposure. The exponential growth started from the fourth day and lasted until the twelfth day of the culture period. After that point, the cyanobacteria growth gradually entered the stationary phase. Two antibiotics stimulated the growth of Synechocystis sp. PCC 6803 and significantly increased (p < 0.05) the biomass (represented by OD730) in the exponential phase and the stationary phase. The specific growth rate in the solvent control was 0.186 ± 0.006 per day, while the growth rates in the SMZ-treated group and TET-treated group were 0.234 ± 0.005 and 0.212 ± 0.019 per day, respectively. Compared with that of the solvent control, the growth rate was increased by 25.8% and 14.0% following exposure to SMZ and TET, respectively. Cyanobacterial biomass was increased by 53.1% and 48.1% following exposure to SMZ and TET, respectively.
The proteomic responses provided insights into the mechanism of action of the target antibiotics in Synechocystis sp. PCC 6803 for growth stimulation (Table S1 in the Supplementary Material). SMZ induced the upregulation of a protein related to cell differentiation (spoIID) and two proteins related to cell division (minE and sepF) and consequently facilitated cell division. TET also stimulated cyanobacterial growth through the upregulation of spoIID and sepF. In addition, 28 differentially expressed proteins related to gene expression were identified in the SMZ-treated group, among which 25 proteins were upregulated. Similarly, 17 proteins related to gene expression and 3 proteins related to the regulation of gene expression were differentially expressed in the TET-treated group, and 17 of the above 20 proteins were upregulated. The above results suggested that both SMZ and TET showed a tendency to stimulate the gene expression process of Synechocystis sp. PCC 6803. Due to the positive correlation between cell division and gene expression (He et al. 2017), SMZ and TET may stimulate cyanobacterial growth through the promotion of gene expression.
In this study, 8 of 9 differentially expressed stress response proteins (rnhB, slr6040, sodB, lexA, slr1694, priA, nth, and sll0729) were upregulated in the SMZ-treated group, and 5 of 6 differentially expressed stress response proteins (sodB, mntB, slr1436, blaOXA-3, and pds) were upregulated in the TET-treated group. The upregulation of stress response proteins also suggested that Synechocystis sp. PCC 6803 showed an adaptive stress response to antibiotic exposure. Previous studies suggested that increased growth was also an adaptive response to the stress induced by antibiotics at low exposure doses, which was characterized as hormesis (Liu et al. 2016; Zou et al. 2013). Similar growth stimulation effects were reported in another cyanobacterium (M. aeruginosa) exposed to spiramycin and ampicillin at a low exposure dose of 300 ng/L (Wang et al. 2019). Furthermore, Aderemi et al. (2018) reported the stimulatory effects of SMZ, erythromycin, and ciprofloxacin in the oil-producing microalga Raphidocelis subcapitata at exposure doses of 2–8 nM. Therefore, low concentrations of antibiotics have the potential to be introduced into the culture of oil-producing cyanobacteria/microalgae to increase biomass accumulation.
Notably, the stimulatory effects of antibiotics in cyanobacteria have an optimum concentration range. For instance, erythromycin was found to stimulate the growth of Synechocystis sp. at a test concentration of 10 μg/L while inhibiting the growth of Synechocystis sp. at test concentrations of 1, 100, and 1000 μg/L (Pomati et al. 2004). Our group also conducted a preliminary experiment to investigate the effects of SMZ and TET in Synechocystis sp. PCC 6803 at different test concentrations, and the most significant stimulatory effects were observed at the test concentration of 100 ng/L. Therefore, only the experimental results at an antibiotic concentration of 100 ng/L are shown in this study. When antibiotics are applied to the oil-producing cyanobacteria/microalgae culture system, the antibiotic concentration is supposed to be the essential operation parameter.
Influence of antibiotics on the photosynthetic activity
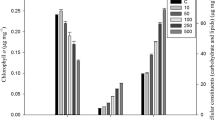
The stimulation of photosynthetic activity is a typical hormesis effect of antibiotics in cyanobacteria. For instance, erythromycin and ofloxacin were found to increase the Fv/Fm and rETRmax values in cyanobacteria at the ng/L concentration level (Deng et al. 2015; Wan et al. 2015). According to Fig. 2, this study also observed increased photosynthetic activity in antibiotic-treated groups. In each test group, the chlorophyll a content increased with increasing culture time and varied with a similar trend as that of the biomass. In the exponential phase and stationary phase, the chlorophyll a content was significantly (p < 0.05) increased by 70.6–87.1% and 49.7–78.3% under exposure to SMZ and TET, respectively. In each test group, Fv/Fm and rETRmax gradually increased in the exponential phase, reached the highest values on the twelfth day, and gradually decreased in the stationary phase. The SMZ and TET treatments significantly (p < 0.05) increased the Fv/Fm and rETRmax values. During the whole exposure period, the Fv/Fm values were significantly increased by 21.5–51.5% and 24.9–48.8% under exposure to SMZ and TET, respectively. The rETRmax values were significantly increased by 28.8–69% and 21.3–59.9% under exposure to SMZ and TET, respectively. These results indicated that both SMZ and TET stimulated photosynthetic activity through the promotion of light energy conversion efficiency (as indicated by Fv/Fm) and photochemical electron transport (as indicated by rETRmax). Furthermore, this study observed that SMZ and TET upregulated 22 and 23 proteins involved in the photosynthetic process, respectively (Table S1). This result was in consistence with a previous study, in which upregulated photosynthesis-related proteins were found to play an important role in tolerance to environmental stresses (Rowland et al. 2010).
Effects of sulfamethoxazole (SMZ) and tetracycline (TET) on a the chlorophyll a content, b the Fv/Fm value, and c the rETRmax value of Synechocystis sp. PCC 6803 (mean ± standard deviation of three replicates are shown for each value; * indicates significant difference between each antibiotic-treated group and the solvent control at p < 0.05 according to Student’s t test)
Overexpression of photosynthesis-related proteins and increased photosynthetic activity may result in an increased energy supply for cyanobacterial growth and fatty acid biosynthesis (Paul 2013).
Influence of antibiotics on biofuel production ability
Previous studies observed that some environmental stresses could increase the lipid content in cyanobacteria. For instance, UV radiation and TiO2 stresses were found to increase the lipid content in Arthrospira (Spirulina) platensis without affecting the biomass (Casazza et al. 2015). Ultrasonic treatment was found to increase the lipid content in oil-producing cyanobacteria, but the influence of ultrasonic treatment on biomass remained unclear (Ellison et al. 2019). The present study observed that antibiotic stress could simultaneously increase the biomass growth and lipid content. According to Table 1, antibiotic exposure significantly increased (p < 0.05) the dry weight of the cyanobacterial cells. This result was in consistence with the responses of the OD730 values. The lipid content (%DW) in the cyanobacterial cells of the solvent control was 14.71%, which was close to the results obtained by Touloupakis et al. (2015). Exposure to SMZ and TET significantly increased (p < 0.05) the lipid content to 19.10% and 16.98%, respectively. Furthermore, the lipid productivity in the SMZ-treated group and TET-treated group was 2.49- and 2.19-fold higher than that in the solvent control, respectively.
According to the proteomic responses to target antibiotics (Table S1), 4 proteins involved in energy derivation by oxidation of organic compounds (sdhB, norB, fumC, and malQ), 3 proteins involved in the catabolic process (gloB, gst1, and sll0654), and one protein involved in carbohydrate metabolic process (sll0529) were differentially expressed in the SMZ-treated group. Four proteins related to carbohydrate metabolic process (slr0453, pgm, fbp and nplT), 2 proteins involved in the catabolic process (sll0828 and gst1), and 2 proteins involved in energy derivation by oxidation of organic compounds (sdhB and norB) were differentially expressed in the TET-treated group. All of the above proteins were downregulated by the target antibiotics, suggesting that the two target antibiotics regulated the proteomic expression profile of Synechocystis sp. PCC 6803 towards an inhibition of carbohydrate catabolism. Furthermore, SMZ downregulated four carbohydrate transport proteins (slr1841, slr1908, sll1550, and slr0042), and TET downregulated six carbohydrate transport proteins (slr0042, sll1271, slr1908, sll0772, sll1550, and slr1841) in Synechocystis sp. PCC 6803. The downregulation of carbohydrate transport proteins may inhibit the transmembrane excretion of carbohydrates. Inhibited carbohydrate catabolism and suppressed carbohydrate excretion might contribute to the accumulation of carbohydrates (including fatty acids) in Synechocystis sp. PCC 6803 and consequently led to increased lipid productivity.
Twenty-six and 25 proteins related to the biosynthetic process were differentially expressed in the SMZ-treated group and the TET-treated group, respectively (Table S1). The proteomic responses suggested that exposure to target antibiotics may cause an alteration in the biosynthetic pattern of cellular substances. The altered composition of fatty acids in each antibiotic-treated group compared with the solvent control further verified this hypothesis. The fatty acids in Synechocystis sp. PCC 6803 are methyl-esterified into FAMEs and quantified by GC/MS, as shown in Table 1. Eight fatty acids were identified in the solvent control, among which palmitic acid (C16:0) and linoleic acid (C18:2) were major components. In each antibiotic-treated group, one more fatty acid, tetradecanoic acid (C14:0), was identified. The proportion of linolenic acid (C18:3) in the three test groups was lower than 12%, which met the requirements for a qualified biofuel according to the European Standards (EN 2004). Compared with the solvent control, SMZ and TET exposure significantly (p < 0.05) increased the proportions of monounsaturated fatty acids (MUFAs), while TET exposure significantly reduced (p < 0.05) the proportion of saturated fatty acids (SFAs) and polyunsaturated fatty acids (PUFAs). The length and unsaturation degree of fatty acid chains and the composition of individual fatty acids are key factors that determine the oxidative stability and physical characteristics of biofuel (Deshmukh et al. 2019a, b; Islam et al. 2013). The biofuel containing more MUFAs and less SFAs and PUFAs presents better combustion performance (Adu-Mensah et al. 2019). In addition, oleic acid (C18:1) is considered to be an optimal fatty acid with good oxidative stability and cold-flow properties (Singh et al. 2019). The production of oleic acid (C18:1) in the cyanobacterial cells was significantly (p < 0.05) stimulated by the two target antibiotics. The above results indicated that both SMZ and TET improved the combustion performance of the biofuel produced by Synechocystis sp. PCC 6803. The above results indicated that antibiotics have good prospects for application in cyanobacterial biofuel production.
Interactions of differentially expressed proteins in the antibiotic-treated groups
SMZ upregulated 83 proteins and downregulated 51 proteins in Synechocystis sp. PCC 6803, whereas TET upregulated 73 proteins and downregulated 44 proteins. There were more upregulated proteins than downregulated proteins, which further verified the adaptive response to the target antibiotics at the proteomic level. Although the two target antibiotics have different antibacterial mechanisms (González-Pleiter et al. 2013), both of them affected 13 biological processes in Synechocystis sp. PCC 6803, namely, biosynthetic process, catabolic process, carbohydrate metabolic process, cell differentiation, cell division, energy derivation by oxidation of organic compounds, nitrogen compound metabolic process, gene expression, photosynthesis, oxidation-reduction process, methylation, transport, and response to stimulus. In addition, 68 differentially expressed proteins were commonly shared by the SMZ-treated group and the TET-treated group. These results suggested that different antibiotics may regulate Synechocystis sp. PCC 6803 in similar patterns.
According to the PPI networks shown in Fig. 3, 129 of the 134 differentially expressed proteins participate in 1235 pairs of interactions in the SMZ-treated group, and 113 of the 117 differentially expressed proteins participate in 935 pairs of interactions in the TET-treated group. In the SMZ-treated group, 27 hub proteins with high connection degrees (≥ 30 interacting proteins) were recognized (Table 2), which were regarded as the most essential proteins in the PPI network. The hub proteins included 20 proteins (74.1%) related to gene expression (rpsD, rpsL, rpsE, efp, rpsG, rplB, rpsS, rpsH, rplQ, rpsO, rsmA, rplP, rpsJ, rplR, nusG, rnc1, rplO, dnaE-N, rplT, and rpmB), 3 proteins (11.1%) involved in stress response (groL1, groS, and sodB), 3 proteins (11.1%) involved in photosynthesis (psaF, petB, and psbH), and one protein (3.7%) involved in transport (secY). Most of the hub proteins (23 of 27) were upregulated. Similar results were also observed in the PPI network of the TET-treated group. Thirteen hub proteins were recognized in the PPI network, namely, 4 proteins (30.8%) involved in the photosynthesis process (psaF, petB, petD, and psbF), 6 proteins (46.1%) involved in gene expression (rpsL, rpsD, rplB, rpsH, rplQ, and rpsG), one protein (7.7%) involved in biosynthetic process (glyA), one protein (7.7%) involved in response to stimulus (sodB), and one protein (7.7%) involved in transport (secY). Twelve of the 13 hub proteins in the TET-treated group were upregulated. Elevated gene expression and stimulated photosynthesis played essential roles in the response to the stress caused by antibiotic exposure.
The interaction networks of the differentially expressed proteins in Synechocystis sp. PCC 6803 exposed to 100 ng/L of a sulfamethoxazole (SMZ) and b tetracycline (TET), constructed according to the STRING database. The protein symbols are shown in Table S1 in the Supplementary Material. The connection degree of each protein is indicated by color
A significantly enriched (p < 0.05) functional module containing highly interconnected proteins was identified in the PPI network of the SMZ-treated group (Fig. S1 in the Supplementary Material). In this module, 20 upregulated proteins involved in the photosynthesis process (psbH, psbF, psbU, psbE, psaL, psaE, psaF, psaJ, psbV, psbY, psaD, petB, petM, petA, petJ, petG, thf1, isiA, slr1739, and ftrC) were closely correlated with an upregulated protein involved in the biosynthetic process (crtB), a downregulated protein involved in the catabolic process (sll0654), and four downregulated carbohydrate transport proteins (sll1550, slr1908, slr1841, and slr0042). Similar to the SMZ-treated group, in the functional module of the TET-treated group, close correlations among 21 upregulated photosynthesis-related proteins (psbL, psaK2, psbH, psaM, psbE, psbU, psbF, psbY, psaL, psaE, psaD, psaF, petF, petD, petB, petA, petJ, petG, cpcA, cpcG, and ftrC), one upregulated biosynthesis-related protein (hemH), and six downregulated carbohydrate transport proteins (slr0042, sll1271, slr1908, sll1550, sll0772, and slr1841) were observed (Fig. S2 in the Supplementary Material). This result indicated that the stimulation of photosynthesis by antibiotics may further trigger the altered pattern of carbohydrate metabolism and transport. The hub proteins and functional modules in the two PPI networks further verified that the two different antibiotics shared similar regulation mechanisms for the biofuel production of Synechocystis sp. PCC 6803 at the proteomic level.
Environmental impact on the utilization of antibiotics in biofuel production
Antibiotics have been widely used in agriculture to promote the growth of plants and animals (Cerqueira et al. 2019; Zhao et al. 2020). However, the discharge of antibiotic residues was proved to induce antibiotic resistance in environmental bacteria (Cerqueira et al. 2019). In this study, SMZ and TET were highly effective in improving the lipid productivity of oil-producing cyanobacteria, which could consequently increase economic efficiency. However, the potential adverse environmental impacts during the utilization of antibiotics should also be considered. Previous studies observed that antibiotic resistance in bacteria correlated with the differential expression of proteins related to carbohydrate metabolism, transport, and biosynthesis (Jones-Dias et al. 2017; Opoku-Temeng et al. 2019; Zuñiga-Navarrete et al. 2019). These findings were in accordance with the proteomic responses of Synechocystis sp. PCC 6803 under exposure to antibiotics. In addition, the overexpression of ribosomal proteins plays an important role in bacterial resistance to antibiotics (Vranakis et al. 2014). In this study, ribosomal proteins also accounted for a large proportion of the upregulated proteins in Synechocystis sp. PCC 6803 under exposure to antibiotics. The above results suggested that cyanobacteria showed antibiotic resistance mechanisms similar to those of bacteria. Recent studies have suggested that antibiotic resistance in cyanobacteria results in an increased bloom-forming ability, which poses a threat to aquatic ecosystems (Le Page et al. 2019). To eliminate the spread of antibiotic resistance in the environment, antibiotic residues should be completely removed from the culture medium after utilization in the culture of oil-producing cyanobacteria. Various advanced oxidation technologies are effective for the removal of antibiotics, and the elimination of antibiotic residues is easy to accomplish (Eniola et al. 2019; Wu et al. 2020). Furthermore, photolysis and hydrolysis of antibiotics could not be avoided during the cyanobacterial culture process (Biošić et al. 2017), and the influence of antibiotic degradation products on oil-producing cyanobacteria and the environment deserves further investigation.
Conclusions
This study observed beneficial effects of sulfamethoxazole and tetracycline on the biomass growth, lipid content, photosynthetic activity, and fatty acid composition of Synechocystis sp. PCC 6803 at the ng/L concentration level. The proteomic responses indicated that upregulation of gene expression-related proteins and photosynthesis-related proteins contributed the most to the stimulatory effects of the target antibiotics. The upregulated photosynthesis-related proteins further triggered the differential expression of proteins related to carbohydrate metabolism and transport, which resulted in increased lipid accumulation and altered fatty acid composition in cyanobacterial cells. The introduction of antibiotic stress into the oil-producing cyanobacterial culture system showed good prospects for increasing biofuel productivity.
References
Aderemi AO, Novais SC, Lemos MFL, Alves LM, Hunter C, Pahl O (2018) Oxidative stress responses and cellular energy allocation changes in microalgae following exposure to widely used human antibiotics. Aquat Toxicol 203:130–139. https://doi.org/10.1016/j.aquatox.2018.08.008
Adu-Mensah D, Mei D, Zuo L, Zhang Q, Wang J (2019) A review on partial hydrogenation of biodiesel and its influence on fuel properties. Fuel 251:660–668. https://doi.org/10.1016/j.fuel.2019.04.036
Anahas AMP, Muralitharan G (2015) Isolation and screening of heterocystous cyanobacterial strains for biodiesel production by evaluating the fuel properties from fatty acid methyl ester (FAME) profiles. Bioresour Technol 184:9–17. https://doi.org/10.1016/j.biortech.2014.11.003
Aslam A, Thomas-Hall SR, Manzoor M, Jabeen F, Iqbal M, Uz ZQ, Schenk PM, Asif TM (2018) Mixed microalgae consortia growth under higher concentration of CO2 from unfiltered coal fired flue gas: fatty acid profiling and biodiesel production. J Photochem Photobiol B 179:126–133. https://doi.org/10.1016/j.jphotobiol.2018.01.003
Ben Y, Fu C, Hu M, Liu L, Wong MH, Zheng C (2019) Human health risk assessment of antibiotic resistance associated with antibiotic residues in the environment: a review. Environ Res 169:483–493. https://doi.org/10.1016/j.envres.2018.11.040
Bharte S, Desai K (2019) The enhanced lipid productivity of Chlorella minutissima and Chlorella pyrenoidosa by carbon coupling nitrogen manipulation for biodiesel production. Environ Sci Pollut Res 26:3492–3500. https://doi.org/10.1007/s11356-018-3757-5
Biošić M, Mitrevski M, Babić S (2017) Environmental behavior of sulfadiazine, sulfamethazine, and their metabolites. Environ Sci Pollut Res 24:9802–9812. https://doi.org/10.1007/s11356-017-8639-8
Bland E, Angenent LT (2016) Pigment-targeted light wavelength and intensity promotes efficient photoautotrophic growth of Cyanobacteria. Bioresour Technol 216:579–586. https://doi.org/10.1016/j.biortech.2016.05.116
Casazza AA, Ferrari PF, Aliakbarian B, Converti A, Perego P (2015) Effect of UV radiation or titanium dioxide on polyphenol and lipid contents of Arthrospira (Spirulina) platensis. Algal Res 12:308–315. https://doi.org/10.1016/j.algal.2015.09.012
Cerqueira F, Matamoros V, Bayona J, Piña B (2019) Antibiotic resistance genes distribution in microbiomes from the soil-plant-fruit continuum in commercial Lycopersicon esculentum fields under different agricultural practices. Sci Total Environ 652:660–670. https://doi.org/10.1016/j.scitotenv.2018.10.268
Chandra R, Amit, Ghosh UK (2019) Effects of various abiotic factors on biomass growth and lipid yield of Chlorella minutissima for sustainable biodiesel production. Environ Sci Pollut Res 26:3848–3861. https://doi.org/10.1007/s11356-018-3696-1
Cordeiro RS, Vaz ICD, Magalhaes SMS, Barbosa FAR (2017) Effects of nutritional conditions on lipid production by cyanobacteria. An Acad Bras Cienc 89:2021–2031. https://doi.org/10.1590/0001-3765201720150707
Deng CN, Pan XL, Zhang DY (2015) Influence of ofloxacin on photosystems I and II activities of Microcystis aeruginosa and the potential role of cyclic electron flow. J Biosci Bioeng 119:159–164. https://doi.org/10.1016/j.jbiosc.2014.07.014
Deshmukh S, Bala K, Kumar R (2019a) Selection of microalgae species based on their lipid content, fatty acid profile and apparent fuel properties for biodiesel production. Environ Sci Pollut Res 26:24462–24473. https://doi.org/10.1007/s11356-019-05692-z
Deshmukh S, Kumar R, Bala K (2019b) Microalgae biodiesel: a review on oil extraction, fatty acid composition, properties and effect on engine performance and emissions. Fuel Process Technol 191:232–247. https://doi.org/10.1016/j.fuproc.2019.03.013
Dineshbabu G, Uma VS, Mathimani T, Prabaharan D, Uma L (2020) Elevated CO2 impact on growth and lipid of marine cyanobacterium Phormidium valderianum BDU 20041–towards microalgal carbon sequestration. Biocatal Agric Biotechnol 25:101606. https://doi.org/10.1016/j.bcab.2020.101606
Ellison CR, Overa S, Boldor D (2019) Central composite design parameterization of microalgae/cyanobacteria co-culture pretreatment for enhanced lipid extraction using an external clamp-on ultrasonic transducer. Ultrason Sonochem 51:496–503. https://doi.org/10.1016/j.ultsonch.2018.05.006
EN (2004) Automotive fuels-fatty acid methyl esters (FAME) for diesel engines-requirements and test methods
Eniola JO, Kumar R, Barakat MA (2019) Adsorptive removal of antibiotics from water over natural and modified adsorbents. Environ Sci Pollut Res 26:34775–34788. https://doi.org/10.1016/j.scitotenv.2015.05.130
Fu W, Li P, Wu Y (2012) Effects of different light intensities on chlorophyll fluorescence characteristics and yield in lettuce. Sci Hortic-Amsterdam 135:45–51. https://doi.org/10.1016/j.scienta.2011.12.004
González-Pleiter M, Gonzalo S, Rodea-Palomares I, Leganés F, Rosal R, Boltes K, Marco E, Fernández-Piñas F (2013) Toxicity of five antibiotics and their mixtures towards photosynthetic aquatic organisms: implications for environmental risk assessment. Water Res 47:2050–2064. https://doi.org/10.1016/j.watres.2013.01.020
Han F, Pei HY, Hu WR, Jiang LQ, Cheng J, Zhang LJ (2016) Beneficial changes in biomass and lipid of microalgae Anabaena variabilis facing the ultrasonic stress environment. Bioresour Technol 209:16–22. https://doi.org/10.1016/j.biortech.2016.02.103
He J, Yang Y, Zhang J, Chen J, Wei X, He J, Luo L (2017) Ribosome biogenesis protein Urb1 acts downstream of mTOR complex 1 to modulate digestive organ development in zebrafish. J Genet Genomics 44:567–576. https://doi.org/10.1016/j.jgg.2017.09.013
Islam MA, Magnusson M, Brown RJ, Ayoko GA, Nabi MN, Heimann K (2013) Microalgal species selection for biodiesel production based on fuel properties derived from fatty acid profiles. Energies 6:5676–5702. https://doi.org/10.3390/en6115676
Jones-Dias D, Carvalho AS, Moura IB, Manageiro V, Igrejas G, Caniça M, Matthiesen R (2017) Quantitative proteome analysis of an antibiotic resistant Escherichia coli exposed to tetracycline reveals multiple affected metabolic and peptidoglycan processes. J Proteome 156:20–28. https://doi.org/10.1016/j.jprot.2016.12.017
Le Page G, Gunnarsson L, Trznadel M, Wedgwood KCA, Baudrot V, Snape J, Tyler CR (2019) Variability in cyanobacteria sensitivity to antibiotics and implications for environmental risk assessment. Sci Total Environ 695:133804. https://doi.org/10.1016/j.scitotenv.2019.133804
Lepage G, Roy CC (1984) Improved recovery of fatty acid through direct transesterification without prior extraction or purification. J Lipid Res 25:1391
Liu Y, Chen S, Zhang J, Gao B (2016) Growth, microcystin-production and proteomic responses of Microcystis aeruginosa under long-term exposure to amoxicillin. Water Res 93:141–152. https://doi.org/10.1016/j.watres.2016.01.060
Liu Y, Chen S, Zhang J, Li X, Gao B (2017) Stimulation effects of ciprofloxacin and sulphamethoxazole in Microcystis aeruginosa and isobaric tag for relative and absolute quantitation-based screening of antibiotic targets. Mol Ecol 26:689–701. https://doi.org/10.1111/mec.13934
Maul JD, Schuler LJ, Belden JB, Whiles MR, Lydy MJ (2006) Effects of the antibiotic ciprofloxacin of stream microbial communities and detritivorous macroinvertebrates. Environ Toxicol Chem 25:1598–1606. https://doi.org/10.1897/05-441R.1
Opoku-Temeng C, Onyedibe KI, Aryal UK, Sintim HO (2019) Proteomic analysis of bacterial response to a 4-hydroxybenzylidene indolinone compound, which re-sensitizes bacteria to traditional antibiotics. J Proteome 202:103368. https://doi.org/10.1016/j.jprot.2019.04.018
Pattanaik B, Englund E, Nolte N, Lindberg P (2020) Introduction of a green algal squalene synthase enhances squalene accumulation in a strain of Synechocystis sp. PCC 6803. Metab Eng Commun 10:e125. https://doi.org/10.1016/j.mec.2020.e00125
Paul MJ (2013) Photosynthetic carbon dioxide fixation. 497-502*497-502. In: Lennarz WJ, Lane MD (eds) Encyclopedia of Biological chemistry, Second edn. Academic Press, Waltham. https://doi.org/10.1016/B978-0-12-378630-2.00050-5
Pomati F, Netting AG, Calamari D, Neilan BA (2004) Effects of erythromycin, tetracycline and ibuprofen on the growth of Synechocystis sp. and Lemna minor. Aquat Toxicol 67:387–396. https://doi.org/10.1016/j.aquatox.2004.02.001
Raheem A, Prinsen P, Vuppaladadiyam AK, Zhao M, Luque R (2018) A review on sustainable microalgae based biofuel and bioenergy production: recent developments. J Clean Prod 181:42–59. https://doi.org/10.1016/j.jclepro.2018.01.125
Rowland JG, William JS, Yoshitaka N, Antoni RS (2010) Differential proteomic analysis using iTRAQ reveals changes in thylakoids associated with Photosystem II-acquired thermotolerance in Synechocystis sp. PCC 6803. Proteomics 10. https://doi.org/10.1002/pmic.200900337
Shuba ES, Kifle D (2018) Microalgae to biofuels: ‘promising’ alternative and renewable energy, review. Renew Sust Energ Rev 81:743–755. https://doi.org/10.1016/j.rser.2017.08.042
Singh D, Sharma D, Soni SL, Sharma S, Kumari D (2019) Chemical compositions, properties, and standards for different generation biodiesels: a review. Fuel 253:60–71. https://doi.org/10.1016/j.fuel.2019.04.174
Sivaramakrishnan R, Incharoensakdi A (2018) Enhancement of lipid production in Synechocystis sp. PCC 6803 overexpressing glycerol kinase under oxidative stress with glycerol supplementation. Bioresour Technol 267:532–540. https://doi.org/10.1016/j.biortech.2018.07.058
Touloupakis E, Cicchi B, Torzillo G (2015) A bioenergetic assessment of photosynthetic growth of Synechocystis sp. PCC 6803 in continuous cultures. Biotechnol Biofuels 8:133. https://doi.org/10.1186/s13068-015-0319-7
Vranakis I, Goniotakis I, Psaroulaki A, Sandalakis V, Tselentis Y, Gevaert K, Tsiotis G (2014) Proteome studies of bacterial antibiotic resistance mechanisms. J Proteome 97:88–99. https://doi.org/10.1016/j.jprot.2013.10.027
Wahlen BD, Willis RM, Seefeldt LC (2011) Biodiesel production by simultaneous extraction and conversion of total lipids from microalgae, cyanobacteria, and wild mixed-cultures. Bioresour Technol 102:2724–2730. https://doi.org/10.1016/j.biortech.2010.11.026
Wan J, Guo P, Peng X, Wen K (2015) Effect of erythromycin exposure on the growth, antioxidant system and photosynthesis of Microcystis flos-aquae. J Hazard Mater 283:778–786. https://doi.org/10.1016/j.jhazmat.2014.10.026
Wang Z, Chen Q, Zhang J, Dong J, Ao Y, Wang M, Wang X (2019) Long-term exposure to antibiotic mixtures favors microcystin synthesis and release in Microcystis aeruginosa with different morphologies. Chemosphere 235:344–353. https://doi.org/10.1016/j.chemosphere.2019.06.192
Wiśniewski JR, Zougman A, Nagaraj N, Mann M (2009) Universal sample preparation method for proteome analysis. Nat Methods 6:359–362. https://doi.org/10.1038/nmeth.1322
Wu K, Zhang C, Liu T, Lei H, Yang S, Jin P (2020) The removal of tetracycline, oxytetracycline, and chlortetracycline by manganese oxide–doped copper oxide: the behaviors and insights of Cu-Mn combination for enhancing antibiotics removal. Environ Sci Pollut Res 27:12613–12623. https://doi.org/10.1007/s11356-020-07810-8
Yang Y, Song W, Lin H, Wang W, Du L, Xing W (2018) Antibiotics and antibiotic resistance genes in global lakes: a review and meta-analysis. Environ Int 116:60–73. https://doi.org/10.1016/j.envint.2018.04.011
Zhao F, Chen L, Yen H, Sun L, Li S, Li M, Feng Q, Yang L (2020) Multimedia mass balance approach to characterizing the transport potential of antibiotics in soil–plant systems following manure application. J Hazard Mater 393:122363. https://doi.org/10.1016/j.jhazmat.2020.122363
Živković SB, Veljković MV, Banković-Ilić IB, Krstić IM, Konstantinović SS, Ilić SB, Avramović JM, Stamenković OS, Veljković VB (2017) Technological, technical, economic, environmental, social, human health risk, toxicological and policy considerations of biodiesel production and use. Renew Sust Energ Rev 79:222–247. https://doi.org/10.1016/j.rser.2017.05.048
Zou X, Lin Z, Deng Z, Yin D (2013) Novel approach to predicting hormetic effects of antibiotic mixtures on Vibrio fischeri. Chemosphere 90:2070–2076. https://doi.org/10.1016/j.chemosphere.2012.09.042
Zuñiga-Navarrete F, Flores-Ramirez G, Danchenko M, Benada O, Skriba A, Skultety L (2019) Proteomic analysis revealed the survival strategy of Coxiella burnetii to doxycycline exposure. J Proteome 208:103479. https://doi.org/10.1016/j.jprot.2019.103479
Acknowledgments
This work was supported by the National Natural Science Foundation of China (51679130) partly by the Fundamental Research Funds of Shandong University (2017WLJH35).
Funding
This study was funded by the National Natural Science Foundation of China (51679130) partly by the Fundamental Research Funds of Shandong University (2017WLJH35).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Ta Yeong Wu
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 376 kb)
Rights and permissions
About this article
Cite this article
Cui, M., Liu, Y. & Zhang, J. Sulfamethoxazole and tetracycline induced alterations in biomass, photosynthesis, lipid productivity, and proteomic expression of Synechocystis sp. PCC 6803. Environ Sci Pollut Res 27, 30437–30447 (2020). https://doi.org/10.1007/s11356-020-09327-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-020-09327-6