Abstract
Widely distributed western mosquitofish (Gambusia affinis) has been used as a new model species for hazard assessment of environmental stressors such as polycyclic aromatic hydrocarbons (PAHs). However, most of the PAH studies using G. affinis rely on targeted biomarker-based analysis, and thus may not adequately address the complexity of the toxic mechanisms of the stressors. In the present study, the whole transcriptional sequencing of G. affinis liver after exposure to a PAH model, benzo[a]pyrene (BaP) (100 μg/L), for 20 days was performed by using the HiSeq XTen sequencers. In total, 58,156,233 and 51,825,467 clean nucleotide reads were obtained in the control and BaP-exposed libraries, respectively, with average N50 lengths of 1419 bp. In addition, after G. affinis was exposed for 20 days, 169 genes were upregulated, and 176 genes were downregulated in liver. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were applied to all the genes to determine the genes’ biological functions and processes. The results clearly showed that the differentially expressed genes were mainly related to immune pathways and metabolic correlation pathways. Interestingly, almost all the pathways related with the immunity were upregulated, while the metabolism pathways were downregulated. Lastly, quantitative real-time PCR (qRT-PCR) was performed to measure expressional levels of twelve genes confirmed through the DGE analysis. These results demonstrate that BaP damages immunity and enhances the consumption of all available energy storage to activate mechanisms of the detoxification in G. affinis. Up until now, the present study is the first time that a whole transcriptome sequencing analysis in the liver of G. affinis exposed to BaP has been reported.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Polycyclic aromatic hydrocarbons (PAHs) are notorious and ubiquitous environmental contaminants with mutagenic and carcinogenic properties to all living organisms. PAHs are primarily generated from the incomplete combustion of the organic matter such as the burning of fossil fuels and oil (Fang et al. 2015; Sushkova et al. 2018). Benzo[a]pyrene (BaP) is one of the widely distributed and well-studied PAHs (Cui et al. 2019b). Previous studies on the toxicities of BaP have showed that BaP has deleterious effects on the reproduction systems, development stages, metabolic pathways, and immunological responses of human and wildlife (Corrales et al. 2014; Zhan et al. 2015; Chen et al. 2018; Yanagisawa et al. 2018; Zhang et al. 2018).
With the advancement of sequencing technology, transcriptomic sequencing has been widely used in toxicology research to analyze whole transcriptional changes after the experimental animals are exposed to toxic compounds. Furthermore, the results from transcriptomic sequencing could serve as a powerful method for developing biomarkers for environmental contaminants and even reveal the toxicological mechanisms of toxic compounds. So far, the changes of transcriptome responses when exposed to BaP have been reported in several species, such as Danio rerio (Fang et al. 2015; Jayasundara et al. 2015), Paralichthys olivaceus (Jung et al. 2018), Ruditapes philippinarum (Wang et al. 2018), Luciola lei (Zhang et al. 2019), Diaphanosoma celebensis (Kim et al. 2018), Chlamys farreri (Cai et al. 2014), Eriocheir sinensis (Yu et al. 2018), Gadus morhua (Yadetie et al. 2018), Oreochromis niloticus (Colli-Dula et al. 2018b; Colli-Dula et al. 2018a), Boreogadus saida (Song et al. 2019), and Perna viridis (Jiang et al. 2016).
The western mosquitofish Gambusia affinis (G. affinis) (Baird and Girard, 1853) was distributed in the freshwaters in the mid-western parts of the USA and Northeastern Mexico (Krumholz 1948; Pyke 2008). To be used for mosquito control, the western mosquitofish is now widely distributed all over the world (Gao et al. 2017). Similar to other Cyprinodontiformes fishes including medaka, guppies, and killifish, the western mosquitofish has been widely utilized for environmental toxicity studies because of mosquitofish’s diversity of characteristics in the environment, a wide geographic distribution, and adaptability to live in a wide range of environmental conditions (Caliani et al. 2009; Wills et al. 2009; Xie et al. 2010; Kamata et al. 2011; Hou et al. 2017; Bao et al. 2018). Although the toxicities of BaP have been extensively reported in various species, the effects of BaP toxicities on the liver of G. affinis are still unclear. Based on the report that the concentration of benzo[a]pyrene in some polluted rivers is close to 100 μg/L (Ekere et al. 2019; Mojiri et al. 2019), in our previous study, we chose adult male G. affinis exposed to BaP (100 μg/L). During the experiment, we found that the movement of the western mosquitofish has decreased and they began to die after exposure to BaP (100 μg/L) for 15 days. After exposure for 20 days, 15% (9/60) of the western mosquitofish died and three quarters of living western mosquitofish showed decreased movement. To investigate the molecular mechanisms that explain how the western mosquitofish responds to BaP, the transcriptional sequencing of G. affinis liver exposed to BaP (100 μg/L) for 20 days was performed. In this study, the goals are as follows: (1) to detect the differentially expressed genes involved in the responses of exposure to BaP; (2) to characterize gene expression on a larger scale. This will facilitate the detailed characterization on genes regulating the toxicological responses to BaP; and (3) to discover the pathway and networks of genes that are enriched for regulating G. affinis resistance to PAHs.
Materials and methods
Fish collection and maintenance
Western mosquitofish G. affinis for laboratory experiments were captured from an artificial lake (23° 09′ 48″ N, 113° 21′ 16″ E) which is located at the South China Agricultural University in Guangzhou City, Guangdong province, China. The fish were maintained for at least 1 month at 25 ± 1 °C in two glass tanks of 80 L before the experiment started, with a light:dark photoperiod of 14 h:10 h. The fish were fed with fairy shrimp twice daily.
Chemicals and the procedures for exposure
Dimethyl sulfoxide (DMSO) was obtained from Sangon Biotech (Shanghai, China). BaP (CAS50-32-8; purity > 96%) was purchased from Aladdin (Shanghai, China). Stock solution of BaP was 10 mg/mL in DMSO. To avoid the effect of sex on the results (Sun et al. 2020), we decided to use only adult male fishes in the experiment where the fish were exposed to BaP. In total, 120 healthy adult male fishes were randomly divided into two groups, with three replicates per group. The experiment of exposing the fish to BaP was performed in a 1-L glass crystallizing dish. There are six dishes, with each dish containing 20 healthy fishes and 0.5 L of exposure medium. The treatment groups were exposed to BaP (100 μg/L) for 20 days, and the control groups were immersed in filtered water with exactly the same proportion of DMSO as that in the treatment groups. DMSO, as the co-solvent, used in each experiment was less than or equal to 0.01% (volume ratio). All the experiments were performed under the static condition and the water was replaced daily. During the experimental period, mosquitofish were fed with fairy shrimp twice daily.
RNA extraction, and library construction and sequencing
At the designed time points, 10 liver tissues from fish per replicate from each group were excised and blended into one sample by pooling an equal amount of liver tissue. To obtain the liver tissues, fish were first anesthetized with 3-aminobenzoic acid ethyl ester methane sulfonate (MS222) (50 mg/L) (Adamas-Beta, Shanghai, China). The combined 10 liver tissues were then snap-frozen in the liquid nitrogen. Subsequently, the samples were homogenized in RNAiso Plus (Takara, Dalian, China) and were stored at − 80 °C until RNA extraction is processed. Total RNA was extracted using the Total RNA Extractor (Trizol) kit (B511311, Sangon, China) and was treated with RNase-free DNase I to remove genomic DNA contamination. The RNA integrity was checked on 1% agarose gels. The RNA purity was examined using the Nano Photometer® spectrophotometer (Implen, CA, USA). The RNA concentration was measured using Qubit® RNA Assay Kit in Qubit®2.0 Flurometer (Life Technologies, CA, USA). The quality and quantity of RNA was assessed using the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA). The high-quality RNA samples were subsequently submitted to the Sangon Biotech (Shanghai) Co., Ltd., for the library preparation and sequencing. A total amount of 2 μg RNA per sample was used as input material for the library preparations. Sequencing libraries were generated using VAHTSTM mRNA-seq V2 Library Prep Kit for Illumina® following manufacturer’s recommendations. The library quality was assessed on the Agilent Bioanalyzer 2100 system and then was sequenced using the HiSeq XTen sequencers (Illumina, San Diego, CA).
Transcriptome assembly and gene annotation
The remaining clean reads were de novo assembled into transcripts using Trinity (version 2.0.6) with default settings. Transcripts with a minimum length of 200 bp were clustered into minimize redundancy. For each cluster (i.e., that is represented the transcriptional complexity for the same gene), the longest sequence was preserved and designated as a unigene. Unigenes were blasted against NCBI non-redundant protein database (NCBI Nr), Swiss-Prot, Translated European Molecular Laboratory (TrEMBL), Conserved Domain Database (CDD), the protein families (PFAM), and eukaryotic Orthologous Groups (KOG) databases (E-value < 1e−5). According to the priority order of the best aligned results of Nr, Swiss-Prot and TrEMBL were used to determine the unigene ORF. And then CDS and the corresponding amino acid sequences were determined according to the codon table. In the meantime, TransDecoder (version 3.0.1) was used to predict CDS sequences of the unaligned unigenes. In addition, Gene Ontology (GO) database was obtained according to the results of transcript annotation in the Swiss-Prot and TrEMBL databases. And Kyoto Encyclopedia of Genes and Genomes (KEGG) Automatic Annotation Server (KAAS, version 2.1) was used for KEGG annotation.
RNA-seq validation by qPCR
Twelve immunity and metabolism-related genes underwent qPCR analysis which was conducted using the same total RNA that was used in the RNA-seq analysis. cDNA synthesis was performed on approximately 1 μg of total RNA in a 20-μl setup using the PrimeScript™ RT reagent kit with gDNA Eraser (Perfect Real Time) (Takara, Beijing, China) according to the manufacturer’s recommendations. qPCR was conducted utilizing the SYBR Green assay and were performed in a Bio-Rad CFX Connect™ Real-Time System (Bio-Rad, USA). The qPCR reactions were carried out in a total volume of 20 μl consisting of 10 μl Thunderbird™ SYBR® qPCR mix without ROX dye (Toyobo, Japan), 0.4 μl each of forward and reverse primers (Table 1), and 1 μl of the cDNA reaction. The PCR conditions were set up as 95 °C for 1 min, followed by 40 cycles of 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 45 s. Each run included blank controls and cDNA controls. Each sample was assayed in triplicate. The data were analyzed according to the 2−ΔΔCT method (Livak and Schmittgen 2001). The β-actin was used as the internal control.
Histologic assessment
The whole fish were harvested and fixed in 4% paraformaldehyde (PFA) for at least 24 h. The fixed fish were immersed in the decalcifying solution (Servicebio, China) for 20 days. The samples were then fixed using optimal cutting temperature (OCT) compounds for embedding tissues and subsequently frozen sections. The samples were cut directly in 3–4-μm sections and stained with Oil Red O (ORO).
Data analysis
The relative expression levels of the selected genes analyzed by qPCR were presented as the fold increase or decrease of the fish that are exposed the BaP compared with that of the control fish. Student’s t test was used to evaluate statistical differences between the exposed, experimental, and non-exposed control fish by IBM SPSS Statistics 20. A probability level of 0.05 is considered statistically significant. The minimum acceptable fold change was set at least 2-fold change.
Results
Sequencing, de novo assembly, and functional annotation
Two parallel samples were used for RNA-seq (two control groups and two experimental groups). We pooled equal quantities of total RNA from the three replicates of the two groups to reduce the errors associated with individual differences. The libraries of BaP-exposed and control groups were analyzed via the Illumina HiSeq XTen sequencing in an average read length 150-nt run which generated, on average, a total of 58,156,233 and 51,825,467 raw nucleotide reads, respectively. After removal of the samples with low-quality reads, the total number of clean reads was 55,084,497 (94.72%) and 48,829,956 (94.22%), respectively. The Q20 percentages were 98.52% and 98.39%, respectively, and the GC percentages of clean reads were 51.11% and 51.42%, respectively. The average number of transcripts assembled from reads was 126,408. The average number of unigenes assembled from these transcripts was 76,876, with the average N50 lengths of 1419 bp. The quality of the assembled transcriptomes of G. affinis was good. A total of 76,876 unigenes were aligned to the CDD, KOG, COG, NR, NT, PFAM, Swiss-Prot, TrEMBL, KEGG, and GO databases. For the Nr database, the distribution analysis showed that 35.93% of unigenes had a strong similarity to the sequences of Xiphophorus maculatus, followed by Poecilia reticulata (14.68%), Poecilia Formosa (12.31%), Poecilia mexicana (11.56%), and Poecilia latipinna (10.88%) (Fig. 1).
Differentially expressed genes
To investigate the significance of the gene expression patterns observed in response to BaP exposure, the DESeq2 (version 1.12.4) was used to determine differentially expressed genes (DEGs) between the treatment and control groups. Genes were considered significant differentially expressed if q-value was less than 0.001 and the absolute value of fold change larger than 2. In total, 345 of the 76,876 (0.45%) unigenes showed significant differential expression. Among the 345 differentially expressed genes, a total of 169 (49.0%) unigenes were upregulated while 176 (51.0%) unigenes were downregulated when the western mosquitofish were exposed to BaP (Fig. 2).
Volcano plot of differently expressed genes (DEGs) from the liver of G. affinis exposed to BaP at 100 μg/L concentration for 20 days. The horizontal and vertical dotted lines show the adjusted p value equal to 0.05 and the minimum acceptable fold change, respectively. The value of log2FoldChange for all genes was analyzed as log2 (exposed/control). Tl means the exposed group and Cl means the control group
Functional analysis of differentially expressed genes
To identify which DEGs were significantly enriched in GO terms or metabolic pathways, the functional enrichment analyses namely GO and KEGG were performed. According to the GO database, the potential functions of DEGs were distributed into three categories, namely biological process, molecular function, and cellular component which was further classified into 50 subcategories (Fig. 3). The biological processes of DEGs were clustered into 22 subcategories. Among them, the most enriched components were “cellular process” (182 unigenes), “metabolic process” (156 unigenes), and “biological regulation” (132 unigenes). In contrast, the cellular components of DEGs were distributed into 16 subcategories. Among them, the major part was in the subcategories of “cell” (185 unigenes), “cell part” (180 unigenes), and “organelle” (136 unigenes). And the molecular functions of DEGs were assigned to 12 subcategories. The most enriched subcategories were “binding” (156 unigenes) and “catalytic activity” (110 unigenes).
The top 30 most significantly enriched up- and downregulated GO categories were presented in Fig. 4 a and b, and Table S1–2. Interestingly, according to the GO functional enrichment analysis results, the GO categories that are upregulated DEGs were mainly enriched in the immune-related pathway (Fig. 4a) and the GO categories that are downregulated DEGs were enriched in the metabolic correlation pathway (Fig. 4b). The 20 immune-related GO categories that are upregulated DEGs included immune system process, defense response, immune response, complement activation, humoral immune response mediated by circulating immunoglobulin, and so on. In addition, the 20 metabolic-related GO categories that are downregulated DEGs were mainly enriched in small molecule metabolic process, response to oxygen-containing compound, lipid metabolic process, carbohydrate metabolic process, and metabolism of fats, sugars, and proteins.
Overview of the significantly upregulated and downregulated pathways obtained from GO analysis. The x-axis indicates the enrichment ratio, while the y-axis indicated the specific pathways. The size of the color dots indicates the number of DEGs involved in each pathway. And the color of the dots indicates the value of false discovery rate. a and b showed that the upregulated and downregulated DEGs were enriched in the top 30 most highly enriched GO categories, respectively
For the KEGG pathway assignment, all the annotated pathways were grouped into five major categories: cellular processes (4 pathways), environmental information processing (3 pathways), genetic information processing (4 pathways), metabolism (12 pathways), and organismal systems (10 pathways). The KEGG pathway analysis of G. affinis transcripts showed that a total of 208 significant DEGs were assigned to these KEGG pathways. The highest number of unigenes was in signal transduction, which contained 27 unigenes. The second- and third-to-the highest were endocrine system (23 unigenes) and lipid metabolism (19 unigenes) (Fig. 5). Similar to the GO enrichment results, the results of functional enrichment analysis of KEGG have showed that the upregulated DEGs were enriched in immune-related pathways (Fig. 6a and Table S3) and the downregulated DEGs were enriched in metabolic-related pathways (Fig. 6b and Table S4). The most significantly upregulated KEGG pathway included toll-like receptor signaling pathway (5 genes) and NF-kappa B signaling pathway (5 genes) (Table S3). In contrast, the most significantly downregulated KEGG pathway included “pancreatic secretion” (10 genes), “protein digestion and absorption” (7 genes), and “fat digestion and absorption” (5 genes) (Table S4). Furthermore, the major significant DEGs involved in immune and metabolic pathways were listed in Tables 2 and 3.
Overview of the significantly upregulated or downregulated pathways obtained from KEGG analysis. The x-axis indicates the enrichment ratio, while the y-axis indicated the specific pathways. The size of the color dots indicates the number of DEGs involved in each pathway. And the color of the dots indicates the value of false discovery rate. a and b showed that the upregulated and downregulated DEGs were enriched in the top 30 most highly enriched KEGG categories, respectively
qPCR validation of differentially expressed genes
qPCR analysis of selected immune- and metabolism-related gene expression in the liver of G. affinis corresponds to RNA-seq results. As shown in Fig. 7 of the selected genes, seven genes encoding NF-kappa-B inhibitor alpha (NFKBIA), v-rel avian reticuloendotheliosis viral oncogene homolog B (RELB), complement component C6-like (C6), complement factor properdin (CFP), p53 apoptosis effector related to PMP-22 (PERP), inter-alpha-trypsin inhibitor heavy chain H3-like (ITIH4), and cytochrome P450 1A (CYP1A) were significantly upregulated, whereas the other five genes were significantly downregulated. The results from qPCR analyses were consistent with RNA-seq results except ITIH4 gene, which was found to be upregulated but downregulated in the RNA-seq result.
Validation of RNA-seq analysis by comparing RNA-seq data with qPCR analysis on the 12 selected genes in G. affinis. Among the 12 selected genes, 11 genes had the same trend between RNA-seq analysis and qPCR analysis. Only inter-alpha-trypsin inhibitor heavy chain H3-like (ITIH4) gene showed the opposite trend. NFKBIA, NF-κB inhibitor alpha. RELB, v-rel avian reticuloendotheliosis viral oncogene homolog B. C6, complement component C6-like. CFP, complement factor properdin. PERP, p53 apoptosis effector related to PMP-22. FADS2, fatty acid desaturase 2. ELOVL6, elongation of very long chain fatty acid protein 6. FASN, fatty acid synthase. CYP1A, cytochrome P450 1A. CELA2A, chymotrypsin-like elastase family member 2A. TRY1, trypsin-1
Lipid metabolism disorder induced by BaP
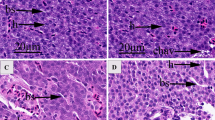
To determine whether the lipid metabolism of G. affinis was disordered or not by BaP, the ORO staining for total lipid content of the livers and the muscle in the control and the treatment groups was performed. Indeed, the histological data revealed that BaP induced a significant decrease in hepatocyte and myocyte lipid droplet contents (Fig. 8).
Discussion
The western mosquitofish, G. affinis, an abundant and widely distributed small fish species, has been used as a new model fish for environmental studies (Hou et al. 2018). A 598.7-Mb high-quality reference genome for G. affinis has been reported last year (Hoffberg et al. 2018). So far, there have been only two reports on the transcriptome of G. affinis: One successfully used the RNA-seq results to characterize a sex-specific marker in G. affinis, while the other revealed the mechanisms underlying female masculinization in G. affinis exposed to progestin (Lamatsch et al. 2015; Hou et al. 2019). To date, the transcriptome of G. affinis exposed to BaP has not yet described. BaP is a ubiquitous organic pollutant that is listed as a group 1 genotoxic carcinogen by International Agency for Research on Cancer (IARC 2017) (Cai et al. 2019). In the present study, we successfully obtained high-quality transcriptome data when G. affinis was exposed to BaP, which was supposed to induce stress to G. affinis. In summary, we found that exposure to BaP for 20 days caused changes in the expression of a variety of genes in the liver of G. affinis. Interestingly, compared with the unexposed group, opposite effects of immune and metabolic responses were observed in the treatment group.
Immunological responses in the liver of G. affinis exposed to BaP
Previous studies have found that BaP is known to cause functional disorder of fish immune responses (Hur et al. 2013). In the present study, we observed an abundance of upregulated genes which are enriched in immune-related KEGG pathways including toll-like signaling, nuclear factor-kappa B (NF-κB), complement and coagulation cascades, osteoclast differentiation, nucleotide oligomerization domain (NOD)-like receptor signaling, retinoic acid-inducible gene I (RIG-I)-like receptor signaling, tumor necrosis factor (TNF) signaling, and T cell receptor (TCR) signaling. Toll-like receptors (TLRs), RIG-I-like receptors (RLRs), and NOD-like receptors (NLRs) are three out of four families of pattern recognition receptors (PRRs) to recognize danger-associated molecular pattern (DAMP) and pathogen-associated molecular pattern (PAMP) for the initiation of innate immune responses (Shaw et al. 2010; Meng et al. 2012; Zhou et al. 2018). On the other hand, TCRs are crucial components in the adaptive immune system. And TCRs are responsible for the recognition and presentation of foreign antigens (Yu et al. 2019). Moreover, TNF and NF-κB are associated with cell inflammation and cell survival (Hacker et al. 2011). In our experiment, NFKBIA, C-X-C motif chemokine 2-like (CXCL2), C-X-C motif chemokine 8-like (CXCL8), mitogen-activated protein kinase kinase kinase 8 (MAP3K8), nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (NFKB2), and RelB are involved in these six pathways. These six genes were significantly upregulated following BaP exposure in the liver when G. affinis was exposed to BaP. However, whether BaP exposure promotes or inhibits these immune-related pathways is controversial. For example, similar to our results, BaP treatment has increased the protein expression of NF-κB in colons of mice (Ajayi et al. 2016). In contrast, some studies showed that BaP exposure inhibits the immune-related pathway in Oryzias melastigma (Cui et al. 2019a), Apostichopus japonicas (Li et al. 2016), and Tegillarca granosa (Su et al. 2017).
The complement components, the first line of defense in the innate immune system, play an important role in anti-bacterial defenses and have a significant effect on pathogenic microorganism invasions (Dang et al. 2016). In this study, the genes that encode the immune factors such as complement component 4 (C4), C6, complement component C7-like (C7), complement component 8, beta polypeptide (C8β), complement factor I (CFI), and complement C1q subcomponent subunit C-like (C1qa) of the classical complement pathways were all upregulated. Similar to our findings, the complement and coagulation cascade pathway was significantly affected by the BaP exposure in Hippocampus erectus (Jiang et al. 2019), Paralichthys olivaceus (Jung et al. 2018), Chlamys farreri (Cai et al. 2014), Cyprinus carpio (Qiu et al. 2016), and Perna viridis (Jiang et al. 2016). However, some of the results showed that the genes involved in the complement system were suppressed, such as the Chlamys farreri, Paralichthys olivaceus, and Perna viridis, while those in Hippocampus erectus were upregulated when the organism was exposed to BaP. Nevertheless, the doses, exposure durations, and exposed organisms were all different across these studies. We speculate that from these reports and our experimental results, the effects on gene expression of the immune system can be affected by any of these factors. For example, in the exposure durations of our experiment, there was no significant difference in fish’s activity and status between the treatment group and the control group at the early stage of the experiment (< 10 days); however, the activity of fish in the treatment group decreased significantly and began to die at the end of the experiment (> 10 days). It has been reported that host resistance against bacterial infection decreased at 200 g/g BW for up to 7 days in Japanese medaka (Carlson et al. 2002). We speculate that BaP inhibits the G. affinis’s immune system at the early exposure time (< 10 days), thus making it vulnerable to infection by external pathogens, which in turn activates immune-related genes at the prolonged exposure time (> 10 days). In a word, our study shows evidence that BaP causes disorder in the immunity of G. affinis.
Metabolism responses in the liver of G. affinis exposed to BaP
The liver, which is known to be one of the major organs for digestion of proteins, carbohydrates, and lipids, and an important detoxifying organ, is related to the metabolism disorders when the animals are exposed to BaP. In the current study, we observed an abundance of downregulated genes in the liver of G. affinis in the enriched KEGG pathways of pancreatic secretion (10 genes), protein digestion and absorption (7 genes), fat digestion and absorption (5 genes), glucagon signaling (5 genes), insulin signaling (5 genes), AMPK signaling (5 genes), PPAR signaling (4 genes), and metabolism of fatty acid (5genes), glycerophospholipid (5 genes), glycerolipid (4 genes), arachidonic acid (3 genes), alpha-linolenic acid (3 genes), and linoleic acid (2 genes). In addition to these downregulated genes, several upregulated genes were enriched in the metabolism related to KEGG pathways. These genes are related to glycolysis/gluconeogenesis (4 genes) and methane metabolism (3 genes). Among these genes, chymotrypsin-like elastase 2A (CELA2A), chymotrypsin-like protease (CTRL), carboxyl ester lipase (CEL), phospholipase A2 group IB (PLA2G1B), carboxypeptidase B1 (CPB1), trypsin1 (TRY1), TRY2, and TRY3 are downregulated and mainly involved in the digestion and absorption of proteins and lipids. Because trypsin is the main digestive enzyme in the hepatopancreas (Shao et al. 2018), low trypsin activity can inhibit the digestion and absorption of protein. In the current experiment, the trypsin was significantly downregulated after G. affinis was exposed to 100 μg/L of BaP for 20 days. Similar to this result, the trypsin-1 showed 6.7-fold significant downregulation when rainbow trout was exposed to 10 parts per billion of tetrachlorodibenzo-p-dioxin (TCDD) at 28 days compared with the control group where rainbow trout was not exposed to TCDD (Liu et al. 2013). Additionally, the result in this study also showed that BaP can disrupt the carbohydrate metabolism. Glucokinase (GCK) is a flux-controlling enzyme for the glycogen synthesis and glycolysis in the liver. It is well-known that hyperglycemia induces insulin secretion, which results in the transcription of sterol regulatory element-binding transcription factor 1c (SREBP1c) and GCK (Hansmannel et al. 2006). In this experiment, SREBF1 and GCK were significantly downregulated. This may indicate that the glycogen storage could have been mobilized after a long exposure (> 10 days).
Moreover, the lipid metabolism was also affected by the BaP exposure. Seven pathways that have been reported to having changes were related to the lipid metabolism. And all the differently expressed genes involved in these seven pathways were downregulated, indicating impairment of lipid metabolism. This observation was consistent with that in the literature. For example, some studies showed that BaP inhibits numerous ER-related genes that affect fat digestion and absorption in Mugilogobius chulae (Cai et al. 2019). And BaP disturbed the lipid metabolism in C57BL/6 mice (Li et al. 2019). Also in Xenopus tropicalis, the lipid metabolism was impaired when BaP was exposed to Xenopus tropicalis, which leads to hepatotoxicity (Regnault et al. 2014). In addition, in our experiment, the BaP exposure induced a significant decrease in hepatocyte and myocyte lipid droplet contents. This may indicate that the energy could be consumed. In the literature, it has been reported that all the available energy storage could be mobilized to activate their detoxification mechanisms in an organism affected by BaP exposure in tilapia (Colli-Dula et al. 2018b). However, the other results in wild-type female mice showed that BaP can increase visceral adiposity and cause hepatic steatosis (Ortiz et al. 2013). Our results indicate that BaP exposed to the male G. affinis could induce metabolic disorders and the energy storage in the liver may be consumed with a prolonged exposure time (> 10 days).
Conclusions
In this study, we have demonstrated that BaP exposure at a relatively high dose (100 μg/L) over a relatively long period of time (> 10 days) exhibited opposite effects on the adult male G. affinis. Using RNA-seq analysis in the liver, we have described a list of molecular changes caused when G. affinis was exposed to BaP. Interestingly, the most salient gene expression alterations were found in immunological and metabolic signaling pathways, which may have an important role in damaging immunity and promoting metabolic disorders in the male G. affinis. Overall, these data add to the increasing evidence on the opposite effects of gene expression when fish was exposed to BaP. And these data provide new insights on the mechanisms that mediate such effects.
References
Ajayi BO, Adedara IA, Farombi EO (2016) Benzo(a)pyrene induces oxidative stress, pro-inflammatory cytokines, expression of nuclear factor-kappa B and deregulation of wnt/beta-catenin signaling in colons of BALB/c mice. Food Chem Toxicol 95:42–51
Bao S, Nie X, Liu Y, Wang C, Liu S (2018) Response of PXR signaling pathway to simvastatin exposure in mosquitofish (Gambusia affinis) and its histological changes. Ecotoxicol Environ Saf 154:228–236
Cai Y, Pan L, Hu F, Jin Q, Liu T (2014) Deep sequencing-based transcriptome profiling analysis of Chlamys farreri exposed to benzo[a]pyrene. Gene 551:261–270
Cai L, Li J, Yu L, Wei Y, Miao Z, Chen M, Huang R (2019) Characterization of transcriptional responses mediated by benzo[a]pyrene stress in a new marine fish model of goby, Mugilogobius chulae. Genes Genomics 41:113–123
Caliani I, Porcelloni S, Mori G, Frenzilli G, Ferraro M, Marsili L, Casini S, Fossi MC (2009) Genotoxic effects of produced waters in mosquito fish (Gambusia affinis). Ecotoxicology 18:75–80
Carlson EA, Li Y, Zelikoff JT (2002) Exposure of Japanese medaka (Oryzias latipes) to benzo[a]pyrene suppresses immune function and host resistance against bacterial challenge. Aquat Toxicol 56:289–301
Chen H, Diao X, Zhou H (2018) Tissue-specific metabolic responses of the pearl oyster Pinctada martensii exposed to benzo[a]pyrene. Mar Pollut Bull 131:17–21
Colli-Dula RC, Fang X, Moraga-Amador D, Albornoz-Abud N, Zamora-Bustillos R, Conesa A, Zapata-Perez O, Moreno D, Hernandez-Nunez E (2018a) Gene expression profile and molecular pathway datasets resulting from benzo(a)pyrene exposure in the liver and testis of adult tilapia. Data Brief 20:1500–1509
Colli-Dula RC, Fang X, Moraga-Amador D, Albornoz-Abud N, Zamora-Bustillos R, Conesa A, Zapata-Perez O, Moreno D, Hernandez-Nunez E (2018b) Transcriptome analysis reveals novel insights into the response of low-dose benzo(a)pyrene exposure in male tilapia. Aquat Toxicol 201:162–173
Corrales J, Thornton C, White M, Willett KL (2014) Multigenerational effects of benzo[a]pyrene exposure on survival and developmental deformities in zebrafish larvae. Aquat Toxicol 148:16–26
Cui Q, Chen F-Y, Chen H-Y, Peng H, Wang K-J (2019a) Benzo[a]pyrene (BaP) exposure generates persistent reactive oxygen species (ROS) to inhibit the NF-κB pathway in medaka (Oryzias melastigma). Environ Pollut 251:502–509
Cui Q, Chen FY, Zhang M, Peng H, Wang KJ (2019b) Transcriptomic analysis revealing hepcidin expression in Oryzias melastigma regulated through the JAK-STAT signaling pathway upon exposure to BaP. Aquat Toxicol 206:134–141
Dang Y, Xu X, Shen Y, Hu M, Zhang M, Li L, Lv L, Li J (2016) Transcriptome analysis of the innate immunity-related complement system in spleen tissue of Ctenopharyngodon idella infected with Aeromonas hydrophila. PLoS One 11:e0157413
Ekere NR, Yakubu NM, Oparanozie T, Ihedioha JN (2019) Levels and risk assessment of polycyclic aromatic hydrocarbons in water and fish of Rivers Niger and Benue confluence Lokoja, Nigeria. J Environ Health Sci Eng 17:383–392
Fang X, Corrales J, Thornton C, Clerk T, Scheffler BE, Willett KL (2015) Transcriptomic changes in zebrafish embryos and larvae following benzo[a]pyrene exposure. Toxicol Sci 146:395–411
Gao J, Ouyang X, Chen B, Jourdan J, Plath M (2017) Molecular and morphometric evidence for the widespread introduction of Western mosquitofish Gambusia affinis (Baird and Girard, 1853) into freshwaters of mainland China. BioInvasions Records 6:281–289
Hacker H, Tseng PH, Karin M (2011) Expanding TRAF function: TRAF3 as a tri-faced immune regulator. Nat Rev Immunol 11:457–468
Hansmannel F, Mordier S, Iynedjian PB (2006) Insulin induction of glucokinase and fatty acid synthase in hepatocytes: analysis of the roles of sterol-regulatory-element-binding protein-1c and liver X receptor. Biochem J 399:275–283
Hoffberg, S.L., Troendle, N.J., Glenn, T.C., Mahmud, O., Louha, S., Chalopin, D., Bennetzen, J.L. and Mauricio, R., 2018. A high-quality reference genome for the invasive Mosquitofish Gambusia affinis using a Chicago library. G3 (Bethesda) 8, 1855-1861
Hou L, Xu H, Ying G, Yang Y, Shu H, Zhao J, Cheng X (2017) Physiological responses and gene expression changes in the western mosquitofish (Gambusia affinis) exposed to progesterone at environmentally relevant concentrations. Aquat Toxicol 192:69–77
Hou LP, Yang Y, Shu H, Ying GG, Zhao JL, Fang GZ, Xin L, Shi WJ, Yao L, Cheng XM (2018) Masculinization and reproductive effects in western mosquitofish (Gambusia affinis) after long-term exposure to androstenedione. Ecotoxicol Environ Saf 147:509–515
Hou L, Chen S, Liu J, Guo J, Chen Z, Zhu Q, Zhang W, Xu G, Liang Y, Wu R, Fang X, Zhang C, Xing K (2019) Transcriptomic and physiological changes in western mosquitofish (Gambusia affinis) after exposure to norgestrel. Ecotoxicol Environ Saf 171:579–586
Hur D, Jeon JK, Hong S (2013) Analysis of immune gene expression modulated by benzo[a]pyrene in head kidney of olive flounder (Paralichthys olivaceus). Comp Biochem Physiol B Biochem Mol Biol 165:49–57
Jayasundara N, Van Tiem Garner L, Meyer JN, Erwin KN, Di Giulio RT (2015) AHR2-mediated transcriptomic responses underlying the synergistic cardiac developmental toxicity of PAHs. Toxicol Sci 143:469–481
Jiang X, Qiu L, Zhao H, Song Q, Zhou H, Han Q, Diao X (2016) Transcriptomic responses of Perna viridis embryo to Benzo(a)pyrene exposure elucidated by RNA sequencing. Chemosphere 163:125–132
Jiang H, Wang K, Fang Y, Chen J, Li Y, Xia G, Zhang Y, Liu Y, Ren C, Lin Q (2019) Sex-biased regulation of respiratory burst, phagocytic activity and plasma immune factors in lined seahorse (Hippocampus erectus) after subchronic benzo[a]pyrene exposure. Fish Shellfish Immunol 86:1162–1168
Jung JH, Moon YS, Kim BM, Lee YM, Kim M, Rhee JS (2018) Comparative analysis of distinctive transcriptome profiles with biochemical evidence in bisphenol S- and benzo[a]pyrene-exposed liver tissues of the olive flounder Paralichthys olivaceus. PLoS One 13:e0196425
Kamata R, Itoh K, Nakajima D, Kageyama S, Sawabe A, Terasaki M, Shiraishi F (2011) The feasibility of using mosquitofish (Gambusia affinis) for detecting endocrine-disrupting chemicals in the freshwater environment. Environ Toxicol Chem 30:2778–2785
Kim BM, Kang S, Kim RO, Jung JH, Lee KW, Rhee JS, Lee YM (2018) De novo transcriptome assembly of brackish water flea Diaphanosoma celebensis based on short-term cadmium and benzo[a]pyrene exposure experiments. Hereditas 155:36
Krumholz LA (1948) Reproduction in the western mosquitofish, Gambusia affinis affinis (Baird & Girard), and its use in mosquito control. Ecol Monogr 18:1–43
Lamatsch DK, Adolfsson S, Senior AM, Christiansen G, Pichler M, Ozaki Y, Smeds L, Schartl M, Nakagawa S (2015) A transcriptome derived female-specific marker from the invasive Western mosquitofish (Gambusia affinis). PLoS One 10:e0118214
Li C, Zhou S, Ren Y, Jiang S, Xia B, Dong X (2016) Toxic effects in juvenile sea cucumber Apostichopus japonicas (Selenka) exposure to benzo[ a ]pyrene. Fish & Shellfish Immunology 59:375–381
Li F, Xiang B, Jin Y, Li C, Li J, Ren S, Huang H, Luo Q (2019) Dysregulation of lipid metabolism induced by airway exposure to polycyclic aromatic hydrocarbons in C57BL/6 mice. Environ Pollut 245:986–993
Liu Q, Rise ML, Spitsbergen JM, Hori TS, Mieritz M, Geis S, McGraw JE, Goetz G, Larson J, Hutz RJ, Carvan MJ 3rd (2013) Gene expression and pathologic alterations in juvenile rainbow trout due to chronic dietary TCDD exposure. Aquat Toxicol 140-141:356–368
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods 25:402–408
Meng Z, Zhang XY, Guo J, Xiang LX, Shao JZ (2012) Scavenger receptor in fish is a lipopolysaccharide recognition molecule involved in negative regulation of NF-kappaB activation by competing with TNF receptor-associated factor 2 recruitment into the TNF-alpha signaling pathway. J Immunol 189:4024–4039
Mojiri, A., Zhou, J. L., Ohashi, A., Ozaki, N. and Kindaichi, T., 2019. Comprehensive review of polycyclic aromatic hydrocarbons in water sources, their effects and treatments[J]. Science of the Total Environment, 696(UNSP 133971)
Ortiz L, Nakamura B, Li X, Blumberg B, Luderer U (2013) In utero exposure to benzo[a]pyrene increases adiposity and causes hepatic steatosis in female mice, and glutathione deficiency is protective. Toxicol Lett 223:260–267
Pyke GH (2008) Plague minnow or mosquito fish? A review of the biology and impacts of introduced Gambusia species. Annu Rev Ecol Evol Syst 39:171–191
Qiu W, Chen J, Li Y, Chen Z, Jiang L, Yang M, Wu M (2016) Oxidative stress and immune disturbance after long-term exposure to bisphenol A in juvenile common carp (Cyprinus carpio). Ecotoxicol Environ Saf 130:93–102
Regnault C, Worms IA, Oger-Desfeux C, MelodeLima C, Veyrenc S, Bayle ML, Combourieu B, Bonin A, Renaud J, Raveton M, Reynaud S (2014) Impaired liver function in Xenopus tropicalis exposed to benzo[a]pyrene: transcriptomic and metabolic evidence. BMC Genomics 15:666
Shao J, Zhao W, Liu X, Wang L (2018) Growth performance, digestive enzymes, and TOR signaling pathway of Litopenaeus vannamei are not significantly affected by dietary protein hydrolysates in practical conditions. Front Physiol 9:998
Shaw PJ, Lamkanfi M, Kanneganti TD (2010) NOD-like receptor (NLR) signaling beyond the inflammasome. Eur J Immunol 40:624–627
Song Y, Nahrgang J, Tollefsen KE (2019) Transcriptomic analysis reveals dose-dependent modes of action of benzo(a)pyrene in polar cod (Boreogadus saida). Sci Total Environ 653:176–189
Su W, Zha S, Wang Y, Shi W, Xiao G, Chai X, Wu H, Liu G (2017) Benzo[a]pyrene exposure under future ocean acidification scenarios weakens the immune responses of blood clam, Tegillarca granosa. Fish Shellfish Immunol 63:465–470
Sun, D., Chen, Q., Zhu, B., Lan, Y. and Duan, S.S., 2020. Long-term exposure to benzo[a]Pyrene affects sexual differentiation and embryos toxicity in three generations of marine medaka (Oryzias melastigma) [J]. International journal of environmental research and public health, 17, doi:10.3390/ijerph17030970
Sushkova S, Deryabkina I, Antonenko E, Kizilkaya R, Rajput V, Vasilyeva G (2018) Benzo[a]pyrene degradation and bioaccumulation in soil-plant system under artificial contamination. Sci Total Environ 633:1386–1391
Wang H, Pan L, Xu R, Miao J, Si L, Pan L (2018) Comparative transcriptome analysis between the short-term stress and long-term adaptation of the Ruditapes philippinarum in response to benzo[a]pyrene. Aquat Toxicol 204:59–69
Wills LP, Zhu S, Willett KL, Di Giulio RT (2009) Effect of CYP1A inhibition on the biotransformation of benzo[a]pyrene in two populations of Fundulus heteroclitus with different exposure histories. Aquat Toxicol 92:195–201
Xie YP, Fang ZQ, Hou LP, Ying GG (2010) Altered development and reproduction in western mosquitofish (Gambusia affinis) found in the Hanxi River, southern China. Environ Toxicol Chem 29:2607–2615
Yadetie F, Zhang X, Hanna EM, Aranguren-Abadia L, Eide M, Blaser N, Brun M, Jonassen I, Goksoyr A, Karlsen OA (2018) RNA-Seq analysis of transcriptome responses in Atlantic cod (Gadus morhua) precision-cut liver slices exposed to benzo[a]pyrene and 17alpha-ethynylestradiol. Aquat Toxicol 201:174–186
Yanagisawa R, Koike E, Win-Shwe TT, Ichinose T, Takano H (2018) Effects of lactational exposure to low-dose BaP on allergic and non-allergic immune responses in mice offspring. J Immunotoxicol 15:31–40
Yu N, Ding Q, Li E, Qin JG, Chen L, Wang X (2018) Growth, energy metabolism and transcriptomic responses in Chinese mitten crab (Eriocheir sinensis) to benzo[alpha]pyrene (BaP) toxicity. Aquat Toxicol 203:150–158
Yu W, Luo Y, Yu Y, Dong S, Yin Y, Huang Z, Xu Z (2019) T cell receptor (TCR) alpha and beta genes of loach (Misgurnus anguillicaudatus): molecular cloning and expression analysis in response to bacterial, parasitic and fungal challenges. Dev Comp Immunol 90:90–99
Zhan S, Zhang X, Cao S, Huang J (2015) Benzo(a)pyrene disrupts mouse preimplantation embryo development. Fertil Steril 103:815–825
Zhang M, Miao Y, Chen Q, Cai M, Dong W, Dai X, Lu Y, Zhou C, Cui Z, Xiong B (2018) BaP exposure causes oocyte meiotic arrest and fertilization failure to weaken female fertility. FASEB J 32:342–352
Zhang QL, Guo J, Deng XY, Wang F, Chen JY, Lin LB (2019) Comparative transcriptomic analysis provides insights into the response to the benzo(a)pyrene stress in aquatic firefly (Luciola leii). Sci Total Environ 661:226–234
Zhou Z, Lin Z, Pang X, Shan P, Wang J (2018) MicroRNA regulation of toll-like receptor signaling pathways in teleost fish. Fish Shellfish Immunol 75:32–40
Funding
This work received the support of the National Natural Science Foundation of China (21806041).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Responsible Editor: Philippe Garrigues
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 30.2 kb)
Rights and permissions
About this article
Cite this article
Feng, Y., Zhou, A., Zhang, Y. et al. Transcriptomic changes in western mosquitofish (Gambusia affinis) liver following benzo[a]pyrene exposure. Environ Sci Pollut Res 27, 21924–21938 (2020). https://doi.org/10.1007/s11356-020-08571-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-020-08571-0