Abstract
Pleurotus ostreatus degrades polychlorinated biphenyls (PCBs) with an increase of laccase activity. Laccases are well known for their detoxifying activity. We show, using reverse transcription polymerase chain reaction and a biochemical assay, that reduction in PCBs (di, tri, tetra, and penta) levels are correlated with an increase in laccase activity. P. ostreatus cultures were obtained from 0 to 30 days in the presence or absence of 7,100 mg/L PCBs (from transformer oil) and a surfactant. After each selected time cultures were withdrawn and remaining PCBs were determined, a maximal removal percentage of PCBs was obtained at 20 (63.5 ± 2.0) and 30 days (63.8 ± 4.6) post-induction. Also, the activity of the enzyme was analyzed and it was found to increase at 10 (6.9-fold) and 20 (6.77-fold) days post-induction in the presence of PCBs, as determined by its activity. Taken together, these data suggest that PCBs induce laccase expression and that laccase catalyzes PCBs removal.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Polychlorinated biphenyls (PCBs) are persistent organic pollutants that were used as dielectric and coolant fluids in transformers and electric motors in the twentieth century. Even if their production and use was banned more than three decades ago, yet 10,000 tons of PCBs persist in the environment (Singer et al. 2000).
PCBs bioaccumulate in individuals and biomagnify in the food chain (Crinnion 2011). In Mexico, there are extensive polluted areas and large amounts of contaminated used transformer oil in storage that require remediation (Rojas-Avelizapa et al. 1999). Bioremedation efficiency of PCBs depends on the nature and extent of contamination. It has been observed that the addition of biphenyl as an inducer and biosurfactants in soil and sediments improve the level of di- and trichlorinated congeners removal (Vasil'eva and Strizhakova 2007). Although the addition of carbon compounds enhances the degradation of PCBs removal patterns, these are significantly different between soils and sediments: soil microorganisms remove more PCBs than the microorganisms present in the sediments, especially PCBs with >4 chlorine atoms (Luo et al. 2008).
Various physicochemical methods to degrade PCBs (Ericson 1999) are available; however, it is necessary to explore economic alternatives technically feasible and environmentally friendly. Therefore, some microorganisms that degrade these pollutants, especially bacteria and fungi have been studied (Moeder et al. 2005; Rodrigues et al. 2006). The white rot fungi (WRF) have shown ability to degrade PCBs: studies with Coriolopsis polyzona, Phanerochaete chrysosporium, Pleurotus ostreatus, and Trametes versicolor have proven that WRF can remove PCBs in vivo, with little specificity of congeners and no correlation between removal rate and degree of chlorination; ligninolytic enzymes such as as laccases, versatile peroxidase, and manganese peroxidase may be involved (Pointing 2001). Although it is known that extracts from WRF or their laccases catalyze the degradation of hydroxylated PCBs, little is known about the in vivo mechanisms of PCBs degradation (Jiang et al. 2008; Fujihiro et al. 2009). Diverse studies have shown that laccases from WRF degrade isolated PCBs congeners (Dietrich et al. 1995) or PCBs in commercial mixes such as Delor 103, Delor 106, and Arochlors 1242, 1254, and 1260 (Moeder et al. 2005; Novotny et al. 1997; Yadav et al. 1995) with some limitation since the higher the chlorinated level the more difficult is the degradation. Most of these studies were carried out with low PCBs concentration (1–2,000 ppm).
In this study we determined the correlation of P. ostreatus laccase activity as confirmed by its transcript expression with the removal of high concentrations (7,100 ppm) of PCBs from Arochlor 1242 in liquid culture. Also, the correlation of PCBs removal ability from P. ostreatus laccases and chlorination level was evaluated.
Materials and methods
Fungal cultures
P. ostreatus (ATCC 38540) was used for this study: the mycelium was grown at 27°C in Erlemeyer flasks containing minimum medium for basidiomycetes (Guillén-Navarro et al. 1998) which composition was (in g/L) glucose 5, yeast extract 5, pH 5.5, and agitated at 200 rpm for 7 days. For the production of laccase, 1 mL of mycelia homogenized (corresponding to 24 mg of dry mass) was harvested and inoculated into a 40-mL of liquid SM medium containing (in g/L) glucose 5, yeast extract 5, wheat straw extract 2.5, CuSO4 0.15, MnSO4 0.05, and pH was adjusted to 6. The cultures were incubated under the same conditions mentioned above, and after 7 days of culture, PCBs (10,000 mg/L) as transformer oil, containing 71% of Arochlor 1242, with a composition of—5.8% penta-, 36.3% tetra-, 46.4% tri-, and 11.5% dichlorinated biphenyls and Tween 80 (3,500 mg/L) were added to each flask and then incubated as long as indicated. In order to monitor any basal laccase activity, two controls were established: (a) without PCBs and (b) abiotic (the culture was sterilized after 7 days of incubation then the PCBs were added); these were also incubated under the same conditions. At least two replicates for each treatment were analyzed for biomass determination, laccase enzymatic activity, reverse transcription-polymerase chain reaction (RT-PCR) assays, and the removal of PCBs at 10, 20, and 30 days post-induction.
All reagents were purchased from Sigma-Aldrich, USA.
Biomass
Assessment of biomass was performed by determining the mycelium dry mass. For this, the mycelium was filtered and washed with hexane–acetone (10:6, v/v) three times and then dried at 50°C to constant mass.
Laccase activity measurement
Laccase activity was determined by using 0.5 mmol/L ABTS [2,2-azinobis (3-ethyl benzthiazoline-6-sulphonate)], in sodium acetate buffer (pH 4.5) and an appropriate volume of the culture supernatant in a range between 10–50 μL. The oxidation of ABTS was monitored spectrophotometrically at 436 nm (Tinoco et al. 2001). One unit of laccase activity is defined as 1 μmol of ABTS oxidized per minute.
Extraction of PCBs
The biomass was filtered and washed with hexane, the washes of mycelium were added to filtrate for quantitative PCBs determination. The PCBs were extracted from culture broth by adding 120 mL of hexane–acetone (10:6, v/v); the organic phase was extracted with 5 mL of concentrated H2SO4 and washed twice with water. The organic phase was transferred to vials where anhydrous sodium sulfate was added to dry the sample. The sample was filtered through a 0.22-μm Millipore membrane and purified on a Florisil cartridge (Ericson 1999). The resulting extract was concentrated to dryness in a rotary evaporator, and then the concentrate was dissolved in hexane. The extraction efficiency was 95% (Novotny et al. 1997). An Agilent model 6890 N gas chromatograph equipped with a 5973 mass selective detector and Agilent HP-5MS (GC/MS) capillary column was used for PCBs quantification. Conditions used were: initial oven temperature 70°C for 2 min, followed by an increase to 150°C at 25 K/min, after to 200°C at 3 K/min, and to 280°C at 8 K/min and an isocratic period of 10 min. Helium was used as the carrier gas at 18 psi pressure and 1.9 mL/min flow. The injector was kept at constant pressure and 250°C temperature, while the temperature of the detector was 270°C. The injection volume was 1 μL in the splitless mode. 1,3,5-trichorobenzene and Arochlor 1242 were used as internal and calibration standards, respectively.
Extraction of RNA and DNA
For RNA and DNA extraction, mycelium of P. ostreatus ATCC 38540 was grown in medium above mentioned, washed and filtered with deionized water, glucose solution and RNaseZap® (Ambion, Inc). For total RNA extraction, samples were transferred to a 1.5-mL Eppendorf tube and stored at −80°C until use. Total RNA was extracted according to the TRIZOL (Invitrogen) protocol, briefly the mycelium was kept frozen by adding liquid nitrogen to a baked mortar, and it was rapidly ground with the pestle. Once completely ground, about 300 μL of an Eppendorf tube was filled and 1 mL of TRIZOL (Invitrogen) was added. Tubes were vortexed and phases separated by adding 200 μL of chloroform and centrifuged to obtain aqueous phase. Total RNA was precipitated with 0.8 mL of isopropanol, then reprecipitated with 100% ethanol and washed with 70% ethanol. Genomic DNA was isolated by a method previously described (Raeder and Broda 1985) with few modifications. Ground mycelium was placed in an Eppendorf tube, with 500 μL of HSD buffer (10 mmol/L HEPES 4-(2-hydroxyethyl)-1-piperazine ethanesulfonic acid, pH 6.9, 0.5 mmol/L sucrose, 20 mmol/L EDTA) and mixed well. Then 50 μL of 10% SDS (sodium dodecyl sulphate) were added and incubated at 65°C during 15 min, after this 500 μL of TED buffer (50 mmol/L Tris, 20 mmol/L EDTA, pH 8) were added. The mixture was extracted with 600 μL of phenol–chloroform–isoamyl alcohol solution (25:24:1, by vol.). The aqueous phase was re-extracted with a chloroform–isoamyl alcohol (24:1, v/v) mixture, centrifuged, and the aqueous phase was precipitated with three sodium acetate pH 5.2 and two volumes of cold ethanol (−20°C). After centrifugation, the pellet was washed with 500-μL 70% ethanol and resuspended in TE (10 mmol/L Tris-HCl, 1 mmol/L EDTA). RNA was removed by incubating the DNA with 5-μL 20 mg/mL RNase A, for 30 min at 37°C. DNA was re-precipitated with ethanol, washed with 70% ethanol, and resuspended in 50 μL of sterile water.
Reverse transcription
In order to compare the relative abundance of the laccase transcript, RT-PCR analysis was performed to make cDNA using Superscript II (Invitrogen) according to manufacturer’s recommendations. Briefly, 8 μg of total RNA (RNA samples from different culture times as indicated), 70 ng of Oligo(dT), and 0.5 mmol/L dNTPs were added up to a final volume of 12 μL, the mixture was incubated at 65°C for 5 min. Then 5× strand buffer and 0.1 mol/L DTT (1,4-dithiothreitol) were added to adjust the final volume to 20 μL with sterile deionized water. The mixture was incubated at 42°C for 2 min, and 1 μL of SuperScript II RNase H-reverse transcriptase was added, to continue incubation for 1 h at 42°C, at the end of the reaction the retrotranscriptase was inactivated at 70°C for 15 min.
PCR
The oligonucleotide primers for laccase were designed and based on the conserved regions of P. ostreatus lccK gene for laccase and POX2 gene for diphenol oxidase (Fig. 1), lately referred as POXC (Palmieri et al. 2000, GenBank accession numbers: AB089612 and Z49075, respectively). P. ostreatus β-tubulin, was used as an internal control, and primers were designed as well (GenBank accession number: AF106146; Table 1). In both cases, the design was made to distinguish between RT-PCR and potential genomic contaminant amplicon products, thus primers were designed to span regions in which predicted introns had been identified. The 50 μL PCR reaction volume contained 1 μL either (50–100 ng) genomic DNA or synthesized cDNA as template, 0.2 mmol/L dNTP mixture, ×1 PCR buffer (Invitrogen), 1.5 mmol/L MgCl2, 1 μmol/L F1 and R1 primers; F and R represent the forward and reverse primers, respectively and 0.25 μL (5 U/μL) Taq-polymerase (Invitrogen). Primers design was carried out with the program Oligo version 4.1 (Primer analysis software). For β-tubulin, initial denaturation at 94°C for 3 min was followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 56°C for 30 s, and elongation at 72°C for 45 s and a final extension for 5 min at 72°C. For laccase, the cycling conditions were; denaturation at 94°C for 3 min, followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 57°C for 30 s, and elongation at 72°C for 2 min. The final extension was for 5 min at 72°C. The reactions were carried out in a Techne TC312 Thermal Cycler (USA). Amplified fragments were analyzed on 1% agarose gel stained with ethidium bromide.
Laccase genes sequences alignment. Different laccase sequences from P. ostreatus were aligned to identify conserved regions where primers were designed (highlighted in gray) for PCR analysis. LCCK, POX2, and POX1 correspond to laccase mRNA sequences with AB089612, Z49075, and Z34847 accession numbers, respectively. Asterisks indicate identity while a dash under an asterisk indicates the beginning of the next exon
Results and discussion
Presence of PCBs increases P. ostreatus biomass
The study was carried out during P. ostreatus growth to compare the production of biomass in the presence and absence of PCBs (transformer oil) in liquid medium at culture times indicated. Biomass production is approximately 2-fold higher in the presence of PCBs. The maximum dry biomass production was 240 mg with PCBs and 110.34 mg without them at 30 days of culture (Fig. 2). We have previously observed that SM supported good growth of the fungus (Gayosso-Canales et al. 2011). Consequently, this medium was selected to evaluate the concomitant production of biomass and enzymatic activity in the presence and absence of PCBs. The growth was better in the presence of PCBs than in their absence suggesting that PCBs along with the surfactant added improve the growth of fungus probably because the surfactant increases bioavailability of PBCs, especially of those congeners with large octanol–water partition coefficients (Moeder et al 2005) thus improving laccase enzyme activity (Nikiforova et al. 2009). Also, it has been reported that surfactants may improve solubility of pollutants with little or none solubility in water, even at concentrations higher than their critical micelle concentration (Robinson et al. 1996).
PCBs modify the laccase activity
While the laccase activity (Fig. 3) was initially repressed with the PCBs addition as compared with the control without PCBs, an increase of enzyme activity was observed after five d of culture presumably due to the presence of transformer oil. Up to 30 days of treatment the enzyme activity was the same. This suggests that, at least initially, the contaminant may have caused a repressive effect, probably due to stress; however after a short-time PCBs seem to increase the laccase activity. The induction levels of laccase activity were 6.9-fold at 10 days and 6.77-fold at 20 days post-induction. It has been shown that laccase activity increases in the presence of CuSO4 and MnSO4 at 1 and 1–5 mmol/L, respectively in P. ostreatus (Stajić et al. 2006), whereas other studies indicate that 150 μmol/L CuSO4 is enough to obtain a laccase induction (Palmieri et al. 2000). In any case, the medium used in the present work contained 0.94 mmol/L CuSO4, and 0.33 mmol/L MnSO4, and it is known that all laccase promoters contain metal responsive elements, which are able to change protein interaction pattern upon metal addition, being copper one of them (Faraco et al. 2003). So, at least initially, a laccase activity was expected.
Thus, CuSO4, MnSO4, and wheat straw extract were added to increase the laccase activity; since previous studies have reported that the addition of lignocellulosic residues and the presence of metals improve the activity of this enzyme (Ullah et al. 2000; Stajić et al. 2004).
Laccase activity is enhanced upon PCBs addition
The presence of PCBs (as transformer oil) improved laccase activity as determined by a chemical assay shown in Fig. 3. To confirm this, relative abundance of laccase transcript was examined using RT-PCR analysis. RNA was isolated from 0-, 10-, 20-, to 30-day cultures in SM medium. β-tubulin amplified products were used as reference. The expression determined as relative abundance of laccase transcript was normalized against the concentration of tubulin obtained from RT-PCR was analyzed, the amplicons obtained corresponded to expected length (Table 1). As a control, amplified genomic DNA was used to amplify PCR products to distinguish them from RNA amplified products (Fig. 4a). These data were quantified and normalized by tubulin expression and plotted as shown in Fig. 4b. Laccase transcript level in the presence of PCBs was higher than that without PCBs. This increase is mainly observed at 20 and 30 days of culture, consistent with their corresponding laccase activity levels. However, there is an inconsistency at 10 days where there is as much laccase activity as that found at 20 days: the transcript levels are lower, indicating either there is an increase rate of laccase synthesis or that the biochemical reaction is not specific.
RT-PCR analyses of P. ostreatus laccase transcript levels. a RNA from fungi grown in SM in the presence and absence of PCBs and at the period of incubation times indicated were retrotranscribed and PCR analyzed. Amplified fragments were resolved in a 1% agarose gel stained with ethidium bromide, a representative gel is shown. Molecular size markers (L) are indicated at the left (above). b Relative transcript abundance of P. ostreatus laccase. Symbols—closed triangles, in presence of transformer oil (PCBs, 7,100 mg/L); closed squares, in its absence; at three culture times. The relative transcript abundance levels are plotted versus culture times (same as indicated for RT-PCT assay). The intensity of the each band from gel A was determined by densitometry, relative intensity of laccase values were normalized with β-tubulin. Each point is an average of two different determinations (below)
Removal of PCBs
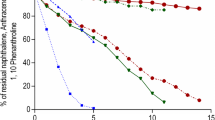
Table 2 shows the removal percentage of PCBs in the medium analyzed; 63.05 at 20 days and 63.8 at 30 days post-treatment, corresponding to 33.3 and 18.9 ppm/mg when normalized by biomass. Figure 5 shows the input, remaining percentage and its corresponding abiotic control per congener identified in the transformer oil after 20 days of culture. From Fig. 5, one can observe that 2,4 and 3,3′ dichloro biphenyls were completely removed; as well as 2,2′,5/2,3′,5/2,4,6 and 2,3,4 trichloro biphenyls. Furthermore, the congeners tetrachlorinated 2,2′,4,5′/2,2′,5,6/2,2′,4,4′/2,3′,5,5′/2,3′,4′,6/2,2′,3,4/3,3′,4,5′/2,3′,4′,5, or 2,3,3′,4/3,3′,5,5′, or 3,3′,4,5′ were eliminated in a 65.88% as well as pentachloro biphenyls 2,2′,3,3′,5, 2,2′,3,4,5′/2,3,3′,4′,6, 2,3,3′,4,4′/2,3′,4,4′,5/2,2′,4,4′,5/2,2′,3,5,5′, or 2,2′,3′,4,5/2,3,3′,4′,6/2,3,3′,4,4′, or 2,3′,4,4′,5/2,3′,4,5′,6 which were removed completely.
Recovery of PCB congeners after 20 days of growth and classification according to their chlorination degree. t 0 time zero, t 20 point taken at day 20, t 20 abiotic abiotic control. Number 1–37 are referred to congeners contained in Table 3
P. ostreatus has shown to be a good biodegradation organism for different contaminants (Palmieri et al. 2005; Rigas et al. 2005). The fungus was able to biodegrade totally the dichlorinated 2 and 3, the trichlorinated 5, 8, 10, and 11, the tetrachlorinated 16, 22, and 24, and the pentachlorinated 29–36 congeners, while it is impossible to determine tetrachlorinated 17 and pentachlorinated congeners enzymatic degradation, as they are removed by the abiotic controls (Table 3; Fig. 5). Our findings indicate that under our conditions highly chlorinated biphenyls were efficiently removed and selectivity in PCB congener degradation was not observed (Table 3) suggesting that P. ostreatus laccase degrades a wide range of congeners. This could be explained, at least in part, because surfactant improved congeners solubility, especially of those that are less soluble, thereby facilitating their biodegradation (Robinson et al. 1996; Paria 2008).
Some previous reports have shown a great resistance of congeners with two chlorine atoms in para positions; the ortho position is preferred for degradation including non planar congeners, and the biodegradation decreases with the increase of chlorination (Beaudette et al. 1998; Kubátová et al. 2001).
The results in present study indicate that the biodegradation of PCBs by P. ostreatus under our culture conditions tested and with the transformer oil type used is independent of number and position of chlorine atoms since both dichlorinated and pentachlorinated congeners, planar and non-planar were degraded. Kamei et al. (2006) also observed that Phanerochaete sp. shows a specific biodegradation of PCBs affected by the mix of congeners.
On the other hand, in several studies the capacity of degradation of polycyclic aromatic hydrocarbons (PAHs) has been associated to the production of laccase (Rodríguez et al. 2004). In our study, the contaminant removal data strongly suggest that P. ostreatus laccase is involved on the removal of highly chlorinated biphenyls contained in transformer oil. This is in contrast to a study where with the results reported in other papers, where a very difficult biodegradation of tetrachlorinated PCBs was reported when P. ostreatus was also used to treat Delor 103 or Delor 106 (Moeder et al. 2005; Novotny et al. 1997). However, these studies were not performed in the presence of a surfactant.
Since laccase activity was confirmed by its specific transcript levels, the pollutant removal activity observed during P. ostreatus growth is due to laccase activity at least under our conditions. The removal activity observed is also consistent with the transcript levels obtained since PCBs elimination and lacccase transcript levels are the highest at 20 and 30 days.
In order to rule out the possibility of having other lignolytic enzymes participating in PCBs removal, versatile peroxidase and manganese peroxidase activities were also determined by chemical assays, however no significant levels were obtained (data not shown); this strongly suggests that the highly chlorinated PCBs removal was only due to laccase activity. However, this does not preclude early involvement of enzymes such as the versatile peroxidase (Pozdnyakova et al. 2010) and cytochrome P450 (Kamei et al. 2006), which might be involved in the oxidation of PCBs prior to their degradation, as it has been demonstrated for PAHs.
Furthermore, PCBs sequestration by lipid vesicles cannot be ruled out as has been previously observed in Fusarium solani studies with PAHs (Verdin et al. 2005).
References
Beaudette LA, Davies S, Fedorak PM, Ward OP, Pickard MA (1998) Comparison of gas chromatography and mineralization experiments for measuring loss of selected polychlorinated biphenyl congeners in cultures of white rot fungi. Appl Environ Microbiol 64:2020–2025
Crinnion WJ (2011) Polychlorinated biphenyls: persistent pollutants with immunological, neurological, and endocrinological consequences. Altern Med Rev 16:5–13
Dietrich D, Hickey WJ, Lamar R (1995) Degradation of 4,4′-dichlorobiphenyl, 3,3′,4,4′-tetrachlorobiphenyl, and 2,2′,4,4′,5,5′-hexachlorobiphenyl by the white rot fungus Phanerochaete chrysosporium. Appl Environ Microbiol 61:3904–3909
Ericson MD (1999) Analytical chemistry of PCBs. CRC Press, Boca Raton
Faraco V, Giardina P, Sannia G (2003) Metal-responsive elements in Pleurotus ostreatus laccase gene promoters. Microbiol-sgm 149:2155–2162
Fujihiro S, Higuchi R, Hisamatsu S, Sonoki S (2009) Metabolism of hydroxylated PCB congeners by cloned laccase isoforms. Appl Microbiol Biotechnol 82:853–860
Gayosso-Canales M, Esparza-García FJ, Bermúdez-Cruz RM, Tomasini A, Ruiz-Aguilar GML, Rodríguez-Vázquez R (2011) Application of 2III7-3 fractional factorial experimental design to enhance enzymatic activities of Pleurotus ostreatus with high concentrations of polychlorinated biphenyls. J Environ Sci Health (Part A) 46:298–305
Guillén-Navarro GK, Márquez-Rocha FJ, Sánchez-Vázquez JE (1998) Production of biomass and ligninolytic enzymes by Pleurotus ostreatus in submerged culture. Rev Iberoam Micol 15:302–306
Jiang GX, Niu JF, Zhang SP, Zhang ZY, Xie B (2008) Prediction of biodegradation rate constants of hydroxylated polychlorinated biphenyls by fungal laccases from Trametes versicolor and Pleurotus ostreatus. Bull Environ Contam Toxicol 81:1–6
Kamei I, Kogura R, Kondo R (2006) Metabolism of 4,4′-dichlorobiphenyl by white-rot fungi Phanerochaete chrysosporium and Phanerochaete sp. MZ142. Appl Microbiol Biotechnol 72:566–575
Kubátová A, Erbanová P, Eichlerová I, Homolka L, Nerud F, Šašek V (2001) PCB congener selective biodegradation by the white rot fungus Pleurotus ostreatus in contaminated soil. Chemosphere 43:207–215
Luo W, D’Angelo EM, Coyne MS (2008) Organic carbon effects on aerobic polychlorinated biphenyl removal and bacterial community composition in soils and sediments. Chemosphere 70:364–373
Moeder M, Cajthaml T, Koeller G, Erbanová P, Šašek V (2005) Structure selectivity in degradation and translocation of polychlorinated biphenyls (Delor 103) with a Pleurotus ostreatus (oyster mushroom) culture. Chemosphere 61:1370–1378
Nikiforova SV, Pozdnyakova NN, Turkovskaya OV (2009) Emulsifying agent production during PAHs degradation by the White Rot Fungus Pleurotus ostreatus D1. Curr Microbiol 58:554–558
Novotny C, Vyas BRM, Erbanova P, Kubatova A, Sasek V (1997) Removal of PCBs by various white rot fungi in liquid cultures. Folia Microbiol 42:136–140
Palmieri G, Giardina P, Bianco C, Fontanella B, Sannia G (2000) Copper induction of laccase Isoenzymes in the ligninolytic fungus Pleurotus ostreatus. Appl Environ Microbiol 66:920–924
Paria S (2008) Surfactant-enhanced remediation of organic contaminated soil and water. Adv Colloid Interface Sci 138:124–158
Pointing SB (2001) Feasibility of bioremediation by white rot-fungi. Appl Microbiol Biotechnol 57:20–33
Palmieri G, Cennamo G, Sannia G (2005) Remazol brillant blue R decolourisation by the fungus Pleurotus ostreatus and its oxidative enzymatic system. Enzyme Microb Technol 36:17–24
Pozdnyakova NN, Nikiforova SV, Turkovskaya OV (2010) Influence of PAHs on ligninolytic enzymes of the fungus Pleurotus ostreatus D1. Cent Eur J Biol 5:83–94
Raeder U, Broda P (1985) Rapid preparation of DNA from filamentous fungi. Lett Appl Microbiol 1:17–20
Rigas F, Dritsa V, Marchant R, Papadopoulou K, Avramides EJ, Hatzianestis I (2005) Biodegradation of lindane by Pleurotus ostreatus via central composite design. Environ Int 31:191–196
Robinson KG, Ghosh MM, Shi Z (1996) Mineralization enhancement of non-aqueous phase and soilbound PCB using biosurfactant. Water Sci Technol 34:303–309
Rodrigues JLM, Kachel CA, Aiello MR, Quensen JF, Maltseva OV, Tsoi TV, Tiedje JM (2006) Degradation of Aroclor 1242 dechlorination products in sediments by Burkholderia xenovorans LB400(ohb) and Rhodococcus sp. Strain RHA1(fcb). Appl Environ Microbiol 72:2476–2482
Rodríguez E, Nuero O, Guillén F, Martínez AT, Martínez MJ (2004) Degradation of phenolic and non-phenolic aromatic pollutants by four Pleurotus species: the role of laccase and versatile peroxidase. Soil Biol Biochem 36:909–916
Rojas-Avelizapa NG, Rodríguez-Vázquez R, Enríquez-Villanueva F, Martínez-Cruz J, Poggi-Varaldo HM (1999) Transformer oil degradation by an indigenous microflora isolated from a contaminated soil. Resour Conserv Recy 27:15–26
Singer AC, Gilbert ES, Luepromchai E, Crowley DE (2000) Bioremediation of polychlorinated biphenyl-contamined soil using carvone and surfactant-grown bacteria. Appl Microbiol Biot 54:838–843
Stajić M, Persky L, Cohen E, Hadar Y, Brčeski I, Wasser SP, Nevo E (2004) Screening of laccase, manganese peroxidase, and versatile peroxidase activities of the genus Pleurotus in media with some raw plant materials as carbon sources. Appl Biochem Biotechnol 117:155–164
Stajić M, Persky L, Hadar Y, Friesem D, Duletić-Laušević S, Wasser SP, Nevo E (2006) Effect of copper and manganese ions on activities of laccase and peroxidases in three Pleurotus species grown on agricultural wastes. Appl Biochem Biotechnol 28:87–96
Tinoco R, Pickard MA, Vázquez-Duhalt R (2001) Kinetic differences of purified laccases from six Pleurotus ostreatus strains. Lett Appl Microbiol 32:331–335
Ullah MA, Kadhim H, Rastall RA, Evans CS (2000) Evaluation of solid substrates for enzyme production by Coriolus versicolor, for use in bioremediation of chlorophenols in aqueous effluents. Appl Microbiol Biotechnol 54:832–837
Vasil'eva GK, Strizhakova ER (2007) Bioremediation of soils and sediments polluted by polychlorinated biphenyls. Mikrobiologiia 76:725–741
Verdin A, Sahraoui AL-H, Newsam R, Robinson G, Durand R (2005) Polycyclic aromatic hydrocarbons storage by Fusarium solani in intracellular lipid vesicles. Environ Pollution 133:283–291
Yadav JS, Wallace RE, Reddy CA (1995) Mineralization of mono- and dichlorobenzenes and simultaneous degradation of chloro- and methyl-substituted benzenes by the white rot fungus Phanerochaete chrysosporium. Appl Environ Microbiol 61:677–680
Acknowledgments
We wish to thank Dr. Germán Cuevas Rodríguez from CIMAV, Chihuahua, Chihuahua, Mexico for allowing us to use the GC/MS.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gayosso-Canales, M., Rodríguez-Vázquez, R., Esparza-García, F.J. et al. PCBs stimulate laccase production and activity in Pleurotus ostreatus thus promoting their removal. Folia Microbiol 57, 149–158 (2012). https://doi.org/10.1007/s12223-012-0106-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-012-0106-9