Abstract
Biodegradable chelant-enhanced phytoremediation offers an alternative treatment technique for metal contaminated soils, but most studies to date have addressed on phytoextraction efficiency rather than comprehensive understanding of the interactions among plant, soil microbes, and biodegradable chelants. In the present study, we investigated the impacts of biodegradable chelants, including nitrilotriacetate, S,S-ethylenediaminedisuccinic acid (EDDS), and citric acid on soil microbes, nitrogen transformation, and metal removal from contaminated soils. The EDDS addition to soil showed the strongest ability to promote the nitrogen cycling in soil, ryegrass tissue, and microbial metabolism in comparison with other chelants. Both bacterial community-level physiological profiles and soil mass specific heat rates demonstrated that soil microbial activity was inhibited after the EDDS application (between day 2 and 10), but this effect completely vanished on day 30, indicating the revitalization of microbial activity and community structure in the soil system. The results of quantitative real-time PCR revealed that the EDDS application stimulated denitrification in soil by increasing nitrite reductase genes, especially nirS. These new findings demonstrated that the nitrogen release capacity of biodegradable chelants plays an important role in accelerating nitrogen transformation, enhancing soil microbial structure and activity, and improving phytoextraction efficiency in contaminated soil.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Chelant-enhanced phytoremediation, applying a chelant to solubilize metals in soils and facilitating plant uptake for the remediation of metal-contaminated soils, has received considerable attention in the past few decades (Kayser et al. 2000; Leštan et al. 2008; Saifullah et al. 2009). The most efficient chelant of its kind is ethylenediaminetetraacetic acid (EDTA), but its low biodegradability, leaching potential, and toxic effect prohibits practical applications in the field (Luo et al. 2006; Meers et al. 2008). In this regard, biodegradable chelants for phytoremediation, e.g., nitrilotriacetate (NTA), S,S-ethylenediaminedisuccinic acid ([S,S]-EDDS), citric acid (CA), etc. have been studied recently as alternative chelants. Recent literature reported their capabilities in rapid degradation and limited leaching from soil (Luo et al. 2005; Quartacci et al. 2007; Cheng et al. 2012; Prieto et al. 2013). The goal of soil phytoremediation process should not only remove the contaminants from the polluted sites but also restore and improve soil quality for future use (Hernandez-Allica et al. 2007). However, knowledge on the impact of chelants, especially for the biodegradable chelants, on the soil quality is limited; hence, there is an urgent need to study the changes of microbial community and nutrient cycling in soil after biodegradable chelant application to soil (Muhlbachova 2011; Yang et al. 2013).
Microbes play an important role in soils and serve as a sensitive indicator to environmental stressors, such as heavy metals and synthetic/natural aminopolycarboxylic acids (Yang et al. 2006; Epelde et al. 2008; Madsen 2011). So far, the majority of studies on the soil microbes toward biodegradable chelants have been conducted using conventional culture methods, including enzyme activities, substrate-induced respiration, mineralizable N, and community-level physiological profiles for environment risk assessment in chelant-enhanced phytoremediation (Epelde et al. 2008; Yang et al. 2013). A few investigations based on these cultures demonstrated less microbial inhibition and higher biosafety in the EDDS-induced phytoremediation in comparison with the EDTA one (Kos and Lestan 2003; Ultra et al. 2005). More recently, a combined analysis using phospholipid fatty acids and PCR-denaturing gradient gel electrophoresis fingerprint was conducted by Yang et al. (2013). They demonstrated that the negative effects on soil enzymatic activity and microbial community structure were present at the first 7 days after the EDDS application, but these effects decreased gradually and disappeared on day 28. However, the mechanisms and dynamics of the processes have not been reported yet in literature.
Nitrogen, a key nutrient for plant growth, can be a key factor in chelant-enhanced phytoremediation because many metal-contaminated sites have barren soil with severe nutrient-deficiency problem. A number of studies have shown that N supply in the form of ammonium (NH4 +), nitrate (NO3 −), or urea can enhance plant biomass yields and phytoextraction efficiency of pollutants from contaminated soils (Loosemore et al. 2004; Zaccheo et al. 2006; Xie et al. 2009; Giansoldati et al. 2012). Biodegradable chelants is rich in carbon and nitrogen, and can provide sufficient nutrition for microorganisms and/or plants if these elements can be transferred to available forms in soil. Therefore, it is hypothesized that external nitrogen supply induced by the application of biodegradable chelants may promote microbial activities and/or plant growth in metal-contaminated soils. These can in turn affect the degradation rate of chelants in soil and the phytoremediation efficiency of metal contaminants from soil.
The present study aims to investigate the interactions among biodegradable chelants, soil microbes, and plant in the phytoremediation process of metal-contaminated soils. We employed various techniques, including conventional culture methods (e.g., Biolog EcoPlates), molecular biology methods (e.g., quantitative real-time PCR), and thermodynamic analysis (e.g., microcalorimetry) in the present research. Three biodegradable chelants with different nitrogen contents, EDDS (28 g mol−1), NTA (14 g mol−1), and CA (0 g mol−1), and ryegrass (Lolium perenne) were used in the pot experiment. The purpose of this study is to characterize the microbial community, nitrogen transformation, plant growth, and metal removal. The results will increase our understanding on the effects of biodegradable chelants to various soil compartments in the whole phytoremediation process.
Materials and methods
Pot experiments
The surface soil samples (0 to 20 cm) were collected in Qingnian Park surrounding activity center of Guangzhou, south China. Five hundred grams of air-dried and sieved soil was packed in cylindrical plastic pots (diameter 11 cm, height 10 cm) in which Rhizon soil moisture samplers (SMS) (Rhizosphere Research Products, Wageningen, Netherlands) were installed. Ryegrass (L. perenne) was used in the pot experiments, which can produce a relatively high biomass and can be cut repeatedly among different croppings. The pot experiments were conducted in a greenhouse under natural light conditions at 10–26 °C. Three different biodegradable chelants (CA, EDDS, and NTA) were used, and the concentrations of chelants were 5 mmol kg−1 soil according to the previous study (Luo et al. 2005). Five tests in this study were labeled as S (only soil), SR (soil + ryegrass), SRC (soil + ryegrass + CA), SRE (soil + ryegrass + EDDS), and SRN (soil + ryegrass + NTA). Each treatment group was performed in triplicate. Pre-germinated seedlings were grown in each pot, and the seedlings were thinned after 10 days of growth to leave 40 plants. Soil moisture was maintained at 70% of water holding capacity by adding deionized water (DIW) to ensure the optimum growth condition for ryegrass. On day 30, chelants were applied to the soil surface in 100 mL solutions at a rate of 5 mmol kg−1 soil. After day 60, the plants were harvested and the soil samples were collected. Soil solutions were collected on day 0, 2, 7, 17, and 30 after adding chelants through the Rhizon SMS. All plant samples were washed with tap water and then rinsed with DIW three times. Clean plant samples were dried at 70 °C in a drying oven for 3 days. After measuring dry weight, the dried plant samples were grounded to about 4 mm size and prepared for acid digestion.
Analysis procedures of soil and plant characterizations
Soil pH, soil organic carbon (SOC), total nitrogen, available nitrogen, phosphorus, and potassium concentration were analyzed according to the procedures described by Lu (2000) and Luo et al. (2005). The soil pH was determined by a pH meter (Model 225 pH Meter; Denver Instrument, USA) with a soil-to-water (non-CO2 DIW) ratio of 1:2.5. SOC was analyzed by dichromate oxidation method. The TOC in soil solution were determined by a TOC analyzer (Shimadzu TOC-5000A, Japan). The Kjeldahl method was used to determine total N in soil. After extraction of the samples with 2.0 mol L−1KCl for 1 h, ammonium nitrogen (NH4 +-N) and nitrate nitrogen (NO3 −-N) were measured by indophenol blue method at 625 nm and ultraviolet colorimetry method at 220 and 275 nm with UV/Vis spectrophotometer (Biochrom Libra S35, England), respectively. Available soil phosphorus was extracted by 0.5 mol L−1 NaHCO3 solution and determined using the molybdenum blue method at 710 nm. Available soil potassium was extracted by 1.0 mol L−1 CH3COONH4 solution and determined by ICP-AES (Perkin-Elmer Optima 3300DV).
The total metal concentrations in soils and plants were determined by ICP-AES (Perkin-Elmer Optima 3300 DV) after strong acid digestion (1:4 concentrated HClO4 and HNO3, v/v) (Li et al. 2001). Certified standard reference materials, NIST 2709 San Joaquin soil and NIST 1573a tomato leaves (National Institute of Standards and Technology, USA), were used in the analysis as part of the Quality Assurance and Quality Control (QA/QC) protocol. The selected physical and chemical properties of the soil samples are shown in Table 1.
Biolog EcoPlate inoculation and incubation
Biolog EcoPlates (Biolog Inc., Hayward, CA, USA) were used to study the substrate utilization pattern of soil microbial community as described by Xue et al. (2008). Briefly, 10 g of fresh soil was placed into an autoclaved triangular flask with 36 mL of 0.85% sterilized NaCl solution. The soil suspensions were sonicated for 1 min and then well mixed on a reciprocal shaker for 30 min. Ten-fold serial dilutions were made, and the 10−3 dilution was used to inoculate the plates. About 150 μL of solution was filled into each well, and all plates were cultivated at 25 °C for 192 h. The optical density at a wavelength of 590 nm was recorded using Microlog 4.01 software (Biolog Inc.) as absorbance daily with an automated plate reader (VMAX; Molecular Devices, Crawley, UK). Average well color development (AWCD) was calculated according to Garland and Mills (1991), i.e., AWCD = Σ(C − R)/n where C is color production within each well, R is the absorbance value of the plate control well, and n is the number of substrates. Mean AWCD values were plotted over time for each sample.
Microcalorimetric measurements
The thermal effect was measured in a TMA III (Thermometric AB, Sweden) multichannel microcalorimetric system to determine variations in enthalpy of the system. This instrument has four channels in which the sample and the reference are introduced simultaneously in a thermostatic cylinder. Before test, the ampoules were sterilized by rinsing in 75% ethanol and sterile DIW for 10 min, and dried under a laminar flow hood. Soil samples were incubated at 25 °C for 24 h before microcalorimetric measurements. Then, 1 g wet weight of soil was introduced in a 4-mL calorimeter stainless steel ampoule. One milliliter of distilled water was used as control. Our initial experiment showed that there was no need to stimulate the microbial activity by the addition of glucose or ammonium sulfate as nutrition. All ampoules, samples, and reference materials were introduced simultaneously in the microcalorimetric channel. The heat flow rate released due to basal respiration in soils was recorded in microwatts per gram of soil per day and represented as power–time lines. The integration of those lines permits to obtain the total heat released by 1 g of soil sample per day, which is termed as soil mass specific heat rate, JQ/S, in Joules per gram of soil per day (Barros et al. 2003).
DNA extraction
Total metagenomic DNA was extracted from 0.5 g of soil using the FastDNA SPIN Kit for Soil (MP Biomedicals, Solon, OH, USA) according to the manufacturer’s instructions. The quality and quantity of DNA extracted were determined in a 1% agarose gel and a NanoDrop spectrophotometer (NanoDrop, Wilmington, DE, USA). DNA concentrations were measured by absorbance at 260 nm (Rodrigues and Tiedje 2007). The extracted DNA for each sample was stored at −20 °C prior to being used in the quantitative real-time polymerase chain reaction (qPCR) assay.
Quantitative real-time PCR
In this study, a total of five bacterial nitrogen metabolism genes, amoA, nosZ, nirS, nirK, and nifH, were selected to investigate their relative change among the treatment groups (Table S1). Thermal cycling and fluorescence detection were conducted in triplicate on a BioRad iCycler with the software iCycler iQ version 3.0 (BioRad, Hercules, CA). Relative quantification was conducted for these genes using the bacterial 16S rRNA gene as a calibrator. The obtained data were analyzed using the 2−ΔΔC T method (Livak and Schmittgen 2001). For QA/QC protocol, the amplification efficiencies for five target genes and one reference gene were tested. Each qPCR reaction was carried out in a 25-μL solution containing 25 ng DNA, 12.5 μL SYBR Green PCR Master Mix (Thermo Scientific), 200 nM of each primer (Table S2), and ddH2O up to 25 μL. The qPCR was performed under the following conditions: 45 cycles of 95 °C for 10 s, 52–58 °C for 30 s, and 72 °C for 60 s. The PCR products were detected by agarose gel electrophoresis, cloning, and Sanger sequencing.
Data analysis
Statistical analyses were carried out by SPSS 17.0 software. The statistical significance of differences and variance (p < 0.05) was determined by one-way ANOVA (LSD).
Results and discussion
Effect of chelants on nitrogen balance in soils and ryegrass tissues
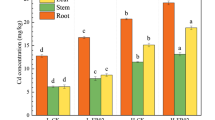
Figure 1 shows the nitrate profile in soils with and without chelant addition. The concentrations of ammonium in all tests (i.e., SR, SRC, SRN, and SRE) were below the detection limit (<0.01 ppm, data not shown), while the nitrate concentration peaked on day 2 with the chelant addition. The concentrations of nitrate on day 2 from the SRC, SRN, and SRE tests increased by a factor of 3.7, 3.6, and 23.5, respectively, compared to the control (SR) one. After day 2, the nitrate concentrations decreased sharply, especially for the SRE, with a decrease to 41 and 21% on day 7 and 30, respectively. Meanwhile, the nitrogen content of the ryegrass corresponds well to the nitrate concentration in soils with chelant application (Fig. 2). The results showed that the nitrogen concentration in the SRE treatment was significantly higher than those in the SRC, SRN, and the control (SR) groups for the shoot and root. In addition, the nitrogen content in ryegrass tissues presented similar variation tendency with the nitrate concentration in soil, mainly derived from the biodegradation of EDDS, and further uptake by the ryegrass within 30 days of growth period. Similar patterns of TOC concentrations could also be found in these experiments (Fig. S1). The TOC concentration (156 mg L−1) in the soil solution on day 7 after EDDS addition was only 18.1% of that on day 2 (860 mg L−1), demonstrating most of the EDDS degradation occurring within the first 7 days of application to soil. Similarly, the reported half-life of EDDS ranges from 2.5 to 8.7 days in most studies (Meers et al. 2008). This degradation can trigger the shoot biomass enhancement in the SRE, SRN, and SRC treatment by 20.1, 12.5, and 12.1%, respectively, in comparison with the control group (without chelant treatment) (Fig. 3). Xie et al. (2009) also demonstrated that supplying N as NO3 − promoted the growth and phytoextraction of Cd and Zn by Thlaspi caerulescens compared with NH4 +. Therefore, the nitrogen content in chelators can be an important factor for plant growth and enhancing the concentration of nitrate in ryegrass.
Effect of chelants on metal distribution in soil and ryegrass tissues
The concentrations of trace metals in soil solutions after the chelant application are presented in Fig. 4. The data showed that the concentrations of metals in all soil solutions reached a peak on day 2 after the addition of chelants, especially for the SRE treatment. Since then, the concentration of trace metals decreased rapidly and reached to a similar level of the control group on day 17. It was observed that Cu release induced by the EDDS addition was the highest among all metals tested. Moreover, the concentrations of Cu on day 7 and 17 after the EDDS application were 44 and 10%, respectively, of that on day 2. As for Pb, the corresponding concentrations were 15 and 2%, respectively. The corresponding concentrations of Zn were 31 and 4%, respectively. After 30 days of the EDDS application, the concentrations of these metals decreased to the initial concentrations before the EDDS application.
Trace metal concentration profile in the soil solutions after adding different biodegradable chelates. SR soil + ryegrass, SRC soil + ryegrass + CA, SRE soil + ryegrass + EDDS, SRN soil + ryegrass + NTA. The Cd concentrations in all treatments were below the detection limit. Error bars represent the standard errors of mean of three replicates (n = 3)
On the other hand, the translocation of metals (Cu, Pb, Zn, and Cd) from roots to shoots was mostly enhanced in the EDDS treatment, as evidenced by identification of the highest translocation factor (shoot concentration/root concentration), being 2.9-, 3.1-, 1.3-, and 1.1-fold higher than the control (Table 2). Regarding the total uptake of metals (Cu, Pb, Zn, and Cd), EDDS application resulted in the highest enhancement of metals in shoots, being 6.4-, 4.4-, 2.1-, and 1.6-fold higher than the control. Hence, higher phytoextraction efficiency was achieved for Cu and Pb in the EDDS treatment. This result is consistent with a previous report that EDDS was most effective in terms of solubilizing soil Cu and Pb for root uptake and translocation into aboveground biomass (Luo et al. 2006, 2015).
Effect of chelants on soil microbial community and metabolic activities
Measurements of substrate utilization can enable the quantification of bacterial metabolic capabilities and the functional diversity of a microbial community. The effect of chelants on microbial metabolic activities on day 30 for carbon substrate utilization by bacteria determined by Biolog EcoPlates is shown in Fig. 5. The bacterial activity occurred during the period of 24 to 120 h, according to its rapid increase of the average well color development (AWCD) values (Fig. 5a). After 30 days of the chelant applications (e.g., NTA, EDDS, and CA), the higher values of the AWCD in comparison with the control (SR) group indicated that chelant addition had a positive effect on metabolic activities in soil. NTA application showed the strongest ability to promote the AWCD values in comparison with other chelants. Additionally, the utilization of various substrates in the Biolog plates by the microorganisms was analyzed (Fig. S2), which further demonstrated that the NTA application showed the strongest ability to promote the utilizations of all substrates among all the treatments. The EDDS application also has a positive effect on increasing the utilizations of amino acids, carbohydrates, amines, and carboxylic acids.
a Variations in AWCD of soil bacterial community for different treatments on day 30 after chelate applications. b Effects of EDDS application on the AWCD values at different times. S only soil, SR soil + ryegrass, SRC soil + ryegrass + CA, SRE soil + ryegrass + EDDS, SRN soil + ryegrass + NTA. SRE-day 2, 10, and 30 represents soil planted with ryegrass on day 2, 10, and 30 after the EDDS application, respectively. SR-day 30 represents soil planted with ryegrass on day 30 without the EDDS application. Error bars represent the standard errors of mean of three replicates (n = 3)
According to the cluster analysis of the community-level physiological profiles (Fig. 6a), the microbial structure in the SRN and SRC treatment have a distinguished difference compared to the control (SR) group and thus indicated that the bacterial community changed drastically with the addition of NTA or CA, and remained different even after 30 days. However, the SR and SRE treatments presented highly correlated microbial profiles and thus demonstrated that the native microbial structure would not be affected with the EDDS addition.
Cluster analysis of community-level physiological profile for different soil treatments (a) and at different times after the EDDS application (b). S only soil, SR soil + ryegrass, SRC soil + ryegrass + CA, SRE soil + ryegrass + EDDS, SRN soil + ryegrass + NTA. SRE-day 2, 10, and 30 represents soil planted with ryegrass on day 2, 10, and 30 after the EDDS application, respectively. SR-day 30 represents soil planted with ryegrass on day 30 without the EDDS application
Figures 5b and 6b present the AWCD outcomes and cluster analysis of the community-level physiological profiles with EDDS addition at different times, respectively. Interestingly, both figures showed similar profiles on day 2 and 10, which may be caused by microbial activity inhibition during the initial EDDS application. Combined with the changes of metal concentrations in soil solutions (Fig. 4), it may be true that the direct toxic effect of the sharply increased metals in this period was the main reason for this inhibition effect on microbial activity. Afterwards, this inhibition disappeared on day 30, as shown from their high correlation between the SRE-day 30 and the SR-day 30 in Fig. 6b. Figure 4 also demonstrates that the bioavailability of metals sharply decreased to the background level on day 30 after the EDDS application. These results strongly suggested that the direct negative effects of metal toxicity on soil microorganisms were closely related to the concentrations of metals in soil solution and the metabolic activities. The bacterial community was able to recover to its original metabolic function after 30 days of the EDDS application. Consequently, the results of AWCD values and the cluster analysis suggested that the EDDS is an environmental-friendly chelator among the three tested even though NTA has the highest response from the AWCD values.
The heat generation from microbial metabolism has been recognized as a key index of microbial activity (Sparling 1981), which could be monitored by microcalorimetry, one of the emerging methods in recent years (Barros et al. 2007; Wang et al. 2009). The microcalorimetrical flows in the soil samples were recorded as power–time lines during the first 24 h (Fig. 7). The soil mass specific heat rate JQ/S (1.49–3.60 J g−1 day−1) was obtained (Fig. S3) by the integration of the microcalorimetrical flows, which was comparative to the ones reported by Barros et al. (2003). The values were in the range of 1.0–6.6 J g−1 day−1 from the soils collected in Galicia (Northwest Spain) and in the Brazilian Amazon (Barros et al. 2003). The order of heat rates on day 30 after chelant application were SRE (3.60) > SRN (3.13) > SRC (2.77) > SR (2.46) > S (1.81), indicating that chelants have a strong positive effect on soil metabolic activities, especially for the EDDS application (Fig. S3a).
a Power–time curves of microbes from different soil treatments. b Effects of EDDS application on the power at different times. S only soil, SR soil + ryegrass, SRC soil + ryegrass + CA, SRE soil + ryegrass + EDDS, SRN soil + ryegrass + NTA. SRE-day 2, 10, and 30 represents soil planted with ryegrass on day 2, 10, and 30 after the EDDS application, respectively. SR-day 30 represents soil planted with ryegrass on day 30 without the EDDS application
Figure S3b presents the effect of EDDS application on the microcalorimetrical flows at different days with the highest value of JQ/S on day 2 (2.06). It is likely that the sharply increased nitrate concentration on day 2 in soil was related to the microcalorimetrical flows due to microbial activities in soil. Barros et al. (2003) also reported a positive linear relation between the soil mass specific heat rate JQ/S and the soil nitrogen availability. Furthermore, the microbial activity was correlated to nitrogen at a higher level of significance than soil organic matter (Kaiser et al. 1992), which further stressed the importance of nitrogen availability for soil microbial metabolism. In the present study, microbial activity reflects from the observation of higher heat in the SRE treatment in comparison with the SRN treatment, which was consistent with the Biolog analysis on the nitrogen content. In addition, the value of JQ/S on day 30 (1.81) after the EDDS application was higher than that on day 10 (1.64) and the control group (1.49), implying the achievement of microbial activities recovery and stimulation in soils on day 30 after the EDDS application (Fig. 5b). Our results implied that the addition of EDDS may help to detoxify the metals and/or to tolerate by increasing its external nitrogen nutrient. This presents an indirect positive effect on the microorganisms. A similar finding was also reported by Yang et al. (2013), which showed that the recovery of soil microbial character by the addition of EDDS could be achieved over time and disappeared on the 28th day. Combined with the dynamic changes of metals and TOC concentrations in soil over time, the gradual recovery ability of metabolic activity from day 2 to day 30 was caused mainly by the released nitrogen due to the degradation of EDDS and the rapidly decreased bioavailability of metals in soils over time.
Effect of chelants on N cycling gene abundances
The abundance of N cycling-associated bacteria in soils is sensitive to the dynamic variations of N (Mueller et al. 2013). A large quantity of nitrogen is released during the degradation of chelants, which could influence the microbial growth, especially for N cycling-associated bacteria. The biogeochemical cycles of nitrogen in the soil–microbe–plant system are the key factors on chelator-regulated phytoremediation. Previous studies showed that the degradation pathway of S,S-EDDS is through S,S-EDDS → N-(2-aminoethyl) aspartic acid (AEAA) → ethylenediamine, and that of NTA is NTA → glycine + glyoxylate (Egli 2001). The final degradation products of these biodegradable chelants are CO2 and NH4 +-N, whereas the NH4 +-N is easily oxidized to NO3 −-N through nitrification. This is consistent with our finding that the NO3 −-N concentration sharply increased on day 2 after the chelant application, and the NH4 +-N concentration were all below the detection limit. There was no significant increase in total nitrogen concentration in soil for chelant application, while the NO3 −-N concentration in the SRE treatment increased significantly (Fig. 1).
Table 3 summarizes the ratios of the N cycling functional genes of the treated soils to the control (SR). The relative quantification of the target genes (amoA, nosZ, nirS, nirK, and nifH) in soils substantially increased after planting of the ryegrass in the pots. A critical finding was that the EDDS application showed the highest abundance ratios of 1.92 in amoA for ammonia oxidation, and 2.84 in nirK and 13.1 in nirS for denitrification, denoting its active nitrogen transformation in the treated soils. The addition of NTA, however, only facilitated limited denitrification, albeit no ammonium oxidation, since the ratios in amoA, nirK, and nirS are 0.72, 1.15, and 3.62, respectively. In contrast, CA application showed no significant effect on increasing the level of N cycling-associated functional genes. The difference was probably caused by the absence of nitrogen in CA, resulting in no additional nitrogen released in soils. Besides, there was no induced nitrogen fixation activity while applying in the chelators, according to the low ratios of one in nifH with SRC, SRE, and SRN. These results indicate that there is a positive correlation between the nitrogen content in chelants and the abundance of amoA for ammonia oxidation and nirS and nirK for denitrification in treated soils.
The reduction of nitrite by denitrifiers can be performed either by a copper nitrite reductase (nirK) or by a cytochrome cd1 nitrite reductase (nirS), and denitrification catalyzes the nitrite (NO2 −) reduction to nitric oxide (NO). Therefore, the sharp increasing abundance of nirS and nirK in the SRE treatment demonstrated the response of N cycling-associated bacteria to nitrogen releasing by the degradation of EDDS. A similar finding was also reported by Yuan et al. (2012), which showed that nirS copy numbers after nitrate addition increased 10-fold in response to nitrate addition in a Chinese rice field soil. The nosZ gene has been treated as the primary marker gene for denitrification in previous studies. It has also been shown that the abundance ratios of nosZ and nirK/S genes influenced N2O emissions (Zhao et al. 2014). We found that the abundance of nosZ was decreased and the ratio of nosZ abundance to the sum of nirK and nirS gene abundances was <1.0 in this research. This indicates that N2O emissions were not accelerated from the test soil (Philippot et al. 2009). Therefore, our results hereby indicated that the EDDS addition made the positive contribution to the soil microbial abundance of denitrification, but insignificant in nitrous oxide emission, in the chelator-induced phytoremediation. However, the transformation process of nitrogen released by the chelator addition in the soil–plant system still lacks comprehensive investigations. Therefore, it would require further investigation of soil nitrogen mineralization and atmospheric nitrogen fixation, and nitrogen distribution by the nitrogen isotope labeling approach.
Conclusions
The nitrogen release through the degradation of chelators showed the enhancement of phytoextraction efficiency and denitrification process in the soil–plant system. There was more nitrogen content in chelators, higher remediation efficiency, and less inhibition on metabolic activity and microbial structure in the chelant-enhanced phytoremediation. In addition, there was more nitrogen content in chelators and higher active denitrification via the sharp increase of nirS in the chelant-enhanced phytoremediation. Hence, the nitrogen content in chelators can be an important factor for the application of chelant-enhanced phytoremediation technology. Collectively, this leads to a firm conclusion that how best to select a BC for phytoremediation lies in its degrading into organics and nitrogen products simultaneously for the soil microbial community mitigation, plant growth enhancement, and metal and nutrient uptake. The above results showed some possible mechanistic/interactions among plant, soil microbes, and chelants in phytoremediation and thus will help stimulate the greater understanding of the influence of the nitrogen cycle and soil microbes in the chelant-induced phytoremediation.
References
Barros N, Feijóo S, Fernández S (2003) Microcalorimetric determination of the cell specific heat rate in soils: relationship with the soil microbial population and biophysic significance. Thermochim Acta 406:161–170
Barros N, Gallego M, Feijóo S (2007) Sensitivity of calorimetric indicators of soil microbial activity. Thermochim Acta 458:18–22
Cheng GL, Ma XF, Sun XB, Zhao SQ (2012) Effects of EDTA, EDDS and citric acid on growth of maize and uptake of lead by maize in contaminated soil. Adv Res Mater Sci Environ Sci 534:277–280
Egli T (2001) Biodegradation of metal-complexing aminopolycarboxylic acids. J Biosci Bioeng 92:89–97
Epelde L, Hernandez-Allica J, Becerril JM, Blanco F, Garbisu C (2008) Effects of chelates on plants and soil microbial community: comparison of EDTA and EDDS for lead phytoextraction. Sci Total Environ 401:21–28
Garland JL, Mills AL (1991) Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Appl Environ Microbiol 57:2351–2359
Giansoldati V, Tassi E, Morelli E, Gabellieri E, Pedron F, Barbafieri M (2012) Nitrogen fertilizer improves boron phytoextraction by Brassica juncea grown in contaminated sediments and alleviates plant stress. Chemosphere 87:1119–1125
Hernandez-Allica J, Garbisu C, Barrutia O, Becerril JM (2007) EDTA-induced heavy metal accumulation and phytotoxicity in cardoon plants. Environ Experimen Bot 60:26–32
Kaiser EA, Mueller T, Joergensen RG, Insam H, Heinemeyer O (1992) Evaluation of methods to estimate the soil microbial biomass and the relationship with soil texture and organic-matter. Soil Biol Biochem 24:675–683
Kayser A, Wenger K, Keller A, Attinger W, Felix HR, Gupta SK, Schulin R (2000) Enhancement of phytoextraction of Zn, cd, and cu from calcareous soil: the use of NTA and sulfur amendments. Environ Sci Technol 34:1778–1183
Kos B, Lestan D (2003) Influence of a biodegradable ([S,S]-EDDS) and nondegradable (EDTA) chelate and hydrogel modified soil water sorption capacity on Pb phytoextraction and leaching. Plant Soil 253:403–411
Leštan D, Luo CL, Li XD (2008) The use of chelating agents in the remediation of metal-contaminated soils: a review. Environ Pollut 153:3–13
Li XD, Poon CS, Liu PS (2001) Concentration and chemical partitioning of road dusts and urban soils in Hong Kong. Appl Geochem 16:1361–1368
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) methods. Methods 25:402–408
Loosemore N, Straczek A, Hinsinger P, Jaillard B (2004) Zinc mobilization from a contaminated soil by three genotypes of tobacco as affected by soil and rhizosphere pH. Plant Soil 260:19–32
Lu RK (2000) Analysis methods of soil agricultural chemistry. China Agricultural Science Press, Beijing (in Chinese)
Luo CL, Shen ZG, Li XD (2005) Enhanced phytoextraction of Cu, Pb, Zn and Cd with EDTA and EDDS. Chemosphere 59:1–11
Luo CL, Shen ZG, Lou LQ, Li XD (2006) EDDS and EDTA-enhanced phytoextraction of metals from artificially contaminated soil and residual effects of chelant compounds. Environ Pollut 144:862–871
Luo CL, Wang SR, Wang Y, Yang RX, Zhang G (2015) Effects of EDDS and plant-growth-promoting bacteria on plant uptake of trace metals and PCBs from e-waste-contaminated soil. J Hazard Mater 286:379–385
Madsen EL (2011) Microorganisms and their roles in fundamental biogeochemical cycles. Current Opinion Biotechnol 22:456–464
Meers E, Tack FMG, Verloo MG (2008) Degradability of ethylenediaminedisuccinic acid (EDDS) in metal contaminated soils—implications for its use soil remediation. Chemosphere 70:358–363
Mueller KE, Hobbie SE, Tilman D, Reich PB (2013) Effects of plant diversity, N fertilization, and elevated carbon dioxide on grassland soil N cycling in a long-term experiment. Glob Chang Biol 19:1249–1261
Muhlbachova G (2011) Soil microbial activities and heavy metal mobility in long-term contaminated soils after addition of EDTA and EDDS. Ecol Eng 37:1064–1071
Philippot L, Čuhel J, Saby NPA, Chèneby D, Chroňáková A, Bru D, Arrouays D, Martin-Laurent F, Simek M (2009) Mapping field-scale spatial patterns of size and activity of the denitrifier community. Environ Microbiol 11:1518–1526
Prieto C, Lozano JC, Rodriguez PB, Tome FV (2013) Enhancing radium solubilization in soils by citrate, EDTA, and EDDS chelating amendments. J Hazard Mater 250:439–446
Quartacci MF, Irtelli B, Baker AJ, Navari-Izzo F (2007) The use of NTA and EDDS for enhanced phytoextraction of metals from a multiply contaminated soil by Brassica carinata. Chemosphere 68:1920–1928
Rodrigues DF, Tiedje JM (2007) Multi-locus real-time PCR for quantitation of bacteria in the environment reveals Exiguobacterium to be prevalent in permafrost. FEMS Microbiol Ecol 59:489–499
Saifullah ME, Qadir M, de Caritat P, Tack FMG, Du LG, Zia MH (2009) EDTA-assisted Pb phytoextraction. Chemosphere 74:1279–1291
Sparling GP (1981) Heat output of the soil biomass. Soil Biol Biochem 13:373–376
Ultra VU, Yano A, Iwasaki K, Tanaka S, Kang YM, Sakurai K (2005) Influence of chelating agent addition on copper distribution and microbial activity in soil and copper uptake by brown mustard (Brassica juncea). Soil Sci Plant Nutri 51:193–202
Wang F, Yao J, Chen HL, Zhou Y, Chen YJ, Chen H, Gai N, Zhuang RS, Tian L, Maskow T, Ceccanti B, Trebse P, Zaray G (2009) Microcalorimetric measurements of the microbial activities of single- and mixed-species with trivalent iron in soil. Ecotoxicol Environ Saf 72:128–135
Xie HL, Jiang RF, Zhang FS, McGrath SP, Zhao FJ (2009) Effect of nitrogen form on the rhizosphere dynamics and uptake of cadmium and zinc by the hyperaccumulator Thlaspi caerulescens. Plant Soil 318:205–215
Xue D, Yao HY, Ge DY, Huang CY (2008) Soil microbial community structure in diverse land use systems: a comparative study using Biolog, DGGE, and PLFA analyses. Pedosphere 18:653–663
Yang YG, Campbell CD, Clark L, Cameron CM, Paterson E (2006) Microbial indicators of heavy metal contamination in urban and rural soils. Chemosphere 63:1942–1952
Yang L, Wang GP, Cheng ZN, Liu Y, Shen ZG, Luo CL (2013) Influence of the application of chelant EDDS on soil enzymatic activity and microbial community structure. J Hazard Mater 262:561–570
Yuan Q, Liu PF, Lu YH (2012) Differential responses of nirK- and nirS-carrying bacteria to denitrifying conditions in the anoxic rice field soil. Environ Microb Rep 4:113–122
Zaccheo P, Crippa L, Pasta VDMP (2006) Ammonium nutrition as a strategy for cadmium mobilization in the rhizosphere of sunflower. Plant Soil 283:43–56
Zhao MX, Xue K, Wang F, Liu SS, Bai SJ, Sun B, Zhou JZ, Yang YF (2014) Microbial mediation of biogeochemical cyclings revealed by simulation of global changes with soil transplant and cropping. ISME J 8:2045–2055
Acknowledgements
This research was financially supported by the National Natural Science Foundation of China (41571314, 41201226).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Zhihong Xu
Electronic supplementary material
ESM 1
(DOCX 215 kb).
Rights and permissions
About this article
Cite this article
Fang, L., Wang, M., Cai, L. et al. Deciphering biodegradable chelant-enhanced phytoremediation through microbes and nitrogen transformation in contaminated soils. Environ Sci Pollut Res 24, 14627–14636 (2017). https://doi.org/10.1007/s11356-017-9029-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-017-9029-y