Abstract
Inter-basin water transfer and source water switching will be increasingly launched due to significant population increase and the shortage of the local water resources in cities around the world. Source water switch may cause physiochemical and microbiological de-stabilization of pipe material, biofilms, and loose deposits in drinking water distribution system (DWDS). Great sulfate alteration during source water switch had been deemed as the main cause of a red water case that occurred in a northern China city. To ascertain the relationship between water quality changing and bacterial communities of biofilms in DWDS and possible bacteria risk in a red water case, water quality changing experiments in simulated DWDSs were conducted for approximately 2 years. Twenty-five corrosion scale samples and eight water samples collected from pipe harvest sites or during experimental periods were analyzed for their bacterial community composition by 454-pyrosequencing technology. Taxonomy results together with redundancy analysis (RDA) or canonical correspondence analysis (CCA) and hierarchical cluster analysis all indicated that bacterial community of samples with groundwater (GW) or surface water (SW) supply history and their variations under high sulfate water were rather different owing to different water source histories and the original pipe scale characteristics. Potential opportunistic pathogens: Burkholderia, Escherichia-Shigella, Mycobacterium, Serratia, Ralstonia, Novosphingobium, Flavobacterium, Sphingomonas, and Sphingopyxis were observed in scale or water samples.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Switching to alternative source water (e.g., inter-basin water source, desalinated water) is launched for solving water shortage problems worldwide due to the population increase in cities as well as the rapid development of economy. Water quality changes owing to the alternative source water switch may cause physiochemical and microbiological de-stabilization of pipe material, biofilms, and loose deposits in the distribution system that have been established over decades and may harbor components that cause health or esthetical issues (Liu et al. 2017). Under regular operations, distribution networks can function as sinks (contaminant accumulation, as in particle sedimentation) or sources of trace contaminants in the bulk water because of additional forces, such as (bio-)aggregation and precipitation, chemical bonds of corrosion products, and bio-adhesion of biofilm (Liu et al. 2014; McNeill and Edwards 2001; Lytle et al. 2004; Flemming and Wingender 2010). Once de-stabilization caused by source water switch occur, pipe material, biofilms, and the above-accumulated harbored material in DWDS will become a major source of contaminants, which may pose health risks to consumers (Liu et al. 2017). The subsequent physiochemical and microbiological water quality problems caused by de-stabilization included water discoloration, such as blue water (high copper concentration) (Edwards et al. 2000) and red water (high iron concentration) (Wang et al. 2009; Li et al. 2010; Yang et al. 2014), the release of heavy metals (Pb, As, Cu, Fe, Mn) (Lytle et al. 2004; Sun et al. 2017; Pieper et al. 2017), and the release of (opportunistic) pathogenic microorganisms (Schwake et al. 2016).
There have been several reports of serious red-water complaints following switches of source water in Southern California, Tucson (Arizona), and Tampa (Florida) in the USA, and a northern city in China (Tang et al. 2006; Yang et al. 2012). Red water issue had occurred in some areas of Beijing, China, soon after 80% of the local source water was replaced by long-distance introduced source water from a neighboring province in October, 2008. It was interesting to find that heavy red water occurred only in areas historically supplying local groundwater, and the areas without red water were historically supplied with local surface waters. The switch in source water caused Fe concentration peaks (> 10 mg L−1) in the first month because of the presence of high SO4 2- concentration compared with that of local groundwater (about 200 vs. 30 mg L-1) (Wang et al. 2009; Yang et al. 2012). Previous studies indicated that sulfate ions had been recognized as a causative agent of red water under source water switch cases, reflected in high values of indices such as the Larson ratio (LR, the ratio of chloride and sulfate to bicarbonate, Eq. (1)) with a higher index indicating a more corrosive water (McNeill and Edwards 2001; Li et al. 2010; Yang et al. 2012). Further investigations about the red water cases in China found that the pipe material and supply-water quality determine the characteristics of the pipe scales, and that different corrosion scales have different adaptability to source water switch (Yang et al. 2012). Dynamical transformation characteristics of iron corrosion products on old cast iron distribution pipes with different source water histories under high sulfate source water switching were also detailedly investigated which illustrated the severe iron release in red water and different adaptability of corrosion scales (Yang et al. 2014).
where, [Cl−], [SO4 2−], and [HCO3 −] are the concentrations of chloride, sulfate, and bicarbonate (mol L−1), respectively.
Study on red water events in Beijing also demonstrated that a diverse array of iron-oxidizing bacteria, including Gallionella, Sideroxydans, Acidothiobacillus ferrooxidans, and Thermomonas were found in red water samples by 16S rRNA cloning library. Quantitative real-time PCR analysis indicated that high levels of Gallionella spp., as well as other neutrophilic iron-oxidizing bacteria in the bulk water, could facilitate the precipitation of iron oxides by converting ferrous to ferric iron, thus contributing to the formation of this red water event (Li et al. 2010). Further investigations about the red water cases in China illustrated that for pipes historically transporting groundwater the high sulfate water source had caused significant increase of sulfur-oxidizing bacteria, sulfate-reducing bacteria, and iron-oxidizing bacteria, which were associated with severe iron release and red water (Yang et al. 2014). The Pb contamination event in the Flint (Michigan, USA) in 2014, which was due to source water switch, triggered a serious public health crisis, as evidenced by high Pb levels in children’s blood, coinciding with increasing water discoloration and high Legionella numbers in premise plumbing (Schwake et al. 2016; Pieper et al. 2017).
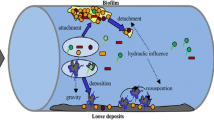
Drinking water-related outbreaks of waterborne infectious diseases pose serious health threat to human beings worldwide. Even multibarrier water treatment processes at water utilities are implemented; it is not possible to remove all microorganisms from source water. Meanwhile, treatment failures and ineffective management of distribution system may lead to severe microbial re-growth in DWDS (Reynolds et al. 2008). Particularly, biofilm formed by microorganisms via adhesion, nucleation, and growth on pipe walls provide resistance to environmental stresses and increase the overall fitness of the microbial community (Liu et al. 2017). Activities of microbial re-growth and the presence of (opportunistic) pathogenic microorganisms have been frequently detected in DWDSs or from customer taps (Vaz-Moreira et al. 2011; Chaves Simões and Simões 2013; Shaw et al. 2015; Douterelo et al. 2016a), thus, their release into water can pose higher health risk to customers than discoloration.
Biofilms generally recognized as the primary source of microorganisms in DWDS have the potential to cause public health issues, such as harboring microorganisms, promoting bacterial re-growth, consuming disinfection agents and causing microbial corrosion problems (Rajasekar et al. 2017; Douterelo et al. 2016b). Therefore, many researchers have focused on biofilm characterization in DWDSs using cultivation-based or molecular biological approaches including clone library, denaturing gradient gel electrophoresis (DGGE), fluorescent in situ hybridization (FISH), quantitative real-time PCR (qPCR), and terminal restriction fragment length polymorphism (T-RFLP) (Teng et al. 2008; Li et al. 2010; White et al. 2011; Douterelo et al. 2016a).
Pyrosequencing, as a new generation of sequencing technology, which allows high-throughput sequencing and avoids the bias of clone libraries has revolutionized the study of microbial diversity and successfully utilized to characterize drinking water biofilms. Both T-RFLP and 454-pyrosequencing were used to reveal the reversible shifts in microbial communities in real DWDS under disinfection transitions between chlorination and chloramination (Hwang et al. 2012). Microbial communities of the biofilm on the aged pipe samples from affected areas of the large-scale red water event in Beijing determined by polymerase chain reaction (PCR), DGGE analysis, and 454-pyrosequencing under different disinfection conditions showed great changes and the new dominant groups promoted more compact corrosion scale formation and higher inhibition of iron release (Li et al. 2016). Pyrosequencing and a new “core-satellite” model were used to analyze the whole community and the dynamics of biofilms owing to seasonal water temperature fluctuation in an urban DWDS, and the findings provided critical insights for developing more targeted water quality monitoring programs and treatment strategies for groundwater-sourced drinking water systems (Ling et al. 2016). The above researches revealed not only the activities of microbial re-growth in DWDS but also the microbial communities’ variations under supply-water quality changes.
The pipe material, supply-water quality, and the water flow condition determine the formation process and physicochemical characteristics of pipe scales and also might affect biofilm composition and morphology in DWDSs (McNeill and Edwards 2001; Douterelo et al. 2016a; Liu et al. 2017). It is well known that microbial activity have great influence on the corrosion behavior of metals, biogeochemical redox cycling of metal oxides, and thus the corrosion by-product release (Iwona and Christine 1999; Teng et al. 2008; White et al. 2011). Considering that microbiological de-stabilization in DWDSs might degrade water quality and safety through the potential hosting of undesirable microorganisms, research on bacterial community alteration under source water switch with red water occurrence is essential to understand the factors that influence their development and the potential microbial risk.
In this study, the bacterial community alteration and the potential functional bacteria transformation under high-sulfate source water switch with red water occurrence were put particular emphasis for understanding the relationship between water quality and bacterial community composition, revealing the potential microbial-related risk and providing valuable information for red water control strategies by 454-pyrosequencing of 16S rRNA gene.
Materials and methods
Test pipes
The cast iron pipes were all approximately 20 years old, with diameter of DN100 and harvested from five different sites in the DWDS of a city in northern China. The morphological and structural characteristics of the corrosion products on these pipes had been investigated previously (Yang et al. 2012). For consistency of the two works, the five studied pipes used in this work were identified using the same symbols as previous, that is, Pipe-SW1, Pipe-SW2, Pipe-SW3, PipeB-GW3, and PipeC-GW3, respectively (Yang et al. 2012, 2014). The information about the water treatment plants and supply-water quality are listed in Tables S1 and S2. A total of 11 DNA samples of pipe scales on those pipes with different source water histories and different physical characteristics were collected and analyzed for comparison.
Pipe loop setup and experimental operation
Pipe loop facilities were set up in parallel using Pipe-SW2 (designated as loop 1 (L1) and loop 2 (L2), the same symbols as in a previous paper for consistency) and PipeC-GW3 (designated as loop 4 (L4)) as test pipe sections for water source switch experiments (Yang et al. 2014). Their schematic diagram and detailed setup can be referred to Fig. S1.
Experimental operation for each pipe loop was divided into five consecutive phases (Table 1). Due to the different harvesting time and the distinct corrosion scale characteristics of test pipes, the operation condition and phase definition for the three pipe loops were not the same. Experimental phase definition, detailed operation condition, and corresponding feed water types for each pipe loop also can be referred to Tables S3, S4, S5, S6 and the supporting information.
Fourteen DNA samples of pipe scales and eight DNA samples of water samples in those three pipe loops under different phases were collected for bacterial community transformation analysis.
Water quality of historic source water and experimental water
Some main water quality parameters of historical supply water for collected pipes, tap water, and simulated water (manually adjusted high sulfate water as new water source) during different experimental phases are provided in Table 1.
As listed in Table 1, the sulfate and chloride contents of SW and tap water were similar, which were higher than that of GW3. GW3, which was treated groundwater had the higher alkalinity and lowest LR value; however, GW2 with highest alkalinity, sulfate, and chloride was different from other GW source. The LR values of SW sources were in the range of 0.48–0.76 and that of the tap water used in the experiment was in the range of 0.5–1.5. The simulated water for L1 and L2 had higher LR values ranging from 2.2 to 4.5 due to the addition of sulfate, while LR of simulated water for L4 were raised consecutively in three steps (from 1.16 to 1.40, and then to 3.43, respectively).
A brief introduction on water quality changes of L1–L4 were given for wholeness and better understanding of subsequent discussion. Before the simulated water was fed into pipe loops, tap water was first introduced to stabilize the newly installed pipe loops since the corrosion scales on pipe surface could be disturbed to some extent during harvest and transportation. Apart from the stagnation phase, pipes on loops 1–4 in chronological order were all undergone source water switching during tap water or simulated water phases compared with the sulfate and LR values of their old source water (Table 1).
Analytical methods and reagents
Water quality was measured according to standard methods (Clesceri et al. 1998). Both the monitored parameters and their analytical methods were summarized in Table S7. Sampling for water quality analysis was performed at a minimum of two times per week, while turbidity, disinfection residuals and total iron were monitored on a more frequent basis. Unless otherwise specified, all chemicals used in this study were analytical reagent (AR) grade.
Sampling and sample description
A total of 25 DNA samples were collected from corrosion scales with different morphologies on collection sites or under different experimental phases in three pipe loops. As soon as the pipe section was obtained, scale samples were removed with a sterile spatula, pulverized in an anaerobic glove box and then vacuum freeze-dried. Eight water samples collected within different experimental phases were immediately filtered with 0.22-μm Millipore GSWP filters and then stored at – 80 °C, pending DNA extraction. Both scale samples and water samples were collected twice for L1 and L2, at the end of phase II (under simulated water phase) and phase III (under simulated water stagnation phase), respectively. For L4, scale samples were collected four times: during phase I and phase III (under tap water phase), phase IV (under simulated water phase), and at the end of phase V (under simulated water stagnation phase); while water samples were collected under phase I, phase II, phase III, and at the end of phase V (Tables 1 and 2, all the above Roman numeral symbols were in accordance with the previous articles). The identification (ID) of each DNA sample and its corresponding serial number, water source type (for site collection samples), or experimental phase (for pipe loop samples) and associated water quality are presented in Table 2.
DNA extraction from water and pipe scale samples
DNA was extracted from 0.5 g fresh-scale powder after anaerobic-glove-box-milled and vacuum freeze-dried or Millipore GSWP filters preserving water suspension using the FASTDNA SPIN Kit for soil (MP Biomedicals, Solon, OH) according to the manufacturer’s instructions. The concentration of extracted nucleic acids was determined photometrically using Nanodrop ND-1000 UV-Vis spectrophotometer (Thermo Scientific, Wilmington, DE, USA). All DNA samples were stored at – 20 °C until further processing.
Pyrosequencing
The 16S rRNA gene of DNA was amplified by PCR using barcoded primers 8F and 533R containing the A and B sequencing adaptors (454 Life Sciences) (Bai et al. 2012). The fusion PrimerA-8F was 5′-cgtatcgcctccctcgcgccatcagAGAGTTTGATCCTGGCTCAG-3′ where the sequence of the A adaptor is shown in lowercase letters. The reverse fusion Primer B-533R was 5′-ctatgcgccttgccagcccgctcagTTACCGCGGCTGCTGGCAC-3′ where the sequence of the B adaptor is shown in lowercase letters. The ten base pair barcodes unique to each sample were embedded in the Primer B-533R set.
The PCR amplification was performed in ABI GeneAmp9700 thermocycler (ABI, Foster City, USA) using the program 95 °C for 2 min; 25 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s; 72 °C for 5 min; and finally kept at 10 °C. Triplicate positive PCR products were pooled and purified with AxyPrep DNA Gel Extraction Kit (Axygen, USA). Prior to sequencing, the amplicons from each reaction mixture were mixed in equal amounts based on concentration and subjected to emulsion PCR, and amplicon libraries were generated as recommended by 454 Life Sciences. Sequencing was performed from the primer B end using the 454/Roche B sequencing primer kit using a Roche Genome Sequencer GS-FLX according to the protocol.
Analysis of pyrosequencing data
Pyrosequencing flowgrams were converted to sequence reads using MOTHUR software (Schloss et al. 2009). Sequence reads were initially filtered and de-noised for removing low quality or ambiguous reads (Bai et al. 2012). Then the treated sequences were subjected to systematic checks to remove replicates, duplicates, barcodes, primer sequences, and low-quality reads. Briefly, high-quality sequences > 150 bp in length, quality score > 25, exact match to barcode and primer, and containing no characters were remained with an average length of 475 bp. Then the remaining high-quality sequences were compared with those in the non-redundant small-subunit (SSU) SILVA111 reference database by kmer searching and the putative chimeric sequences were excluded for further analysis (http://www.arb-silva.de/). Sequences that passed quality control and chimeras’ sequences removal were clustered into OTUs using MOTHUR program, with 97% sequence identity threshold. The species richness estimators (abundance-based coverage estimator (ACE) and Chao1), the Shannon index (SHIN), as well as Simpson’s diversity index (SIM) were generated in MOTHUR for each sample. Shannon-Wiener curves were performed on R (http://www.r-project.org/). Sequences were phylogenetically assigned to taxonomic classifications using an RDP Classifier with a confidence threshold of 80%. After phylogenetic allocation of the sequences down to the phylum, class, and genus level, relative abundance of a given phylogenetic group was set as the number of sequences affiliated with that group divided by the total number of sequences per sample.
Statistical analysis
In order to reveal the relationship between the bacterial composition, and samples’ water environment, their species matrixes were tested under a detrended correspondence analysis (DCA) model using CANOCO 4.5.1 (Biometris-Plant Research International, Wageningen, The Netherlands). After preliminary trials, a linear model (redundancy analysis (RDA)) or unimodal model (canonical correspondence analysis (CCA)) was constructed for L1, L2, or L4 samples according to their DCA analysis. Hierarchical cluster of biofilm samples with different bacterial community were performed by using the SPSS 16.0 (SPSS Inc., USA) for sample classification and comparison.
Results
Water quality variations during source water switch experiments
The detailed discussion about iron releases, turbidity changes, the scale chemical characteristics varieties, and their inner-relationship under high sulfate water switch were already published (Yang et al. 2014); the bacteria community transformation and potential pathogenic bacteria risk in red water occurrence resulted from high sulfate water switch was the main focus in this work. Therefore, the iron release was briefly explained here.
The total iron release of L1 and L2 effluents (Tables S4 and S5) during the source water switch phases were all below 0.3 mg L−1 (the upper limit of drinking water standard for total iron in China) and therefore no red water happened in L1 or L2. According to the previous studies, scales of pipes with SW supply history possessed much higher adaptability to high sulfate concentration or other water quality changes due to the larger proportion of stable constituents (mainly Fe3O4) in their shell layer (Yang et al. 2014).
However, for L4, most total iron release levels during source water switch phases were above 0.3 mg L−1and even reached a much higher level (0.33–1.76 mg L−1) owning to the substantial LR elevation (Table S6). The average turbidity values of L4 effluent were from 2.3 to 21.9 NTU. The rather thin and uniform non-layered corrosion scales on groundwater history pipes contained larger proportion of less stable iron oxides (e.g., β-FeOOH, FeCO3, and green rust) and had weaker adaptability to high sulfate water, thus the effluents of L4 always exhibited yellowish color under high sulfate water condition. This “red water” occurred in simulated drinking water system provided opportunity for investigating the bacterial community of both aqueous phase and pipe scales under high sulfate water switch.
Bacterial diversity and richness
A total of 324,943 valid sequences and 23,906 OTUs at 97% similarity level from 25 scale samples were obtained through 454-pyrosequencing analysis (Table 3). For water samples, a total of 62,112 valid sequences and 4170 OTUs at 97% similarity level were tested. The Good’s coverage revealed that these libraries represented the majority of bacterial 16S rRNA sequences presented in each scale samples, with values ranging from 0.87 to 0.98. The richness (ACE and Chao1) and diversity (SHIN and SIM) indices were calculated at a 3% width, as shown in Table 3. The SHIN for all samples ranged from 2.43 to 5.21, the values of which were comparable with some derived from river water or even soil, indicating that the bacterial species diversities were high in DWDSs (Li et al. 2010). The bacterial diversity index (SHIN or SIM) of L1 or L2 scale samples under high sulfate water or stagnation phases only had a little higher level than those of their original Pipe-SW2 scales upon collection (about 4.8 (average value) vs. 4.47). But the variation trends of L4 scale samples were different. The diversity of L4 scale samples under tap water phase was higher than those of their original scales upon collection and their subsequent samples under high sulfate water phases. Water samples of L1, L2, and L4 had much lower bacterial richness and diversity values than those of their correspondent scale samples under any experimental phases except from L4 samples in high sulfate water stagnation period.
Taxonomic composition of scale and water samples
All sequences were classified from phylum to genus according to the program MOTHUR, and 38 different bacterial phyla were identified across all the 33 samples via SILVA database. The bacterial community in scale samples of collection site pipes and experimental pipes on L1, L2, and L4 under experimental periods were analyzed and compared separately in Figs. 1 and 2. The most predominant phylum in the 25 scale samples was Proteobacteria, with high fraction ranging from 81.2 to 87.6% (pipes with SW supply history and L1–L2) and 69.1 to 96.6% (pipes with GW supply history and L4), respectively (Figs. 1a and 2 (A1, B1)).Other bacterial phyla with lower fraction for samples of pipes with different source water histories were Firmicutes with abundance of 8.6% (pipes with SW supply history) and 1.9% (pipes with GW supply history), respectively; Actinobacteria with abundance of 16.2% (pipes with GW supply history) (shown in Fig. 1a). For L1, L2, and L4 and their original collected pipes, Firmicutes and Actinobacteria were the abundant phyla inferior to Proteobacteria (Fig. 2 (A1, B1)).
Bacterial composition of the scale samples on pipes with different source water histories (a: in phylum level; b: in class level). Sequences that could not be classified into any known group and the other smaller phyla (or classes) in amount were assigned as “Others.” The relative abundance was the average values of the scale samples with different morphology on pipes with SW or GW supply history
Bacterial composition of the scale samples on pipe-SW2 and their related L1-L2 pipes (A) and PipeC-GW3 and their related L4 pipes (B) (A1, B1: in phylum level; A2, B2: in class level) under water quality changing experiment. The relative abundance transformation of above six kinds of potential corrosive microorganisms in biofilm of scale samples on Pipe-SW2 and L1-L2 (A3) and PipeC-GW3 and L4 (B3). Sequences that could not be classified into any known group and the other smaller phyla (classes) in amount were assigned as “Others.” The relative abundance was the average values of the scale samples with different morphology on pipes with SW or GW supply history or under water quality changing experiment
Based on the class taxonomy, in the scale samples with GW supply history, the relatively abundances of Gammaproteobacteria (21.5%) were lower than those in the scale samples with SW supply history (78.4%), but the relative abundances of Actinobacteria (15.6%) and Alphaproteobacteria (14.3%) of scale samples with GW supply history were higher than those in samples with SW supply history (1.0–5.4%). In addition, scale samples with GW supply history harbored higher fraction of Betaproteobacteria (37.3%), while scale samples with SW supply history had lower proportion (8.0%) (Fig. 1b). While L1, L2, and L4 experienced similar tap water or high sulfate water period, the bacterial classes’ transformation trends of those scale samples were distinct. The relative abundances of the major classes including Bacilli, Betaproteobacteria, and Gammaproteobacteria in samples of L1–L2 did not have much variations compared with their collection site samples. However, under high sulfate-simulated water, the class Alphaproteobacteria (decreasing from 17.9 to 1.7%), Gammaproteobacteria (decreasing from 7.6 to 2.8%), Betaproteobacteria (fluctuating from 36.3 to 87.3%), Clostridia (fluctuating from 0 to 19.8%), and Deltaproteobacteria (fluctuating from 0.1 to 8.2%) in scale samples of L4 had great content variety (Fig. 2 (A2, B2)).
Studies on the bacterial community in the scale samples of DWDS disclosed that bacterial metabolic activity influences the corrosion behavior of metals, biogeochemical redox cycling of metal oxides and thus the corrosion by-product release (Iwona and Christine 1999; Teng et al. 2008; White et al. 2011). Based on evaluation at the genus level, some typical corrosive microorganisms with considerable abundances were chosen and classified into iron-oxidizing bacteria (IOB), iron-reducing bacteria (IRB), sulfur-oxidizing bacteria (SOB), sulfate-reducing bacteria (SRB), nitrate-reducing bacteria (NRB), acid-producing bacteria (APB), etc. Corrosive microorganisms and their related class and phylum classification are summarized in Table 4.
Corrosive microorganisms with relatively high abundances were identified in both scale and water samples according to the previous published reports: IOB, anaerobic nitrate-dependent Fe(II)-oxidizing bacteria (classified into IOB by its function) (Straub et al. 1996), SOB, SRB, NRB, and APB (Iwona and Christine 1999; Emde et al. 1992; Weber et al. 2006). A large number of sequences related to the genera Bacillus, Clostridium, Escherichia-Shigella, Pseudomonas, Geothrix, Thermincola, and Shewanella were identified in scale samples. Previous research reported the following species, Bacillus sp. Clostridium sp., Escherichia coli, Pseudomonas sp., Geothrix sp., Thermincola, and Shewanella sp. had the ability to reduce ferric iron to ferrous iron, known as IRB (Emde et al. 1992; Weber et al. 2006; Herrera and Videla 2009). But the identification to the species level may not be accurate enough with the relatively short reads of 454-pyrosequencing.
In Pipe-SW2, L1, and L2 scale samples, the dominant class Gammaproteobacteria mainly consisted of Escherichia-Shigella and Pseudomonas (IRB) and Serratia, the other class Bacilli mainly consisted of Bacillus (IRB) and Streptococcus (APB), and the minor community Betaproteobacteria mainly consisted of Aquabacterium (IOB). In addition, under source water switching their relative abundance did not change much and maintained at their original level (IRB, 50.3–57.3%; APB, 1.8–2.30%; IOB, 0.6%–1.46% shown in Fig. 2 (A3)). However, the corrosive genera and their relative abundances in PipeC-GW3 and L4 scale samples had undergone much variation when the sulfate elevated (Fig. 2 (B3)). The collection site scale samples of PipeC-GW3 harbored a high fraction of Alphaproteobacteria mainly consisted of Ochrobactrum (NRB), Betaproteobacteria mainly consisted of Delftia (NRB) (NRB, 54.5% for PipeC-GW3); a low fraction of IOB with a value of 0.6% (mainly consisted of Acidovorax, Aquabacterium, Bradyrhizobium). Under high sulfate-simulated water, the NRB abundances of scale samples on L4 fluctuated from 54.5 to 8.0% and the IOB abundances of scale samples on L4 varied from 0 to 6.2%, respectively. After source water switch, new IOB genera–Gallionella was identified in L4 scale samples. In some L4 scale samples, under tap or simulated water phases, there were a relatively high proportion of Desulfovibrio (1.4–22.1%), Desulfosporosinus (1.0–2.5%), and Sulfuricella (8.4%), which were negligible in L1 and L2 scale samples. Besides, scale samples of L4 had increased fraction of IRB including Bacillus, Escherichia-Shigella, Pseudomonas, Geothrix, and Therminco (increasing from 2.2 to 44.2%) but varied fraction of SRB (Desulfosporosinus, Desulfovibrio, and Desulfurivibrio). Under tap water period, L4 scale samples had increased fraction of APB including Massilia, Nocardioides, Propionivibrio, Propionibacterium, Streptococcaceae_Streptococcus, or Streptococcus (ranging from 0.7 to 10.8%).
For the eight water samples, the majority of the sequences were assigned to Proteobacteria, with a high fraction of 83.3–98.4% (L1–L2) and 78.2–94.7% (L4), respectively. Analysis of the phylum Proteobacteria at the class level showed that most water samples were dominated by Betaproteobacteria and Alphaproteobacteria (Fig. S2).
Relationship between bacterial community and water quality
At class level, RDA results of scale samples with different source water histories demonstrated that Betaproteobacteria, Actinobacteria, and Alphaproteobacteria were negatively related with pH, SO4 2−, and LR, but positively related with HCO3 −; however, Gamaproteobacteria and Bacilli positively related with SO4 2−, pH, and LR, but negatively related with HCO3 − (Fig. 3 (A1)). The above situation consistent with the community analysis previously stated that SW with higher SO4 2−, pH, and LR promoted scale sample with SW history hold the unique dominant bacterial group, and GW with higher HCO3 − promoted scale sample with GW history also hold the preferred dominant bacterial group.
Redundancy analysis (RDA) for bacterial communities at class level of scale samples on collection site pipes with different source water histories (A1); RDA for bacterial communities at class level of L1–L2 (A2) and L4 (B2) scale samples affected by main water quality parameters; RDA for bacterial communities at genus level of L1–L2 scale samples (A3) and canonical correspondence analysis (CCA) for bacterial communities at genus level of L4 (B3) scale samples affected by main water quality parameters
Two predominant genera—Escherichia-Shigella and Serratia in L1–L2 scale samples were ranged from 38.1 to 59.2% and 17.4 to 24.7%, respectively, which had no strong relationship with SO4 2−, pH, LR, and Fe (Fig. 3 (A2, A3)). RDA/CCA analysis (Fig. 3 (B2, B3)) showed that Alphaproteobacteria (for example, Ochrobactrum) and Betaproteobacteria (for example, Delftia) in L4 scale samples were negatively related with pH, but positively related with HCO3 −; besides the abundance of Gamaproteobacteria (for example, Pseudomonas, Escherichia-Shigella) or Bacilli in L4 scale samples were positively related with pH. Thus, it could be seen that major genera in L4 scale samples all had some relationships with the primary water quality parameters.
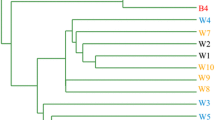
Hierarchical cluster analysis based on bacterial community
Hierarchical cluster analysis (Fig. 4) based on class composition in 11 scale samples of pipes with different water sources showed that the pipes could be divided into two categories, category I was composed of two groups: Pipe-SW1, Pipe-SW2, and Pipe-SW3 scale samples had more similar bacterial community than those of PipeC-GW3; category II was composed of PipeB-GW3 scale (Fig. 4a). Thus, the water quality discrepancy between SW and GW had obvious effects on the bacterial community of pipe scale samples with different water sources. The water samples of L1–L2 and the scale samples of Pipe-SW2 and L1-L2 were clearly divided into two groups for differences (Fig. 4b). However, some water samples of L4 and some scale samples of PipeC-GW3 and L4 were classified into the same groups for their similar bacterial community composition (Fig. 4c).
Hierarchical cluster analysis for bacterial community of scale samples on pipes with different source water histories (a), scale and water samples of L1–L2 (b), and L4 (c) under water quality changing experiment (the codes in the left of (a)–(c) were the serial numbers of the scale or water samples which can be referred in Table 2)
Potential bacterial pathogens
Several potential waterborne pathogenic genera (WHO 2008) were simultaneously identified from the scale and water samples by the pyrosequencing method. Evaluation at the genus level showed that members of the genera Burkholderia (0–59.3%), Escherichia-Shigella (0–59.2%), Mycobacterium (0–53.7%), Serratia (0–24.7%), and Ralstonia (0–10.1%) possessed a large amount of bacterial populations in scale samples with different water source history, together with minor populations from Sphingopyxis (0–4.2%), Flavobacterium (0–0.5%), and Sphingomonas (0–0.6%) (Table 5). Different from scale samples, only genera Burkholderia (0–7.2%), Ralstonia (0–1.1%), Sphingobium (0.2–4.2%), Novosphingobium (2.7–88.7%), and Sphingopyxis (0.1–0.9%) were detected of L1–L2 water samples and Novosphingobium (0.3–24.6%) and Sphingopyxis (0.2–11.2%) were identified in L4 water samples.
Figure 5 shows the cumulative occurrence profiles for eight potential bacterial pathogens of scale samples on pipes with different water source histories. Previous studies suggested that specific dominant groups in locations with high disinfectant concentrations are likely to be disinfection-resistant bacteria groups. Therefore, the Burkholderias existing in scale samples might be able to tolerate high levels of disinfectant (Shaw et al. 2015). The profiles of Escherichia-Shigella and Serratia across the entire range for pipes receiving SW were similar, and their concentrations were higher than those for pipes receiving GW. The concentrations of Mycobacteria for pipes receiving GW were much higher above the 40th percentile. Mycobacteria were generally tolerant to disinfectors due to the higher ability to form biofilms and to survive in nutrient-poor conditions and noted at the ends of the distribution lines, and pipe sediments or tuberculation accumulated (Gomez-Smith et al. 2015). The concentrations of Ralstonia for pipes receiving GW were higher above the 70th percentile. The family Sphingomonadaceae is ubiquitous bacteria frequently found in DWDS, and recognized opportunistic pathogens. They can survive in chlorinated waters due to the oligotrophic character of these bacteria and their production of biofilms (Vaz-Moreira et al. 2011). Novosphingobium, Sphingobium, and Sphingopyxis belonging to Sphingomonadaceae were detected in relatively high proportions in some postdisinfection water samples.
Discussion
Our previous investigations on red water events in Beijing found that the pipe material and supply-water quality determine the characteristics of the pipe scales and that different corrosion scales have different adaptability to high sulfate source water switch. Results showed that iron release increased markedly and even “red water” occurred for pipes historically transporting groundwater or blended water (alternant supply by groundwater and surface water) when feed water sulfate elevated abruptly. Meanwhile, the iron release of pipes historically transporting surface water just changed slightly without noticeable color appearing and even the feed water sulfate was elevated in multiple times. The pipes of surface water history with thick-layered corrosion scale (or densely distributed tubercles) possessed much higher stability due to the larger proportion of stable constituents (mainly Fe3O4) in its shell layer of corrosion scales; instead, the rather thin and uniform non-layered corrosion scales on groundwater history pipes contained larger proportion of less stable iron oxides (e.g., β-FeOOH, FeCO3, and green rust) (Yang et al. 2012, 2014). This work was the continuation of the above study and focused on the bacterial community alteration and the potential microbial-related risk under high-sulfate source water switch with red water occurrence.
The community richness indicator CHAO (or ACE) values of L1, L2, and L4 scale or water samples after high sulfate water phases were all increased to different extents compared with their original pipe scale upon collection. For the unstable L4 loop with pipes possessing thin and uniform non-layered corrosion scales, source water switch had great influence on the bacterial richness and diversity of their water samples and scale samples, but these impacts gradually attenuated after they had reached peaks. The bacterial richness and diversity changing trends of pipe loop scale samples and water samples were consistent with the variation trends of bacterial community structure discussed afterwards.
Class taxonomy results together with RDA/CCA and hierarchical cluster analysis all indicated that the water quality parameters can shape biofilm formation, taxonomy composition, and their amounts, and hence managing water quality is critical for the best operation of DWDS. Scale samples (SAs1-SAs5, SBs1, SCs1) with SW supply history, which is denoted by its higher sulfate and LR had two predominant groups: Gamaproteobacteria and Bacilli; while samples (GAs1, GBs1–GBs3) on pipes with GW supply history, which is denoted by its lower sulfate and higher HCO3 − had three predominant groups: Betaproteobacteria, Alphaproteobacteria, and Actinobacteria. The dominant classes or genera fraction for pipes with SW or GW supply history were different due to their old water sources, and their different transformation trends for L1, L2, and L4 under high sulfate-simulated water might be also attributed to their original scale structure and bacterial composition. The thin-unstable scales on pipes with GW history or thick-stable scales on pipes with SW history all experienced high sulfate water switching, but the iron release of each type scale was very different as discussed in “Water quality variations during source water switch experiments.” Because of the corrosion inhibition effect of abundant existing IRB and thick-stable scales of L1–L2 pipes, bacterial community in scale and water samples of L1–L2 under source water switch were all maintained stable (Yang et al. 2014). The SRB corrosive effect and nitrification may promote corrosion (Iwona and Christine 1999; White et al. 2011); whereas, SOB and IRB could inhibit the SRB-induced corrosion, therefore, the synergistic effect of SOB, SRB, IOB, IRB in L4 scales promoted iron release, thus red water happened. The pervasion of high sulfate through thin-unstable corrosion scale of L4 greatly influenced the bacteria community structure of L4 scale samples. Under high sulfate water, the unstable scales of L4 may release some bacteria to aqueous phase so that some water samples of L4 had similar bacterial community composition with their scale samples which hierarchical cluster (Fig. 4) results demonstrated.
Some potential pathogenic genera were identified at some locations along the DWDSs and in pipe loops under source water switch experiment. Burkholderia, Escherichia-Shigella, Mycobacterium, Serratia, Ralstonia, Novosphingobium, Flavobacterium, Sphingomonas, and Sphingopyxis were all identified in various samples throughout the systems. Some Burkholderia sequences were closely related to Burkholderia sp. and Burkholderia fungorum. Burkholderia fungorum belonging to the Burkholderia cepacia complex had been reported to cause bacteria and invasive infection; besides, Burkholderia strains had been found to possess higher iron weathering efficacy in soil of Scleroderma citrinum ectomycorrhizosphere (Cohn et al. 1999). Besides, Burkholderias taxa were demonstrated to be disinfection-resistant bacteria groups and also contain pathogenic species (Shaw et al. 2015). Escherichia-Shigella sequences were shown to have a high similarity to Escherichia coli, which was an indicator of fecal contamination from warm-blooded animals (Cohn et al. 1999). Escherichia coli had been used as the biological indicator of water treatment safety as their presence indicates an undesirable contamination of water systems due to treatment deficiencies or lack of water system integrity, or a possible fecal contamination originated from the source water (Lee et al. 2010). Genera of Flavobacterium and Serratia were also identified as potential opportunistic bacterial pathogens (Cohn et al. 1999; Geldreich and Lechevallier 1999). The genus Flavobacterium was closely related to Flavobacterium sp. and Serratia sequences were shown high similarity to Serratia sp. and Serratia symbiotica. Mycobacterium had been frequently found in DWDSs and members of them were reported as potential opportunistic pathogens which can often cause severe diseases in newborns, the elderly and other individuals with weakened immune systems. Many of Mycobacterium sequences were closely related to Mycobacterium chelonae, Mycobacterium frederiksbergense, Mycobacterium gadium, Mycobacterium gordonae, and Mycobacterium neoaurum. Mycobacterium chelonae and Mycobacterium gordonae may cause nosocomial pulmonary and systemic infections in the elderly and the immunosuppressed. Researchers also reported cutaneous infections caused by Mycobacterium frederiksbergense and bloodstream infection caused by Mycobacterium neoaurum (Gomez-Smith et al. 2015; Geldreich and Lechevallier 1999). Although Mycobacterium species and Burkholderia species’ pathogenicity requires further research and their resistance to chlorine and chloraimine remains unclear, the potential pathogenic Mycobacteria and Burkholderia in DWDSs should still arouse more authorities’ attention. Most of the Ralstonia sequences were closely related to Ralstonia pickettii, which had gained substantial interests as a nosocomial infections agent in water, water system components, distilled facilities, and potable water dispenser in international space station (Lee et al. 2010). The genera Novosphingobium, Sphingomonas, and Sphingopyxis which belonged to the family of Sphingomonadaceae and the class of Alphaproteobacteria were recognized as potential opportunistic pathogens and demonstrated to be antibiotic-resistant bacteria which may pose a health threat if these strains are opportunistic pathogens or serve as donors of the resistant factor to other bacteria that could be pathogens (Vaz-Moreira et al. 2011). In scale samples, there were a relatively low proportion of Sphingopyxis (0–4.2%) and Sphingomonas (0–0.6%), however Sphingobium (0.1–4.2%), Novosphingobium (0.3–88.7%), and Sphingopyxis (0–11.2%) were identified in large proportions in L1, L2 and L4 water samples. The fact that Sphingomonadaceae are recognized opportunistic pathogens, antibiotic resistance reservoirs, and chlorination-resistant bacteria in DWDS makes such ubiquity potentially hazardous to public health. Despite the presence of these genera, it must be stressed that due to the short reads of 454-pyrosequencing we could not precisely confirm the presence of potential pathogenic strains and also provide information on the viability or infectivity of the taxa detected. It can be anticipated that some of these populations could serve as inoculum and proliferate in DWDS if suitable growth conditions are provided.
Although the multifaceted approaches that water utilities employ to control microbial re-growth within DWDS, the systems still harbor substantial quantities of viable bacteria especially when red water cases occurred under source water switch. Furthermore, corrosion scales in drinking water system can serve as an environmental reservoir for pathogenic microorganism and represent a potential source of water contamination, resulting in a potential health risk for humans. The corrosive bacteria, disinfectant-resistant bacteria, potential pathogens and other organisms detrimental to DWDS found in water samples and scale samples in this study should arouse authorities’ attention for healthy, high-quality DWDS. In the future, pyrosequencing analysis should be associated with other techniques (real-time quantitative PCR, metabolic pathways, taxonomic assignments of functional genes, quorum-sensing technique and so on) to foster a more detailed community profile, diagnose problems within DWDS, and design effective treatment strategies for safeguarding human health. To minimize the occurrence of waterborne disease resulted from drinking water system, effective flushing programs, elimination of static water areas, targeted water quality monitoring program, corrosion control, and maintenance of an effective disinfectant residual must be employed throughout the distribution system.
Conclusions
Investigation on the transformation characteristics of bacterial community in DWDS under high-sulfate source water switching is important for drinking water safety and protection from waterborne infectious diseases. The main findings of this study include:
The richness estimators (ACE and Chao1), diversity indicators SHIN, and SIM values of all samples demonstrated that changing to high sulfate water as well as stagnation increased the bacterial richness and diversity of pipe loop water samples and scale samples. The most predominant phylum in the 25 scale samples and 8 water samples was Proteobacteria in the range of 69.1 to 96.6% and 78.2–98.4%, respectively. The most prominent population in scale samples on pipes with GW supply history were Actinobacteria (1.6–43.8%), Alphaproteobacteria (10.1–26.6%), Betaproteobacteria (15.3–70.0%), and Gammaproteobacteria (4.8–10.3%), but the corresponding class populations in scale samples on pipes with SW supply history were 1.0–1.8%, 0.7%–1.3%, 6.5–10.4%, and 72.%–81.1%, respectively. The RDA/CCA analysis and hierarchical cluster analysis for bacterial community of scale samples on pipes with different source water histories were consistent with the above results that bacterial community diversity was dependent on their original water sources and scale physicochemical property. Through the background survey database about the bacterial community of corrosion scales collected from the real DWDS and pipe loops under source water switch experiment, water utilities can regulate the key water parameters for pipe with particular physicochemical property scales in order to keep the corrosion scale stable under water quality changes.
The dominant corrosive group on scales of pipes with SW supply history was IRB, while that on scales of pipes with GW supply history was NRB. Under source water switching, bacterial community and their abundances for L1 and L2 scale sample changed little, whereas IOB, IRB, SOB, SRB, and NRB abundances in L4 scale samples had great fluctuation. In all, the pervasion of high sulfate through thin-unstable corrosion scale of L4 greatly influenced the bacteria community structure of L4 scale samples; while the corrosion inhibition effect of abundant existing IRB and thick-stable scales of L1 and L2, bacterial community in scales, and water samples of L1 and L2 under source water switch were all maintained stable. Hierarchical cluster analysis also indicated that the thin-unstable scale on pipes with GW supply history released some bacteria into water phase, thus their bacterial community could be classified into similar groups. Due to the high sulfate source water switch, pipes with GW supply history had the release of cells or even (opportunistic) pathogenic microorganisms, which were associated with severe iron corrosion scale release.
The potential opportunistic pathogens: Burkholderia, Escherichia-Shigella, Mycobacterium, Serratia, Ralstonia, Novosphingobium, Flavobacterium, Sphingomonas, and Sphingopyxis were observed in scale or water samples. The abundance of Mycobacterium tolerating to disinfectors was higher in groundwater scale samples. Preventing the potential physiochemical and microbiological de-stabilization of DWDS, re-conditioning the treated water (such as applying corrosion inhibitors, enhancing disinfection measures, and so on), pre-cleaning of the pipe, monitoring water quality parameters, and other strategies should be carefully evaluated by simulated source water switch experiment, and then developing appropriate plans for safe source-water switching.
References
Bai YH, Shi Q, Wen DH, Li ZX, Jefferson WA, Feng CP, Tang XY (2012) Bacterial cmmunities in the sediments of Dianchi lake, a partitioned eutrophic waterbody in China. PLoS One 7:e37796
Chaves Simões L, Simões M (2013) Biofilms in drinking water: problems and solutions. RSC Adv 3(8):2520–2533
Clesceri LS, Greenberg AE, Eaton AD (1998) Standard method for the examination of water and wastewater. American Public Health Association, Washington, DC
Cohn PD, Cox MMPH, Berger PS (1999) Health and aesthetic aspects of water quality. In: AWWA, Edzwald JK (eds) Water quality and treatment: a handbook of community water supplies. McGraw-Hill Companies, New York
Douterelo I, Husband S, Loza V, Boxall J (2016a) Dynamics of biofilm re-growth in drinking water distribution systems. Appl Environ Microbiol 82:4155–4168
Douterelo I, Jackson M, Solomon C, Boxall J (2016b) Microbial analysis of in situ biofilm formation in drinking water distribution systems: implications for monitoring and control of drinking water quality. Appl Microbiol Biot 100:3301–3311
Edwards M, Jacobs S, Taylor RJ (2000) The blue water phenomenon. Am Water Works Assoc J 92(7):72
Emde KME, Smith DW, Facey R (1992) Initial investigation of microbially influenced corrosion (MIC) in a low temperature water distribution system. Water Res 26:169–175
Flemming HC, Wingender J (2010) The biofilm matrix. Nat Rev Microbiol 8(9):623–633
Geldreich EE, LeChevallier M (1999) Microbiological quality control in distribution systems. In: AWWA and Edzwald JK (ed) Water quality and treatment: a handbook of community water supplies, 5th edn. McGraw-Hill Companies, New York, pp 1–49
Gomez-Smith CK, Lapara TM, Hozalski RM (2015) Sulfate reducing bacteria and mycobacteria dominate the biofilm communities in a chloraminated drinking water distribution system. Environ Sci Technol 49:8432–8440
Herrera LK, Videla HA (2009) Role of iron-reducing bacteria in corrosion and protection of carbon steel. Int Biodeterior Biodegrad 63:891–895
Hwang C, Ling F, Andersen GL, LeChevallier MW, Liu WT (2012) Microbial community dynamics of an urban drinking water distribution system subjected to phases of chloramination and chlorination treatments. Appl Environ Microbiol 78:7856–7865
Iwona BB, Christine CG (1999) Recent advances in the study of biocorrosion-an overview. Rev Microbiol 30:177–190
Lee J, Lee CS, Hugunin KM, Maute CJ, Dysko RC (2010) Bacteria from drinking water supply and their fate in gastrointestinal tracts of germ-free mice: a phylogenetic comparison study. Water Res 44:5050–5058
Li D, Li Z, JW Y, Cao N, Liu RY, Yang M (2010) Characterization of bacterial community structure in a DWDS during an occurrence of red water. Appl Environ Microbiol 76:7171–7180
Li XX, Wang HB, XX H, Hu C, Liao LF (2016) Characteristics of corrosion sales and biofilm in aged pipe distribution systems with switching water source. Eng Fail Anal 60:166–175
Ling F, Hwang C, Lechevallier MW, Andersen GL, Liu WT (2016) Core-satellite populations and seasonality of water meter biofilms in a metropolitan drinking water distribution system. The ISME Journal 10:582–595
Liu G, Bakker G, Li S, Vreeburg J, Verberk J, Medema G, Liu W, Van Dijk J (2014) Pyrosequencing reveals bacterial communities in unchlorinated drinking water distribution system: an integral study of bulk water, suspended solids, loose deposits, and pipe wall biofilm. Environ Sci Technol 48(10):5467–5476
Liu G, Zhang Y, Knibbe WJ, Feng CJ, Liu WS, Medema G, Van der Meer W (2017) Potential impacts of changing supply-water quality on drinking water distribution: a review. Water Res 116:135–148
Lytle DA, Sorg TJ, Frietch C (2004) Accumulation of arsenic in drinking water distribution systems. Environ Sci Technol 38(20):5365–5372
McNeill LS, Edwards M (2001) Iron pipe corrosion in distribution systems. J Am Water Works Assoc 93(7):88–100
Pieper KJ, Tang M, Edwards MA (2017) Flint water crisis caused by interrupted corrosion control: investigating “ground zero” home. Environ Sci Technol 51(4):2007–2014
Rajasekar A, Wang X, Sethuraman M, Parthipan P, Elumalai P (2017) Airborne bacteria associated with corrosion of mild steel 1010 and aluminum alloy 1100. Environ Sci Pollut R 24:8120–8136
Reynolds KA, Mena KD, Gerba CP (2008) Risk of waterborne illness via drinking water in the United States. In: Whitacre DW (ed) Reviews of environmental contamination and toxicology. Springer, New York, pp 117–158
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Schwake DO, Garner E, Strom OR, Pruden A, Edwards MA (2016) Legionella DNA markers in tap water coincident with a spike in legionnaires’ disease in Flint, MI. Environ Sci Technol Lett 3(9):311–315
Shaw JL, Monis P, Weyrich LS, Sawade E, Drikas M, Cooper AJ (2015) Using amplicon sequencing to characterize and monitor bacterial diversity in drinking water distribution systems. Appl Environ Microbiol 81:6463–6473
Straub KL, Benz M, Schink B, Widdel F (1996) Anaerobic, nitrate-dependent microbial oxidation of ferrous iron. Appl Environ Microbiol 62:1458–1460
Sun HF, Shi BY, Yang F, Wang DS (2017) Effects of sulfate on heavy metal release from iron corrosion scales in drinking water distribution system. Water Res 114:69–77
Tang Z, Hong S, Xiao W, Taylor J (2006) Characteristics of iron corrosion scales established under blending of ground, surface, and saline waters and their impacts on iron release in the pipe distribution system. Corros Sci 48(2):322–342
Teng F, Guan YT, Zhu WP (2008) Effect of biofilm on cast iron pipe corrosion in DWDS: corrosion scales characterization and microbial community structure investigation. Corros Sci 50:2816–2823
Vaz-Moreira I, Nunes OC, Manaia CM (2011) Diversity and antibiotic resistance patterns of Sphingomonadaceae isolates from drinking water. Appl Environ Microbiol 77:5697–5706
Wang Y, Zhang X, Chen C, Pan A, Xu Y, Liao P, Zhang S, Gu J (2009) Case study of red water phenomenon in drinking water distribution systems caused by water source switch. Environ Sci Chin 30(12):3555–3561
Weber KA, Achenbach LA, Coates JD (2006) Microorganisms pumping iron: anaerobic microbial iron oxidation and reduction. Nat Rev Microbiol 4:752–764
White C, Tancos M, Lytle DA (2011) Microbial community profile of a lead service line removed from a DWDS. Appl Environ Microbiol 77:5557–5561
WHO (2008) Microbial aspects. In: Guidelines for Drinking-water Quality, 3rd edn. World Health Organization, Geneva, p 121–144
Yang F, Shi BY, JN G, Wang DS, Yang M (2012) Morphology and composition characteristics of iron corrosion scales formed under different water source histories in a drinking water distribution system. Water Res 46:5423–5433
Yang F, Shi BY, Bai YH, Sun HF, Lytle DA, Wang DS (2014) Effect of sulfate on the transformation of corrosion scale composition and bacterial community in cast iron water distribution pipes. Water Res 59:46–57
Acknowledgements
We would like to recognize and thank the Beijing Waterworks Group for assistance in sampling and providing water quality monitoring data.
Funding information
This work was financially supported by the National Natural Science Foundation of China (Nos. 51308392, 51378493). Also, the project was funded by the Research Fund of Tianjin Key Laboratory of Aquatic Science and Technology. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Responsible editor: Diane Purchase
Electronic supplementary material
ESM 1
(DOCX 77 kb)
Rights and permissions
About this article
Cite this article
Yang, F., Shi, B., Zhang, W. et al. Pyrosequencing analysis of source water switch and sulfate-induced bacterial community transformation in simulated drinking water distribution pipes. Environ Sci Pollut Res 24, 28220–28238 (2017). https://doi.org/10.1007/s11356-017-0370-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-017-0370-y