Abstract
The rapid development of antibiotic-resistant bacteria (ARB) has been of concern worldwide. In this study, antibiotic resistance genes (ARGs) were investigated in antibiotic-resistant Escherichia coli isolated from surface water samples (rivers, n = 17; Taihu Lake, n = 16) and from human, chicken, swine, and Egretta garzetta sources in the Taihu Basin. E. coli showing resistance to at least five drugs occurred in 31, 67, 58, 27, and 18 % of the isolates from surface water (n = 665), chicken (n = 27), swine (n = 29), human (n = 45), and E. garzetta (n = 15) sources, respectively. The mean multi-antibiotic resistance (MAR) index of surface water samples (0.44) was lower than that of chicken (0.64) and swine (0.57) sources but higher than that of human (0.30) and E. garzetta sources (0.15). Ten tetracycline, four sulfonamide, four quinolone, five β-lactamase, and two streptomycin resistance genes were detected in the corresponding antibiotic-resistant isolates. Most antibiotic-resistant E. coli harbored at least two similar functional ARGs. Int-I was detected in at least 57 % of MAR E. coli isolates. The results of multiple correspondence analysis and Spearman correlation analysis suggest that antibiotic-resistant E. coli in water samples were mainly originated from swine, chicken, and/or human sources. Most of the ARGs detected in E. garzetta sources were prevalent in other sources. These data indicated that human activities may have contributed to the spread of ARB in the aquatic environment.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Antibiotics have been widely used in medical settings and animal feed to treat infections and increase production capacity, respectively. Application of antibiotics can directly alter the microbiota by selecting antibiotic-resistant bacteria (Akiyama and Savin 2010). Many antibiotics can induce cell-wall-deficient bacteria and therefore enhance the horizontal gene transfer that contributes to the transmission of antibiotic resistance genes (ARGs) among bacteria via the genetic mobile elements (plasmids, transposons, and integrons) (Baquero et al. 2008; Woo et al. 2003). Residual antibiotics released from these environments not only result in chemical pollution but also threaten the health of human and animals by contributing to the development of antibiotic resistance in non-antibiotic-producing microorganisms (Cummings et al. 2011; Knapp et al. 2010; Martinez 2008). Thus, it is necessary to evaluate and track the development of antibiotic resistance among bacteria to reduce their deleterious effects.
E. coli commonly inhabit the intestines of warm-blooded animals. A number of E. coli strains are important pathogens that can be responsible for colibacillosis in poultry, and some of them can cause severe human diseases such as the hemolytic uremic syndrome (Momtaz et al. 2012; Zhao et al. 2009). Various antibiotic-resistance phenotypes and/or genotypes of E. coli have been found in rivers, lakes, beaches, sediments, soils, and wastewaters (Hu et al. 2008; Li et al. 2014; Ram et al. 2007; Zhang et al. 2009a). Previous studies have shown that surface waters facilitate the dissemination of antibiotic-resistant organisms, ARGs, and antibiotics among human and animal populations, and throughout the natural environment (Baquero et al. 2008; Hu et al. 2008; Tacão et al. 2014). Therefore, E. coli from nonpoint sources as well as humans, farm animals, wildlife, and pets can affect water quality of catchments or lakes (Ibekwe et al. 2011; Wang et al. 2013). However, the propagation mechanism of ARGs in E. coli and its impacts on the environment remain unclear, especially in a wide ecosystem (Sayah et al. 2005).
Taihu Lake, the third largest freshwater lake in China, has approximately 2250 km2 of surface area and 100 tributaries. The population density is over 1200 per square kilometer. Approximately six million pigs and/or chickens are raised each year in the Taihu Lake Basin. Although most wastewaters are treated following certain standards before being discharged into receiving water bodies, the discharge from wastewater treatment plants may be an important reservoir of antibiotic resistance bacteria (Zhang et al. 2009d). To date, most wastewaters from feedlots are not or inadequately treated in this region. Therefore, the aquatic ecosystems in this basin are affected. The multi-antibiotic-resistant bacteria in this area have been posing a serious environmental issue (Zhang et al. 2009b; Zhao et al. 2009). The phenotype and genotype characterization of antibiotic-resistant E. coli strains isolated from surface water, human, swine, chicken, and wildlife present in the Taihu Lake Basi can be particularly interesting for understanding the effect of anthropogenic activities on the emergence and prevalence of antibiotic resistance at a basin level.
Fecal contamination indicator E. coli was employed in this study to characterize the antibiotic resistance and ARGs in E. coli strains isolated from surface water and from human, feedlot, and wildlife sources, and to study the source of antibiotic-resistant E. coli in surface water and its potential impact on wildlife and/or human health in the Taihu Lake basin.
Materials and methods
Sample collection
Surface water samples were collected from Taihu Lake in December 2009 and May 2010; and from 17 tributary rivers in Taihu Lake basin in May 2010 (Supplementary information, Fig. S1 and Table S1). Water samples were obtained from the effluents of biological treated wastewater from chicken and swine feedlots and from wastewater treatment plant (three sites) (Supplementary information) to represent chicken, swine, and human sources, respectively (May, 2010). Fresh fecal samples were collected from three nests of the wild heron, Egretta garzetta, on an island in Taihu Lake in April–July 2010.
E. coli isolation and antibiotic susceptibility testing
Colony counts were performed by spreading 100 ml of each diluted sample of feces or water samples on MacConkey agar plates at 42 °C for 24 h. The colonies suspected to be E. coli were further purified and identified by standard methods for all samples (Zhang et al. 2009c). E. coli isolates were screened for susceptibility to a panel of nine antibiotics on Mueller-Hinton agar (Oxoid) by a disk diffusion method according to Clinical and Laboratory Standards Institute (CLSI) 2005 guidelines. Ampicillin (AM, 10 μg), oxytetracycline (OT, 30 μg), tetracycline (TET, 30 μg), gentamicin (GT, 10 μg), streptomycin (30 μg, SM), sulfamethoxazole/trimethoprim (ST, 23.75 μg/1.25 μg), cefoperazone (CF, 75 μg), levofloxacin (LE, 5 μg), and nalidixic acid (NA, 30 μg) disks were used in this study. E. coli ATCC25922 was used as a reference strain. The diameter of inhibition zones surrounding the antibiotic disks was interpreted according to the CLSI 2005 guidelines. The isolates were recorded if they were resistant to antibiotics. The total frequencies of antibiotic-resistant E. coli were estimated by calculating the percentages of the isolates that resisted to at least one antibiotic versus the total numbers of isolates from the sample. The MAR index of the samples were calculated based on the equation a/(b × c), where a is the total antibiotic resistance numbers of all isolates, b is the number of antibiotics, and c is the number of isolates in the same sample (Hu et al. 2008).
If E. coli isolates from the same sample showed the same antibiotic resistance phenotype, they were further characterized using repetitive sequence-based PCR (REP-PCR) with primers BOXA1R according to the method described by Hu et al. (2008). Isolates representing similar fingerprint (85 % cutoff values) were discarded and only one of them was recorded. After removing the duplicates, 15 isolates for each surface water sample were obtained, and 27, 29, 45, and 15 isolates of E. coli were respectively isolated from chicken, swine, human, and E. garzetta source, respectively (Supplementary information; Tables S2 and S3).
Detection of ARGs
Specific primers (Supplementary information; Table S4) were used to detect 39 ARGs and integrons I in antibiotic-resistant E. coli isolates was randomly selected from water samples (Supplementary information; Table S5) or in all antibiotic-resistant isolates from chicken, swine, human, and E. garzetta sources (Supplementary information; Table S6). The details for PCR assays are provided in Supplementary information.
The PCR products of representative ARGs detected in this study and conserved sequence of class I integrons (Levesque et al. 1995) were cloned into plasmid pGEM-T Easy vector (Promega Corporation, Madison, WI), and E. coli TOP10 was used for transformation. To check the correct insertion of the expected size, colony-direct PCR was performed using T7 primer (5′-TAATACGACTCACTATAGGG-3′) and SP6 primer (5′-GATTTAGGTGACACTATAG-3′). Plasmids were extracted for sequencing. Online similarity searching was performed using the Basic Local Alignment Search Tool (BLAST) at the National Center for Biotechnology Information Web site (NCBI, http://www.ncbi.nlm.nih.gov/).
Nucleotide sequence accession numbers and phylogenetic analysis of qnr (B)
DNA sequences of the seven qnr (B) and gene cassettes of integron Class (I) were submitted to GenBank under accession numbers from HE575214 to HE575220 and from FR875291 to FR875305, respectively. Nucleotide sequences of qnr (B) were aligned and a neighbor-joining tree was constructed using MEGA 5 (Tamura et al. 2011). The statistical significance of branching was evaluated by bootstrap analysis involving the construction of 1000 trees from resampled data. Predicted amino acids were also compared by using MEGA 5.
Statistical analyses
Chi square (χ 2) tests were performed for each antimicrobial agent to analyze the distribution of antimicrobial-resistant E. coli between the sample sites in Lake (eight groups) or river (seventeen groups) (Supplementary information; Table S2). The Spearman correlation analysis was performed to determine correlation between the ARGs detected in E. coli isolates from surface water and from chicken, swine, and human sources. Multiple correspondence analyses (MCA) were performed using XLSTAT-pro (version 7.5.2) to group the source (human, surface water, swine, and chicken) using seven ARGs (tet (A), tet (E), sul (I), sul (II), bla TEM, bla CTX-M, and Int-I). These ARGs occurred at a high frequency among the E. coli isolates (Tables S5 and S6). The sources of these isolates and ARGs were used as supplementary variables and active variables, respectively.
Results
Characterization of antibiotic-resistant E. coli from surface water sample
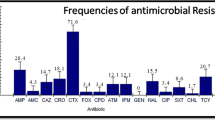
In total, 665 bacterial strains from 33 surface water samples (16 from Taihu Lake and 17 from rivers) were identified as E. coli and characterized for their resistance against nine antibiotics. About 49 % of these isolates showed resistance to at least four antibiotics (Fig. 1). TET, OTC, GT, AM, SM, LE, NA, TS, and CF resistance occurred in 42.71, 39.25, 43.76, 41.35, 30.53, 20.75, 44.81, 67.52, and 37.89 % of the 665 E. coli isolates from surface water samples (Table S3), respectively. MAR indexes of Taihu Lake samples ranged from 0.33 (Site c# and f#) to 0.49 (Site b#) in December 2009 and from 0.36 (Site d#) to 0.43 (Site g#) in May 2010 (Table S3). MAR index ranged from 0.33 (Site 7#) to 0.64 (Site 2#) in the river samples, and their mean value was 0.44. Two hundred twelve resistance phenotypes were detected in the isolates from surface water sample (Fig. 1).
In total, 27, 29, 45, and 15 strains of E. coli were respectively isolated from chicken, swine, human, and E. garzetta sources, and only the isolates showing resistance to at least one drug were further analyzed. For the same antibiotic, the resistance rate varied significantly among the four sample sources (except for TS). The mean MAR indexes of E. coli from chicken, swine, human, and E. garzetta sources (Table S4) were 0.64, 0.57, 0.30, and 0.15, respectively.
Characterization of ARGs in antibiotic-resistant E. coli
The presence of 39 ARGs were detected in randomly selected antibiotic-resistant E. coli strain from surface water (n = 164), swine (n = 28), human (n = 27), and E. garzetta (n = 7) sources and all the isolates from chicken (n = 27). Ten (seven efflux- and three ribosomal protection-related) of 21 tet genes were detected in the 75, 15, 20, and 18 TET-resistance isolates from surface water (n = 81), chicken (n = 17), swine (n = 21), and human (n = 20) sources, respectively (Table 1 and Fig. 2a). tet (A) was detected in the highest frequency and followed by tet (E), tet (B), tet (M), and tet (O). Combinations containing more than two tet genes were detected in 56, 12, 14, and 10 TET-resistant isolates from surface water, chicken, swine, and human sources, respectively (Table 1). Most of the tet patterns in strains from chicken, swine, and human sources occurred in the surface water isolates. All the TET-resistant strains were characterized by the presence of at least two tet genes, and tet (A + E), tet (A + B), and tet (A + D) were detected more frequently than other combinations. Combinations with three tet genes were detected in the isolates from surface waters (n = 11), chicken (n = 3), swine (n = 5), and humans (n = 4). The TET resistance genes tet (A) and tet (E) were detected in the isolates from E. garzetta sources.
Among the four sul genes targeted, sul (I) (ranged from 73 to 90 %) was the most prevalent genes in SFT-resistant isolates, followed by sul (A), sul (III), and sul (II) (Fig. 2b). Fifty-three, seven, two, and 12 isolates harboring at least two sul genes were detected from the surface water, chicken, swine, and human source. Combinations sul (A + I + II) were detected in surface, chicken, human, and E. garzetta source, while sul (I + II + III) and sul (A + I + III) were only detected in the surface water and human samples, respectively (Table 1).
Among the four quinolone resistance genes, qnr (B), qnr (S), and aac (6′)-Ib were detected in quinolone-resistant (LE and/or NA) strains from surface water, chicken, swine, and human sources (Fig. 2b). At least one quinolone resistance gene was detected in 63.5, 43.5, 60, 38.5, and 16.7 % of quinolone-resistant isolates from surface water (n = 96), chicken (n = 23), swine (n = 20), humans (n = 13), and E. garzetta (n = 6), respectively (Table 1). Among the sequenced 27 partial nucleic acid sequences of qnr (B), seven unique sequences were new qnr (B) variants and clustered into two groups (Fig. S2).
Five of the eight β-lactamase genes were detected in AM- and/or CF-resistant isolates from the surface water, chicken, swine, and human sources (Fig. 2c and Table 1). β-Lactamase genes like bla TEM, bla OXA, bla OXY, bla SHV, and bla CTX-M occurred respectively in 51.3, 14.9, 12.3, 8.7, and 17.4 % of AM- and/or CF-resistance isolates in the Taihu region. Two β-lactamase genes coexisted in isolates from chicken, swine, and human sources were found in isolates from surface water except that combination bla OXA + bla CTX-M was only detected in the chicken sources. Combination bla TEM + bla OXY + bla CTX-M was only detected in the surface water and swine sources. Str (A) and str (B) were detected in streptomycin-resistant E. coli isolates from all the sources (Fig. 2c and Table 1). Occurrence of str (A) (68 %) in streptomycin-resistant isolates was higher than that of str (B) (51 %). Combination str (A + B) was detected in 95, 93, 94, and 90 % of SM-resistant isolates from surface water, chicken, swine, and human sources, respectively.
Characterization of integron I
Class I integron was detected in 68, 85, 86, 67, and 57 % of MAR E. coli isolates from surface water (n = 164), chicken (n = 27), swine (n = 28), human (n = 27), and E. garzetta (N = 7) sources, respectively (Tables S5 and S6). Variable region of class I integrons was detected in 156 of 181 integron-positive isolates and grouped into four classes, largely based on the length of the amplified products. The most prevalent length of gene cassettes of class I integrons was at 1.6–1.8 kb, followed by 1.0–1.2 kb. It was noted that 60 of 156 isolates contained at least two different lengths of gene cassettes. Eight patterns of gene cassettes were detected in class I integrons (Table 2). Genes encoding dihydrofolate reductase (drfA) and adenylyltransferase (aadA) conferring resistance respectively to trimethoprim and spectinomycin/streptomycin were detected in the variable regions of class I integrons. Class I integrons from E. coli isolates mainly carried with 1.6–1.8 kb of variable regions, which contained gene combinations like dfrA17-aadA5 and dfrA14-aadA6/aadA10 (Table 2).
MCA and Spearman correlation analysis of ARGs in E. coli from various sources
MCA software was used to analyze the relationship between the surface water and animal source of E. coli based on the presentence or absence of ARGs. However, most of these ARGs could not represent strains from surface water or animal source samples because of low detection frequency. Therefore, six sets of ARGs and class I integrons were used. Results from MCA showed that total variance has no statistical interpretation within the framework of MCA and first two eigenvalues were selected to make the map of MCA (Fig. 3). River, lake, chicken, swine, and human sources were clustered into the same group. Furthermore, using the percentage values of 24 ARGs in their corresponding antibiotic-resistant isolates from these sources, the correlation coefficients (p < 0.01; n = 24) between surface water and swine, chicken, and human sources were 0.65, 0.83, and 0.77, respectively.
Discussion
Aquatic ecosystems are constantly subjected to anthropogenic activities such as pollution from urban, agricultural, and industrial sources, and therefore, they may provide an ideal setting for the acquisition and spread of antibiotic-resistant bacteria (Baquero et al. 2008; Sidrach-Cardona et al. 2014). The rates of drug resistance in bacterial strains from the surface water of Taihu Lake basin were somewhat lower than that from a lake in India (Abhirosh et al. 2011). In total, 212 antibiotic resistance phenotypes were detected in 650 E. coli strains in surface water samples. E. coli showing resistance to several antibiotics (such as sulfonamides, tetracycline, ampicillin, and streptomycin) have been detected in the surface water of other geographic regions in China and other countries (Abhirosh et al. 2011; Hu et al. 2008; Olaniran et al. 2009; Sayah et al. 2005). Results from χ 2 test and MAR indexes showed that the distribution of antibiotic-resistant E. coli varied more among samples from the river than other sources (Table S3). Taken together, these data demonstrated that abundant E. coli isolates were associated with different sources (such as humans and poultry sources) (Abhirosh et al. 2011; Martinez 2008; Olaniran et al. 2009).
The presence of ARBs (including pathogens) in aquatic ecosystems may be harmful not only to public health but also to wildlife, which do not expose to antibiotics directly. Mean MAR index and resistance rate of E. coli from E. garzetta were obviously lower than that from surface water, chicken, swine, and human sources (Table 1). Kozak et al. (2009) also found that E. coli isolates from wild small mammals living on farms have higher rates of resistance and are more frequently multi-resistant than E. coli isolates from non-farm environments. Recent reports showed that wild animals could acquire antimicrobial agent-resistant bacteria through contact with humans and domestic animals (Kozak et al. 2009; Zhang et al. 2009d). However, E. garzetta is a type of waterfowl and mainly eats fish, insects, amphibians, crustaceans, and reptiles, indicating that waterfowl may be exposed to E. coli from the natural environment since they were not exposed to antibiotics directly. Alali et al. (2008) found that the prevalent MAR E. coli isolates from human and swine sources showed no significant temporal trends although a high variability was observed among seasonal samples over the 3-year period.
Among the seven TET resistance genes, tet (A), tet (B), and tet (E) were detected at high frequencies, which is consistent with the findings of previous studies on TET-resistant E. coli isolates from other parts of China (Hu et al. 2008; Wang et al. 2013) and on isolates from human and animal sources (Soufi et al. 2009; Zhang et al. 2014). The tet (E) originally gene, isolated from E. coli, was detected in this study, but not in the isolates from Wenyu River basin (Hu et al. 2008). tet (J) found in the chromosome of Proteus mirabilis (Magalhaes et al. 1998) was detected in E. coli isolates from all the surface water, swine, and chicken sources (Table 1). Similar to our results, 90.6 % (77/85) of the E. coli isolates encoded two or more tet genes (Zhang et al. 2014).
Genes in the sulfonamide-resistance classes, such as sul (A), sul (I), sul (II), and sul (III), encoding dihydropteroate synthase confer resistance to SFT. The four sul class genes can explain about 91 % (136/150), 95 % (19/20), 100 % (21/21), and 96 % (21/22) of ST-resistant E. coli isolates from the surface water, chicken, swine, and human sources (Table 1). This is in agreement with previous reports (Hu et al. 2008; Wang et al. 2013). Among the four sul genes, sul (I) occurred at the highest frequency (Fig. 2b), while sul (II) dominated in the SFT-resistant E. coli isolates from Dongjiang River, South China (Su et al. 2012). Additionally, the frequency of sul (A) and sul (II) was lower in Taihu basin than in Jinsha Lake (Wang et al. 2013). Among the two-sul combination, sul (I + II) was detected at the highest frequency, and all the two-sul combinations from the surface water, chicken, swine, and human sources were also found in the other surface waters, indicating that the source of E. coli is a highly mixed watershed (Ibekwe et al. 2011). The frequencies of three-sul combinations were lower in these samples than the samples from Jinsha Lake, China (Wang et al. 2013) and from clinical isolates from patients exposed to ST (Koljalg et al. 2009).
Quinolone resistance is always associated with the accumulation of mutations in the fluoroquinolone resistance genes. These mutant-encoded proteins can block the action of ciprofloxacin and is associated with the protection of DNA gyrase and topoisomerase IV from quinolone inhibition (Manuel Rodriguez-Martinez et al. 2011; Robicsek et al. 2006). In agreement with the report about quinolone resistance genes in E. coli isolated from companion and food-producing animals (Ma et al. 2009), qnrB, qnrS, and aac (6′)-Ib were also detected in quinolone-resistant (LE and/or NA) isolates from the surface water, chicken, swine, and human sources except that qnr (A) was not detected (Fig. 2b). The transfer of STR resistance was closely related to multiple plasmids (Zhang et al. 2014). Only one quinolone resistance gene was detected in most of the quinolone-resistant isolates, which can be explained by the fact that qnr was always carried by different plasmids that cannot coexist in the same cell (Manuel Rodriguez-Martinez et al. 2011; Wang et al. 2013). Several new variants of qnr (B) were obtained (Fig. S2). qnr (B) is a plasmid-mediated quinolone resistance gene, and the number of qnr (B) variants is greater than for qnr (A) and qnr (S) (Manuel Rodriguez-Martinez et al. 2011). Reports from a previous study (Manuel Rodriguez-Martinez et al. 2011) have shown that mutations on quinolone resistance occurred sequentially rather than simultaneously, and the appearance rates of high-level quinolone resistance in bacteria exposed to low level of quinolone were higher than those exposed to therapeutic level of quinolone. Taken together, these data indicated that quinolone resistance could be ascribed to the mutations and the transmission of qnr among bacteria via movable elements plasmids (Manuel Rodriguez-Martinez et al. 2011).
Five β-lactamase genes were detected in randomly selected AM- and/or CF-resistant strains. TEM occurred at the highest frequency in AM- and/or CF-resistant isolates (Fig. 2c and Table 1), which is similar to reports of Henriques et al. (2006) and Wang et al. (2013). β-Lactamase is mainly the derivatives of TEM or SHV β-lactamases enzymes and confers resistance to ampicillin, penicillin, monobactams, and cephalosporins (Chen et al. 2010; Sharma et al. 2010; Wang and Schaffner 2011). Fourteen genotypes of AM and/or CF resistance were detected and nine of them consisted of at least two β-lactamase genes. The three β-lactamase gene combinations were detected in the isolates from the surface water and chicken sources, while combinations with three to five genes had been detected in E. coli isolates obtained from patients (Kiiru et al. 2012). In general, the frequency of strA was higher than that of strB in streptomycin-resistant isolates (Table 1). The frequencies of str (A + B) were lower in Taihu Lake basin than other parts of China (Wang and Schaffner 2011). The combination of str (A) and str (B) is probably involved in conferring high-level resistance to streptomycin (Sunde and Norstrom 2005). The transmission of str (A + B) was associated with many plasmids such as plasmid R2418S and RSF1010 (Ben-Mahrez and Sioud 2010).
At least 57–86 % of class I integrons were detected in antibiotic-resistant E. coli isolates from several sources (Tables S5 and S6), and the frequency of class I integrons was higher in MAR E. coli from the swine and chicken sources than those from human and E. garzetta sources. Recent reports showed that class I integrons was detected in 65.2 % of coliforms isolated from the Yangtze River basin in Chongqing, China (Chen et al. 2010). Integrase genes were detected in 58.7 % (101/172), 60.4 % (64/106), and 26.1 % (6/23) of E. coli isolates from swine, chickens, and farm workers, respectively (Zhang et al. 2009c). Similar to our results, dfrA17-aadA5 was found the most prevalent in antibiotic-resistant E. coli isolates from farm workers, swine, and chickens (Zhang et al. 2009c). In fact, class I integrons not only contain drfA and aadA but also carry many other ARGs like sul, aac (6′)-Ib, and chloramphenicol resistance gene, cmlA1 (Hussein et al. 2009; Ma et al. 2009). Reports from Chen et al. (2011) showed that class I integrons positive isolates had a significantly higher probability of resistance to several antibiotics than isolates with no class I integrons. Horizontal transfer of class I integrons has been reported between the E. coli isolates from farm workers and animals (Zhang et al. 2009c) and among the isolates from storm water and bovine feces (Nagachinta and Chen 2008). Consequently, the horizontal transfer of integrons might contribute to the wide dissemination of antimicrobial resistance in E. coli isolates from human and animal sources (Zhang et al. 2009c). The spread of integron elements may have contributed to the spread of resistance among bacteria since the ARGs could persist in the environments even in the absence of selection pressure (Tamminen et al. 2011).
Alien bacteria like E. coli possessing ARGs can be spread to surface waters not only via point sources (e.g., wastewater treatment plants and animal feeding operations) but also from nonpoint sources, including runoff from manure or bio-solids applied agricultural fields (Allen et al. 2011; Storteboom et al. 2010). Normally, it is difficult to track the contaminated source in a big basin, especially when data are limited and the resistance phenotypes and genotype of E. coli vary significantly among the surface water samples (Table S3). The MCA is a multivariate technique used to examine, describe, and summarize the geometric relations of qualitative variables (Kich et al. 2011). Though data about ARGs in antibiotic-resistant E. coli were limited, the results from MCA analysis indicated that the ARBs in surface water were mainly originated from humans, swine, and chickens. This was further supported by the fact that the percentage values of 24 ARGs in the corresponding antibiotic-resistant isolates from surface water were positively (p < 0.01) correlated with that from swine, chicken, and human sources except for E. garzetta sources. Moreover, most of the ARGs detected in E. coli isolates were prevalent in the isolates from surface water samples.
In conclusion, 212 antibiotic resistance phenotypes were detected in 650 isolates from the surface water samples of 17 tributary rivers and Taihu Lake, and the distribution of antibiotic-resistant E. coli had more variation among the samples from rivers, as compared to the lake. Mean MAR index and the frequency of antibiotic resistance in E. coli from E. garzetta sources were obviously lower than that from the surface water, chicken, swine, and human sources. Twenty-four ARGs and class I integrons could explain most of the corresponding antibiotic resistance of E. coli and indicated that both horizontal gene transfer and gene mutations contribute to the prevalence of ARGs among E. coli. The presence of E. coli in surface water samples of Taihu Lake could be ascribed to the rivers, which carried the E. coli from the human, swine, and chicken sources. The data on fecal contamination indicator E. coli indicates that the prevalence of ARBs in the surface water is potentially a public health threat and may negatively affect the wildlife in this region.
References
Abhirosh C, Sherin V, Thomas AP, Hatha AAM, Mazumder A (2011) Potential public health significance of faecal contamination and multidrug-resistant Escherichia coli and Salmonella serotypes in a lake in India. Public Health 125:377–379
Akiyama T, Savin MC (2010) Populations of antibiotic-resistant coliform bacteria change rapidly in a wastewater effluent dominated stream. Sci Total Environ 408:6192–6201
Alali WQ, Scott HM, Harvey RB, Norby B, Lawhorn DB, Pillai SD (2008) Longitudinal study of antimicrobial resistance among Escherichia coli isolates from integrated multisite cohorts of humans and swine. Appl Environ Microbiol 74:3672–3681
Allen SE, Boerlin P, Janecko N, Lumsden JS, Barker IK, Pearl DL, Reid-Smith RJ, Jardine C (2011) Antimicrobial resistance in generic Escherichia coli isolates from wild small mammals living in swine farm, residential, landfill, and natural environments in southern Ontario, Canada. Appl Environ Microbiol 77:882–888
Baquero F, Martinez JL, Canton R (2008) Antibiotics and antibiotic resistance in water environments. Curr Opin Biotechnol 19:260–265
Ben-Mahrez K, Sioud M (2010) Presence of STRA-STRB linked streptomycin-resistance genes in clinical isolate of Escherichia coli 2418. Arch Inst Pasteur Tunis 87:77–81
Chen H, Shu W, Chang X, J-a C, Guo Y, Tan Y (2010) The profile of antibiotics resistance and integrons of extended-spectrum beta-lactamase producing thermotolerant coliforms isolated from the Yangtze River basin in Chongqing. Environ Pollut 158:2459–2464
Chen B, Zheng W, Yu Y, Huang W, Zheng S, Zhang Y, Guan X, Zhuang Y, Chen N, Topp E (2011) Class 1 integrons, selected virulence genes, and antibiotic resistance in Escherichia coli isolates from the Minjiang River, Fujian Province, China. Appl Environ Microbiol 77:148–155
Cummings DE, Archer KF, Arriola DJ, Baker PA, Faucett KG, Laroya JB, Pfeil KL, Ryan CR, Ryan KRU, Zuill DE (2011) Broad dissemination of plasmid-mediated quinolone resistance genes in sediments of two urban coastal wetlands. Environ Sci Technol 45:447–454
Henriques IS, Fonseca F, Alves A, Saavedra MJ, Correia A (2006) Occurrence and diversity of integrons and beta-lactamase genes among ampicillin-resistant isolates from estuarine waters. Res Microbiol 157:938–947
Hu J, Shi J, Chang H, Li D, Yang M, Kamagata Y (2008) Phenotyping and genotyping of antibiotic-resistant Escherichia coli isolated from a natural river basin. Environ Sci Technol 42:3415–3420
Hussein AIA, Ahmed AM, Sato M, Shimamoto T (2009) Characterization of integrons and antimicrobial resistance genes in clinical isolates of Gram-negative bacteria from Palestinian hospitals. Microbiol Immunol 53:595–602
Ibekwe AM, Murinda SE, Graves AK (2011) Genetic diversity and antimicrobial resistance of Escherichia coli from human and animal sources uncovers multiple resistances from human sources. Plos One 6(6):e20819
Kich JD, Coldebella A, Mores N, Nogueira MG, Cardoso M, Fratamico PM, Call JE, Fedorka-Cray P, Luchansky JB (2011) Prevalence, distribution, and molecular characterization of Salmonella recovered from swine finishing herds and a slaughter facility in Santa Catarina, Brazil. Int J Food Microbiol 151:307–313
Kiiru J, Kariuki S, Goddeeris BM, Butaye P (2012) Analysis of beta-lactamase phenotypes and carriage of selected beta-lactamase genes among Escherichia coli strains obtained from Kenyan patients during an 18-year period. BMC Microbiol 12:155
Knapp CW, Dolfing J, Ehlert PAI, Graham DW (2010) Evidence of increasing antibiotic resistance gene abundances in archived soils since 1940. Environ Sci Technol 44:580–587
Koljalg S, Truusalu K, Vainumaee I, Stsepetova J, Sepp E, Mikelsaar M (2009) Persistence of Escherichia coli clones and phenotypic and denotypic antibiotic resistance in recurrent urinary tract infections in childhood. J Clin Microbiol 47:99–105
Kozak GK, Boerlin P, Janecko N, Reid-Smith RJ, Jardine C (2009) Antimicrobial resistance in Escherichia coli isolates from swine and wild small mammals in the proximity of swine farms and in natural environments in Ontario, Canada. Appl Environ Microbiol 75:559–566
Levesque C, Piche L, Larose C, Roy PH (1995) PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother 39:185–191
Li X, Watanabe N, Xiao C, Harter T, McCowan B, Liu Y, Atwill ER (2014) Antibiotic-resistant E. coli in surface water and groundwater in dairy operations in Northern California. Environ Monit Assess 186:1253–1260
Ma J, Zeng Z, Chen Z, Xu X, Wang X, Deng Y, Lue D, Huang L, Zhang Y, Liu J, Wang M (2009) High prevalence of plasmid-mediated quinolone resistance determinants qnr, aac (6′)-Ib-cr, and qepA among ceftiofur-resistant Enterobacteriaceae isolates from companion and food-producing animals. Antimicrob Agents Chemother 53:519–524
Magalhaes VD, Schuman W, Castilho BA (1998) A new tetracycline resistance determinant cloned from Proteus mirabilis. Biochim Biophys Acta Gene Struct Expr 1443:262–266
Martinez JL (2008) Antibiotics and antibiotic resistance genes in natural environments. Science 321:365–367
Momtaz H, Rahimi E, Moshkelani S (2012) Molecular detection of antimicrobial resistance genes in E. coli isolated from slaughtered commercial chickens in Iran. Vet Med 57:193–197
Nagachinta S, Chen J (2008) Transfer of class 1 integron-mediated antibiotic resistance genes from Shiga toxin-producing Escherichia coli to a susceptible E. coli K-12 strain in storm water and bovine feces. Appl Environ Microbiol 74:5063–5067
Olaniran AO, Naicker K, Pillay B (2009) Antibiotic resistance profiles of Escherichia coli isolates from river sources in Durban, South Africa. World J Microbiol Biotechnol 25:1743–1749
Ram S, Vajpayee P, Shanker R (2007) Prevalence of multi-antimicrobial-agent resistant, Shiga toxin and enterotoxin producing Escherichia coli in surface waters of river Ganga. Environ Sci Technol 41:7383–7388
Robicsek A, Strahilevitz J, Sahm DF, Jacoby GA, Hooper DC (2006) qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrob Agents Chemother 50:2872–2874
Rodriguez-Martinez JM, Eliecer Cano M, Velasco C, Martinez-Martinez L, Pascual A (2011) Plasmid-mediated quinolone resistance: an update. J Infect Chemother 17:149–182
Sayah RS, Kaneene JB, Johnson Y, Miller R (2005) Patterns of antimicrobial resistance observed in Escherichia coli isolates obtained from domestic- and wild-animal fecal samples, human septage, and surface water. Appl Environ Microbiol 71:1394–1404
Sharma J, Sharma M, Ray P (2010) Detection of TEM & SHV genes in Escherichia coli & Klebsiella pneumoniae isolates in a tertiary care hospital from India. Indian J Med Res 132:332–336
Sidrach-Cardona R, Hijosa-Valsero M, Marti E, Balcazar JL, Becares E (2014) Prevalence of antibiotic-resistant fecal bacteria in a river impacted by both an antibiotic production plant and urban treated discharges. Sci Total Environ 488:220–227
Soufi L, Abbassi MS, Saenz Y, Vinue L, Somalo S, Zarazaga M, Abbas A, Dbaya R, Khanfir L, Ben Hassen A, Hammami S, Torres C (2009) Prevalence and diversity of integrons and associated resistance genes in Escherichia coli isolates from poultry meat in Tunisia. Foodborne Pathog Dis 6:1067–1073
Storteboom H, Arabi M, Davis JG, Crimi B, Pruden A (2010) Tracking antibiotic resistance genes in the South Platte River basin using molecular signatures of urban, agricultural, and pristine sources. Environ Sci Technol 44:7397–7404
Su H-C, Ying G-G, Tao R, Zhang R-Q, Zhao J-L, Liu Y-S (2012) Class 1 and 2 integrons, sul resistance genes and antibiotic resistance in Escherichia coli isolated from Dongjiang River, South China. Environ Pollut 169:42–49
Sunde M, Norstrom M (2005) The genetic background for streptomycin resistance in Escherichia coli influences the distribution of MICs. J Antimicrob Chemother 56:87–90
Tacão M1, Moura A, Correia A, Henriques I (2014) Co-resistance to different classes of antibiotics among ESBL-producers from aquatic systems. Water Res 48:100–107
Tamminen M, Karkman A, Lohmus A, Muziasari WI, Takasu H, Wada S, Suzuki S, Virta M (2011) Tetracycline resistance genes persist at aquaculture farms in the absence of selection pressure. Environ Sci Technol 45:386–391
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Wang HH, Schaffner DW (2011) Antibiotic resistance: how much do we know and where do we go from here? Appl Environ Microbiol 77:7093–7095
Wang C, Gu XC, Zhang SH, Wang PF, Guo C, Gu J, Hou J (2013) Characterization of antibiotic-resistance genes in antibiotic resistance Escherichia coli isolates from a lake. Arch Environ Contam Toxicol 65:635–641
Woo PCY, To APC, Lau SKP, Yuen KY (2003) Facilitation of horizontal transfer of antimicrobial resistance by transformation of antibiotic-induced cell-wall-deficient bacteria. Med Hypotheses 61:503–508
Zhang T, Zhang M, Zhang X, Fang HH (2009a) Tetracycline resistance genes and tetracycline resistant lactose-fermenting Enterobacteriaceae in activated sludge of sewage treatment plants. Environ Sci Technol 43:3455–3460
Zhang X, Wu B, Zhang Y, Zhang T, Yang L, Fang HHP, Ford T, Cheng S (2009b) Class 1 integronase gene and tetracycline resistance genes tetA and tetC in different water environments of Jiangsu Province, China. Ecotoxicology 18:652–660
Zhang XY, Ding LJ, Yue J (2009c) Occurrence and characteristics of class 1 and class 2 integrons in resistant Escherichia coli isolates from animals and farm workers in northeastern China. Microb Drug Resist 15:323–328
Zhang YL, Marrs CF, Simon C, Xi CW (2009d) Wastewater treatment contributes to selective increase of antibiotic resistance among Acinetobacter spp. Sci Total Environ 407:3702–3706
Zhang W-J, Xu X-R, Schwarz S, Wang X-M, Dai L, Zheng H-J, Liu S (2014) Characterization of the IncA/C plasmid pSCEC2 from Escherichia coli of swine origin that harbours the multiresistance gene cfr. J Antimicrob Chemother 69:385–389
Zhao L, Chen X, Zhu X, Yang W, Dong L, Xu X, Gao S, Liu X (2009) Prevalence of virulence factors and antimicrobial resistance of uropathogenic Escherichia coli in Jiangsu province (China). Urology 74:702–707
Acknowledgments
This work was supported by the National Foundation for the Natural Science Foundation of JiangSu Province (BK2012413), Innovative Research Groups (NO. 51421006), National Natural Science Foundation of China (NO. 51379063), and for Distinguished Young Scholars (NO. 51225901) and Research Fund for innovation team of Ministry of Education (IRT13061).
Compliance with ethical standards
There are no potential conflicts of interest. All the data in this manuscript have not been published previously (partly or in full). All the authors agreed with this submission.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Responsible editor: Robert Duran
Rights and permissions
About this article
Cite this article
Zhang, S.H., Lv, X., Han, B. et al. Prevalence of antibiotic resistance genes in antibiotic-resistant Escherichia coli isolates in surface water of Taihu Lake Basin, China. Environ Sci Pollut Res 22, 11412–11421 (2015). https://doi.org/10.1007/s11356-015-4371-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-015-4371-4