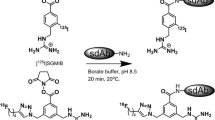
Abstract
Purpose
Molecular imaging of tumor HER2 expression may allow patient selection for HER2-targeted therapies. Our aim was to introduce hexahistidine (His6) peptides into pertuzumab Fab to enable labeling with the [99mTc(CO)3(H2O)3]+ complex and study these radioimmunoconjugates for microSPECT/CT imaging of HER2-positive tumor xenografts in mice.
Procedures
Fab were produced by papain digestion of pertuzumab and reacted with sulfo-SMCC for conjugation to His6-containing peptides (CGYGGHHHHHH). His6-peptide conjugation was measured by a radiometric assay. His6-pertuzumab Fab were labeled at 0.4–1.0 MBq/μg with [99mTc(CO)3(H2O)3]+ for 1 h at 37 °C. HER2 immunoreactivity was assessed in a direct (saturation) binding assay using HER2-overexpressing SK-BR-3 human breast cancer (BC) cells. MicroSPECT/CT and biodistribution studies were performed in NOD/SCID mice with HER2-positive s.c. SK-OV-3 human ovarian cancer, or MDA-MB-361 or MDA-MB-231 human BC xenografts at 4 or 24 h post i.v. injection of [99mTc]His6-pertuzumab Fab (29–49 MBq, 70 μg). The specificity of tumor uptake was assessed by comparison to irrelevant [99mTc]Fab 3913 in SK-OV-3 tumor-bearing mice.
Results
SDS-PAGE analysis demonstrated cleavage of pertuzumab to produce Fab, which eluted as a single peak with a retention time of 13.8 min on SE-HPLC. Fab were conjugated to 2.1 ± 0.5 His6 peptides and labeled with [99mTc(CO)3(H2O)3]+ to a radiochemical purity of 92–97 % at 0.4–0.8 MBq/μg. [99mTc]His6-pertuzumab Fab exhibited saturable and specific binding to SK-BR-3 cells with a KD = 51.3 ± 5.2 × 10−9 M and Bmax = 3.5 ± 0.1 × 106 receptors/cell. SK-OV-3 tumors were imaged at 4 and 24 h p.i [99mTc]His6-pertuzumab Fab. Tumor uptake at 24 h p.i. was 4.1 ± 0.6 %ID/g, which was 13-fold significantly greater than [99mTc]Fab 3913 (0.3 ± 0.0 %ID/g; P < 0.01). MicroSPECT/CT imaged HER2-overexpressing MDA-MB-361 tumors but not MDA-MB-231 tumors with low HER2 expression. Tumor uptake was 5.2-fold significantly greater at 24 h p.i. in MDA-MB-361 than MDA-MB-231 tumors (3.2 ± 0.1 %ID/g vs. 0.8 ± 0.1 %ID/g; P < 0.05).
Conclusions
MicroSPECT/CT with [99mTc]His6-pertuzumab Fab imaged tumors in NOD/SCID mice that exhibited intermediate or high HER2 expression, but not tumors with low HER2. [99mTc]His6-pertuzumab Fab is promising for SPECT imaging of tumor HER2 expression.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The human epidermal growth factor receptor 2 (HER2) is overexpressed in breast, stomach, and ovarian cancer, as well as other cancers [1, 2]. Overexpression of HER2 in breast cancer (BC) confers a more aggressive phenotype and a poor prognosis [3], but the introduction of HER2-targeted therapies, trastuzumab (Herceptin®, Roche), pertuzumab (Perjeta®, Roche), and trastuzumab emtansine (T-DM1; Kadcyla®, Roche) has remarkably improved the outcome of patients with HER2-positive BC [2, 4, 5]. More recently, trastuzumab deruxtecan (DS-8201), an antibody-drug conjugate (ADC) which links trastuzumab to a potent cytotoxic topoisomerase I inhibitor was shown to be effective in patients with metastatic disease previously treated with T-DM1 [6]. HER2 positivity is assessed ex vivo by immunohistochemical staining (IHC) of a tumor biopsy to detect HER2 protein overexpression or by in situ hybridization to identify amplification of the ERBB2 gene which encodes HER2 [7]. Molecular imaging (MI) which includes single photon emission computed tomography (SPECT) and positron emission tomography (PET) offers a promising complementary tool to non-invasively probe HER2 expression in tumors throughout the body, which may aid in selecting patients for HER2-targeted therapies. Importantly, MI overcomes the challenges of biopsy techniques by providing the capability to detect receptor heterogeneity between primary tumors and metastatic lesions, which has been found in up to 20 % of patients with HER2-positive BC [8]. Further, HER2 expression may change throughout the course of the disease, causing resistance to treatment [9]. Thus, MI may be useful to monitor temporal changes in HER2 expression to provide surveillance for resistance to HER2-targeted therapies [10]. Ulaner et al. showed that PET with [89Zr]DFO-pertuzumab was able to assess the HER2 positivity of metastatic lesions in patients with BC [11]. PET with [89Zr]DFO-trastuzumab detected heterogeneity in HER2 expression in metastatic lesions in patients with BC in whom the primary tumor was HER2-positive and predicted response to T-DM1 [12]. PET with [89Zr]DFO-trastuzumab has further been shown to predict response to T-DM1 in patients with lung cancer expressing a mutant or amplified HER2 [13].
HER2 expression in tumors has been previously imaged with monoclonal antibodies (mAbs), antibody fragments [e.g., F(ab′)2 and Fab], affibodies, and nanobodies labeled with 111In, 177Lu, 99mTc for SPECT or 89Zr, 64Cu, 68Ga, 18F for PET [14]. Despite the higher sensitivity and spatial resolution of PET, in Canada, SPECT remains the most readily available, lowest cost, and most commonly used imaging modality in nuclear medicine [15]. Moreover, 99mTc is an attractive radioisotope for SPECT due to abundant γ-photons (140 keV, 99 %), a short physical half-life (t1/2,p = 6 h) which minimizes the radiation dose to the patient, low cost production via the 99Mo/99mTc generator, and efficient metal chelation radiochemistry. The short half-life of 99mTc is feasible for labeling F(ab′)2 and Fab fragments which accumulate in tumors and are rapidly eliminated from the blood and most normal tissues (except the kidneys) resulting in high tumor/blood (T/B) ratios at early time points [< 24 h post-injection (p.i.)].
We previously reported a method to introduce His6 peptides into Fab to enable labeling using the [99mTc]-(I)-tricarbonyl complex [99mTc(H2O)3(CO)3]+, which is prepared from a kit (IsoLink® kit, Paul Scherrer Institute) [16]. This technique permits 99mTc labeling of Fab as reported by Waibel et al. [17] which relies on reaction of the [99mTc(H2O)3(CO)3]+ complex with a His6 tag often present in recombinant proteins. Pharmaceutical quality mAbs do not incorporate a His6 tag, but this can be introduced by conjugation of Fab to His6 peptides (CGYGGHHHHHH) that present a terminal cysteine for reaction with maleimide functional groups inserted into the Fab by reaction with sulfosuccinimidyl 4-(N-maleimidomethyl) cyclohexane-1-carboxylate (sulfo-SMCC). In our previous report, Fab of the anti-EGFR mAb, panitumumab (Vectibix®; Amgen), were conjugated to His6 peptides for labeling with [99mTc(H2O)3(CO)3]+ [16]. MicroSPECT/CT imaging at 4 and 24 h p.i. of [99mTc]His6-panitumumab Fab visualized tumors in non-obese diabetic severe combined immunodeficiency (NOD/SCID) mice with subcutaneous (s.c.) EGFR-positive human BC, pancreatic cancer, or head and neck cancer xenografts. We now report here the first application of this approach to labeling pertuzumab Fab with [99mTc(H2O)3(CO)3]+ for microSPECT/CT imaging of HER2-positive human BC and ovarian cancer xenografts in NOD/SCID mice.
Materials and Methods
Cell Culture and Tumor Xenografts
The human BC cell lines MDA-MB-231, SK-BR-3, and MDA-MB-361 and human ovarian cancer cell line SK-OV-3 were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA) and were cultured at 37 °C with 5 % CO2. MDA-MB-231 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Dublin, Ireland) supplemented with 10 % fetal bovine serum (FBS, Gibco) and 1 % penicillin streptomycin (Gibco). SK-BR-3 and SK-OV-3 cells were cultured in RPMI-1640 medium (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10 % FBS and 1 % penicillin streptomycin. MDA-MB-361 cells were cultured in Leibovitz L15 media (Sigma-Aldrich) supplemented with 20 % FBS and 1 % penicillin streptomycin. MDA-MB-231, MDA-MB-361 and SK-BR-3 cells express 5.4 × 104, 5.1 × 105, and 1.3 × 106 HER2/cell, respectively [18], while SK-OV-3 cells display 1.2 × 106 HER2/cell [19]. Tumor xenografts were established in the hind flank of female NOD/SCID mice by s.c. inoculation of 1–2 × 106 SK-OV-3, 1 × 105 MDA-MB-361, or 1 × 105 MDA-MB-231 cells suspended in 200 μL of a 1:1 ratio of Matrigel (BD Biosciences, San Jose, CA, USA) and phosphate-buffered saline (PBS, pH 7.4). At 24 h prior to MDA-MB-361 cells inoculation, mice were implanted with a 0.72 mg, 60-day sustained release 17β-estradiol pellet (Innovative Research of America, Sarasota, FL, USA). Tumors were allowed to grow to 5–10 mm in diameter for imaging and biodistribution studies. All applicable institutional and/or national guidelines for the care and use of animals were followed. All animal studies were conducted in accordance with the Canadian Council on Animal Care (CCAC) guidelines and under an animal use protocol (AUP 4336.2) approved by the Animal Care Committee of the University Health Network.
Pertuzumab Fab
Fab were generated by digestion of pertuzumab IgG (Perjeta®, Roche, Mississauga, ON, Canada) with papain. Approximately 1.25 mg of immobilized papain (Agarose Resin, ThermoFisher Scientific, Mississauga, ON, Canada) was activated with cysteine digestion buffer (2 mM NaH2PO4, 11 mM EDTA, 88 mM L-cysteine, pH 7.0). To the activated papain slurry, 2.0 mL of pertuzumab (5.0 mg/mL in cysteine digestion buffer) was added and incubated for 20 h at 37 °C, with shaking at 200 rpm on an Excella E24 mixer (New Brunswick Scientific, Edison, NJ, USA). The slurry was repeatedly washed with PBS, and the immobilized papain was sedimented by centrifugation at 200×g for 10 min using an Eppendorf centrifuge (Model 5804R). The supernatant containing the digestion product was isolated by filtration through a 0.22 μm Millex-GV PVDF filter (MilliporeSigma, Darmstadt, Germany). The product was concentrated and purified by ultrafiltration using an Amicon ultra centrifugal filter unit (30 kDa molecular weight cut-off, MWCO; Millipore-Sigma). Fab were concentrated and buffer-exchanged into 0.1 M NaHCO3 (pH 9.0) and stored at 2–8 °C. The Fab concentration was 4–7 mg/mL, measured spectrophotometrically at 280 nm, assuming an absorbance of 1.40 (mg/mL)−1 cm−1 based on the amino acid composition of pertuzumab Fab [20].
Pertuzumab Fab-His6
Pertuzumab Fab were modified with peptides incorporating a hexahistidine (His6) motif (CGYGGHHHHHH; BioBasic, Markham, ON, Canada), and a terminal thiol for reaction with maleimide groups is introduced into the Fab by reaction with sulfo-SMCC (ThermoFisher Scientific). A 20-fold molar excess of sulfo-SMCC (0.1 M sulfo-SMCC in 0.1 M NaH2PO4 buffer) was reacted for 75 min at room temperature (RT) with 1–3 mg of Fab (1–4 mg/mL in PBS). The product was purified by ultrafiltration using a 10 kDa MWCO Amicon ultra centrifugal filter unit (Millipore-Sigma). Then, 1–3 mg of SMCC-modified Fab in PBS were reacted with a 100-fold molar excess of His6 peptides (20 mg/mL in deionized H2O) overnight at 4 °C. Pertuzumab Fab-His6 were further purified by ultrafiltration using a 10 kDa MWCO Amicon ultra device to a final concentration of 2–8 mg/mL, determined spectrophotometrically by the absorbance at 280 nm, assuming an absorbance of 1.38 (mg/mL)−1 cm−1 based on the amino acid sequence of pertuzumab Fab conjugated to two His6 peptides. The number of His6 peptides conjugated to Fab were quantified using a previously reported radiometric method [21]. In this method, 123I-labeled His6 peptides were incorporated into the reaction with SMCC-modified Fab under identical conditions. Fab-[123I]His6 were purified by ultrafiltration, and 123I incorporated into the Fab measured in a γ-counter (PerkinElmer Wizard 3). The number of [123I]His6 peptides conjugated to Fab was calculated based on the specific activity (SA) of the [123I]His6 peptides.
Radioimmunoconjugates
Pertuzumab Fab-His6 were labeled with [99mTc(CO)3(H2O)3]+ complex as previously reported [16]. [99mTc(CO)3(H2O)3]+ was prepared by adding 740 MBq (1 mL) of [99mTc] sodium pertechnetate (Radiopharmacy, University Health Network, Toronto, ON, Canada) to an IsoLink® kit (Paul Scherrer Institute, Villigen, Switzerland) and incubating in a water bath for 20 min under a gentle boil. After cooling to RT, the pH of the kit solution was adjusted to 7.0 by adding 0.14 mL of 0.1 M HCl. [99mTc(CO)3(H2O)3]+ (40–560 MBq; 0.07–0.9 mL) was then reacted with 0.1–0.6 mg of Fab-His6 (0.01–4 mL) for 1 h at 37 °C with shaking at 200 rpm on an Excella E24 mixer. Fab 3913, an irrelevant control Fab which binds Ebola virus, was provided by Dr. Sachdev Sidhu [Toronto Recombinant Antibody Center (TRAC), Toronto, ON, Canada]. Fab 3913 incorporates an endogenous His6 and was labeled by reacting 0.7 mg of the Fab with 407 MBq of [99mTc(CO)3(H2O)3]+ for 1 h at 37 °C. The radiochemical purity (RCP) of 99mTc-labeled Fab was determined by silica gel-instant thin layer chromatography (ITLC-SG) developed in 0.1 M sodium citrate buffer, pH 5.5. The Rf values for 99mTc-labeled Fab and free [99mTc(CO)3(H2O)3]+ are 0.0 and 1.0, respectively [16].
SDS-PAGE and SE-HPLC
Pertuzumab Fab and Fab-His6 were analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) on a 4–20 % Tris-HCl gel (Bio-Rad, Mississauga, ON, Canada) under reducing [dithiothreitol (DTT)] and non-reducing conditions. The gels were stained with Coomassie Blue G-250 (Bio-Rad), and the bands were visualized with a BioRad ChemiDoc™ gel imaging system. The approximate protein molecular weights were estimated by comparison to a 10–250 kDa molecular weight (MW) ladder (Precision Plus Protein™ Prestained Standards, Bio-Rad), and approximate purity estimated by densitometry with Image Lab software (Ver. 6.0; BioRad). The band purity was determined as the density relative to that of all bands detected in the lane. The purity of pertuzumab Fab and Fab-His6 was assessed by size exclusion high-performance liquid chromatography (SE-HPLC; PerkinElmer HPLC, Waltham, MA, USA) on a BioSep™ SEC-s2000 column (Phenomenex, Torrance, CA, USA) eluted with 0.1 M NaH2PO4, pH 7.0 at a flow rate of 0.8 mL/min. A 20 μL aliquot of the sample (0.25 mg/mL) was analyzed. A diode array detector (PerkinElmer) monitored the UV280 signal. The MW of pertuzumab Fab is estimated by SE-HPLC by reference to a calibration curve of log10MW (Da) vs. Ve/Vo, where Ve are the elution volumes of a series of analyzed MW standards (29–200 kDa) and Vo is the void volume of the SEC-s2000 column determined with blue dextran (2000 kDa; see Supplementary Material).
HER2 Immunoreactivity of [99mTc]His6-Pertuzumab Fab
A direct (saturation) radioligand binding assay was used to estimate the dissociation constant (KD) for binding of [99mTc]His6-pertuzumab Fab to HER2-overexpressing SK-BR-3 cells. Briefly, 1 × 106 SK-BR-3 cells were incubated with increasing concentrations of [99mTc]His6-pertuzumab Fab (0.07–300 × 10−9 M) for 3 h at 4 °C in 1.5-mL Eppendorf tubes. The tubes were centrifuged at 1200×g for 5 min with a microcentrifuge (Model 5424, Eppendorf) to separate the cell pellet and supernatant. The cell pellet was resuspended in 0.5 mL PBS twice and then recentrifuged and the supernatant was removed. Finally, the radioactivity of the pellet was measured in a γ-counter to determine the total binding (TB). The assay was repeated in the presence of a 50-fold molar excess of pertuzumab IgG to determine non-specific binding (NSB). Specific binding (SB) was obtained by subtraction of NSB from TB. The cell-bound radioactivity was plotted vs. the concentration of [99mTc]His6-pertuzumab Fab (nM), and the resulting curves were fitted to a one-site receptor-binding model using Prism Ver. 4.0 software. The KD and number of binding sites (Bmax) were calculated from the SB curve. Bmax was converted from counts per minute (CPM) to the number of receptors per cell.
MicroSPECT/CT Imaging and Biodistribution Studies
MicroSPECT/CT imaging and biodistribution studies were performed in NOD/SCID mice with s.c. SK-OV-3 tumor xenografts that overexpress HER2 (1.2 × 106 HER2/cell) [18]. Groups of 5 tumor-bearing mice were injected i.v. (tail-vein) with 36–37 MBq (70 μg) of [99mTc]His6-pertuzumab Fab or 25–30 MBq (70 μg) of irrelevant [99mTc]Fab 3913 in 0.16 mL of PBS, pH 7.4. Representative mice from each group (n = 2) were imaged under anesthesia (1.0–1.5 % isoflurane in O2) at 4 h and 24 h post-injection (p.i.) on NanoSPECT (Bioscan, Washington, DC) and CT (eXplore Locus Ultra Preclinical CT scanner, GE Healthcare) systems at the Spatio-Temporal Targeting and Amplification and Radiation Response (STTARR) facility (University Health Network). Tumor-bearing mice (n = 4) were sacrificed at 24 h p.i., and blood, tumor, and selected normal tissues were obtained, rinsed with PBS, and weighed. The radioactivity was measured by γ-counting and converted to percent-injected dose per gram of tissue (%ID/g) and expressed as mean ± SEM %ID/g.
In separate studies, biodistribution analyses were performed in NOD/SCID mice with s.c. MDA-MB-231 or MDA-MB-361 human BC tumors that have low or intermediate HER2 expression (5.4 × 104 and 5.1 × 105, respectively) [18] at 4 or 24 h p.i. of 29–49 MBq (70 μg, 0.16 mL in PBS, pH 7.4) of [99mTc]His6-pertuzumab Fab. Representative mice from each group (n = 2) were imaged at 24 h p.i. on a NanoScan SPECT/CT/PET trimodality system (Mediso, Budapest, Hungary). At 4 h or 24 h p.i., all mice were sacrificed, and the tumor and samples of blood and other normal tissues were obtained. These tissues were weighed, and the radioactivity in each was measured in a γ-counter and expressed as %ID/g. The mean ± SEM %ID/g was compared for [99mTc]His6-pertuzumab Fab in mice with s.c. MDA-MB-361 and MDA-MB-231 tumors. The details for all SPECT and CT imaging parameters are provided in the Supplementary Material.
Statistical Analysis
Significant differences were calculated by an unpaired Student’s t test (P < 0.05).
Results
Pertuzumab Fab and Pertuzumab Fab-His6
SDS-PAGE shows that pertuzumab IgG migrated as a single band under non-reducing conditions corresponding to a protein at 150 kDa (Fig. 1a; lane 2), while Fab (lane 3) migrated as one major band at 42 kDa corresponding to ~ 80 % of total protein by gel densitometry. In addition, there were minor bands observed at 52, 30, and 16 kDa. The band at 52 kDa may represent Fc impurity. Under reducing conditions, pertuzumab IgG (lane 5) migrated as two bands at 49 kDa and 25 kDa representing dissociated heavy and light immunoglobulin chains. One major band at 25 kDa corresponding to the VH-CH1 and VL-CL chains was observed for pertuzumab Fab under reducing conditions (lane 6), and a faint minor band was present at 30 kDa. Modification of Fab with SMCC and conjugation to His6 peptides resulted in an upward shift in the major band to 46 kDa (lane 4). This increase in MW agreed with 2.1 ± 0.5 His6 peptides incorporated per Fab measured in a radiometric assay, since each peptide has a MW = 1.32 kDa. Under reducing conditions, SMCC-modified Fab exhibited one major band at 26 kDa and two minor bands at 30 and 44 kDa (lane 7). There was a slight upward band shift after conjugation of Fab to His6 peptides (lane 8). SE-HPLC with UV detection at 280 nm revealed a single peak for pertuzumab Fab (Fig. 1b) and pertuzumab Fab-His6 (Fig. 1c) with retention times (tR) = 13.8 min and 13.9 min, respectively. Calibration of the SE-HPLC column using a series of MW standards indicates that the MW of pertuzumb Fab was 38 kDa (Supplementary Material Fig. S1).
a SDS-PAGE analysis on a 4–20 % Tris-HCl gel stained with Coomassie blue G-250 under non-reducing conditions (lanes 2–4) or reducing conditions (lanes 5–8). Lanes: MW, molecular weight markers; 2 and 5, pertuzumab IgG; 3 and 6, pertuzumab Fab; 7, pertuzumab Fab reacted with sulfo-SMCC; 4 and 8, pertuzumab Fab-His6. The slight upward band shift for pertuzumab Fab-His6 in lanes 4 and 8 compared to pertuzumab Fab in lanes 3 and 6 is due to conjugation of His6-peptides to the Fab. b SE-HPLC analysis of pertuzumab Fab and c pertuzumab Fab-His6 on a BioSep-SEC-s2000 column with 0.1 M NaH2PO4, pH 7.0 at a flow rate of 0.8 mL/min with UV detection at 280 nm.
Radioimmunoconjugates
Pertuzumab Fab-His6 were labeled with [99mTc(CO)3(H2O)3]+ over a range of SA (Table 1). Labeling efficiency (LE) was 92–97 % for a SA of 0.4–0.8 MBq/μg, while at a SA of 1.0 MBq/μg, LE decreased slightly to 85 %. Fab 3913 were labeled with [99mTc(CO)3(H2O)3]+ at a SA = 0.58 MBq/μg and LE = 95 %. No post-labeling purification was performed.
HER2 Immunoreactivity
Saturable binding of [99mTc]His6-pertuzumab Fab to HER2-overexpressing SK-BR-3 cells (1.3 × 106 HER2/cell) [22] was observed, and the binding was competed with a 50-fold molar excess of unlabeled pertuzumab IgG (Fig. 2). Fitting the SB curve from duplicate binding assay experiments to a 1-site receptor-binding model revealed that the KD for binding to HER2 on SK-BR-3 cells was 51 ± 5 × 10−9 M and the Bmax = 3.5 ± 0.1 × 106 receptors/cell.
Binding of [99mTc]His6-pertuzumab Fab to HER2-overexpressing SK-BR-3 human breast cancer cells. TB, total binding; NSB, non-specific binding; SB, specific binding. Data shown are the mean ± SEM (n = 2). Fitting of the SB to a 1-site receptor-binding model using GraphPad Prism Ver. 4.0 software revealed that the KD for binding HER2 on SK-BR-3 cells was 51.3 ± 5.2 nM and the Bmax was 3.5 ± 0.1 × 106 receptors/cell.
MicroSPECT/CT Imaging and Biodistribution Studies
Representative microSPECT/CT images at 4 h and 24 h p.i. of [99mTc]His6-pertuzumab Fab or irrelevant [99mTc]Fab 3913 in NOD/SCID mice with s.c. SK-OV-3 human ovarian cancer xenografts with high HER2 expression are shown in Fig. 3. Tumors were visualized at both time points with [99mTc]His6-pertuzumab Fab but were more clearly imaged at 24 h p.i. Tumors were not visualized with [99mTc]Fab 3913. The normal organs which exhibited the highest uptake of [99mTc]His6-pertuzumab Fab or [99mTc]Fab 3913 were the liver, spleen, and particularly the kidneys, with some bladder radioactivity observed at 4 h p.i. Biodistribution studies at 24 h p.i. (Table 2) revealed that the tumor uptake of [99mTc]His6-pertuzumab Fab (4.1 ± 0.6 %ID/g) was 13.7-fold significantly greater than [99mTc]Fab 3913 (0.3 ± 0.0 %ID/g; P < 0.01). Radioactivity in the blood was 8.9-fold significantly lower for [99mTc]Fab 3913 (0.1 ± 0.0 %ID/g) than [99mTc]His6-pertuzumab Fab (1.3 ± 0.2 %ID/g), but the T/B ratio was 1.4-fold significantly higher for [99mTc]His6-pertuzumab Fab (3.2 ± 0.2 vs. 2.2 ± 0.2; P < 0.01). Muscle uptake was 0.6 ± 0.1 %ID/g for [99mTc]His6-pertuzumab Fab and 0.2 ± 0.0 %ID/g for [99mTc]Fab 3913 (P < 0.001), with a 4.3-fold higher T/M ratio for [99mTc]His6-pertuzumab Fab (6.9 ± 1.2 vs. 1.6 ± 0.3; P < 0.01). There was significantly lower uptake of [99mTc]Fab 3913 in all other tissues, with the exception of the spleen (P = 0.20). Liver uptake of [99mTc]His6-pertuzumab Fab was 2.1-fold higher than [99mTc]Fab 3913 (11.3 ± 0.8 %ID/g vs. 5.5 ± 0.7 %ID/g, respectively; P < 0.001). The kidneys exhibited the highest normal organ uptake for both radioimmunoconjugates with slightly but significantly greater uptake observed for [99mTc]His6-pertuzumab Fab than [99mTc]Fab 3913 (53.1 ± 1.1 %ID/g vs. 48.2 ± 1.4 %ID/g, respectively; P < 0.05).
Representative microSPECT/CT maximum intensity projections (MIPs) of female NOD/SCID mice bearing s.c. HER2-expressing SK-OV-3 tumor xenografts at (a) 4 h and (b) 24 h p.i. of 70 μg (36–37 MBq) of [99mTc]His6-pertuzumab Fab or (c) 4 h and (d) 24 h p.i. of irrelevant [99mTc]Fab 3913. The tumor is indicated by the white arrowhead while the liver, kidneys and bladder are indicated by blue, green, and red arrowheads, respectively. Images were normalized to the same intensity for each timepoint. The intensity scale is shown on the right of each image panel.
MicroSPECT/CT and biodistribution studies were also performed in NOD/SCID mice with s.c. MDA-MB-231 or MDA-MB-361 tumors with low or intermediate HER2 expression, respectively (5.4 × 104 and 5.1 × 105 HER2/cell), injected with [99mTc]His6-pertuzumab Fab. MDA-MB-231 tumors were not visualized at 24 h p.i. of [99mTc]His6-pertuzumab Fab (Fig. 4a), while MDA-MB-361 tumors are clearly visualized (Fig. 4b). Biodistribution studies in mice with MDA-MB-361 tumors (Table 2) revealed that tumor uptake increased by 1.8-fold from 3.2 ± 0.1 %ID/g at 4 h p.i. to 5.6 ± 1.4 %ID/g at 24 h p.i. This was accompanied by a 4.6-fold decrease in blood radioactivity from 3.3 ± 0.4 %ID/g at 4 h.p.i. to 0.7 ± 0.0 %ID/g at 24 h, resulting in an increase in the T/B ratio from 1.0 ± 0.1 at 4 h p.i. to 7.8 ± 1.9 at 24 h (P < 0.05). There was a similar reduction in other normal tissue uptake, except for a 2.6-fold increase in uptake in the kidneys from 4 to 24 h p.i. The T/M ratio increased from 3.7 ± 0.8 at 4 h p.i. to 15.8 ± 4.1 at 24 h. The tumor uptake of [99mTc]His6-pertuzumab Fab at 24 h p.i in mice with MDA-MB-231 xenografts was 7.1-fold significantly lower than in mice with MDA-MB-361 xenografts (0.8 ± 0.1 %ID/g vs. 5.6 ± 1.4 %ID/g; P < 0.05; Table 2). There was also high kidney uptake (20.0 ± 1.9 %ID/g) and moderate liver uptake (9.5 ± 1.4 %ID/g) of [99mTc]His6-pertuzumab Fab in mice with MDA-MB-231 tumors. Radioactivity in the blood in these mice was low (0.7 ± 0.0 %ID/g). The T/B ratio was 1.1 ± 0.1, and the T/M ratio was 3.8 ± 0.4. The T/M ratio was 4.2-fold lower than in mice with MDA-MB-361 tumors (P = 0.062). The tumor uptake in mice with s.c. SK-OV-3 xenografts was 5.2-fold significantly higher than in mice with MDA-MB-231 tumors (P < 0.001), and the T/B ratio was 2.8-fold significantly higher (P < 0.0001). There were no significant differences in the tumor uptake (P = 0.37), T/B (P = 0.095), and T/M (P = 0.12) ratios between mice with s.c. MDA-MB-361 or SK-OV-3 tumors.
Representative microSPECT/CT maximum intensity projections (MIPs) of female NOD/SCID mice bearing (a) MDA-MB-231 tumor xenografts with low HER2 expression (5.4 × 104 receptors/cell) or (b) MDA-MB-361 with intermediate HER2 expression (5.1 × 105 HER2/cell) at 24 h p.i of [99mTc]His6-pertuzumab Fab (70 μg, 29–49 MBq). The tumor is indicated by the white arrowhead while the liver, kidneys and bladder are indicated by blue, green, and red arrowheads, respectively. Images were normalized to the same intensity for each timepoint. The intensity scale is shown on the right of each image panel.
Discussion
We describe here labeling of pertuzumab Fab modified with His6 peptides by reaction with the [99mTc(CO)3(H2O)3]+ complex and their evaluation for microSPECT/CT imaging of HER2-positive tumors in NOD/SCID mice. Fab were prepared by digestion of pertuzumab IgG (Perjeta®, Roche) using immobilized papain and purified by ultrafiltration. SDS-PAGE demonstrated one major band for pertuzumab Fab accounting for ~ 80 % of all protein bands by gel densitometry (Fig. 1a), but a single peak was found by SE-HPLC analysis (Fig. 1b). Pertuzumab Fab were reacted with sulfo-SMCC to introduce maleimide groups for conjugation to the terminal cysteine of His6-containing peptides [CGYGGHHHHHH]. Fab-His6 were then labeled with the [99mTc(CO)3(H2O)3]+ complex at a SA up to 0.8 MBq/μg and achieving ≥ 92 % RCP (Table 1). A higher SA (1.0 MBq/μg) resulted in a slightly lower RCP (85 %). The [99mTc(CO)3(H2O)3]+ complex was prepared using the IsoLink® Kit (Paul Scherrer Institute). This method of labeling pertuzumab Fab was adapted from our previous report in which Fab of the anti-EGFR mAb, panitumumab (Vectibix®, Amgen) were modified with His6 peptides to enable labeling with [99mTc(CO)3(H2O)3]+ [16]. Thus, the method is versatile and was extended here to another pharmaceutical quality mAb, pertuzumab, that did not harbor an endogenous His6 tag. An irrelevant Fab 3913 that incorporated an endogenous His6 tag was labeled with [99mTc(CO)3(H2O)3]+ and without conjugation to exogenous His6 peptides.
[99mTc]His6-pertuzumab Fab demonstrated saturable binding to SK-BR-3 human BC cells that was displaced by an excess of pertuzumab IgG, indicating HER2-specific binding (Fig. 2). The KD was 51 ± 5 × 10−9 M and the Bmax was 3.5 ± 0.1 × 106 receptors/cell. In previous studies, we found that [111In]DTPA-pertuzumab and [64Cu]NOTA-pertuzumab F(ab′)2 exhibited -20-25-fold higher affinity than [99mTc]His6-pertuzumab Fab for binding to HER2-expressing SK-BR-3 cells: KD = 2.0 ± 1 × 10−9 M [22] and 2.6 ± 0.3 × 10−9 M [23], respectively. The lower HER2-binding affinity for [99mTc]His6-pertuzumab Fab may be due to monovalent binding of Fab to HER2 compared to the bivalent binding of pertuzumab IgG or F(ab′)2. [99mTc]His6-panitumumab Fab similarly exhibited a lower binding affinity for EGFR (KD = 55 ± 4 × 10−9 M [16]) than [177Lu]DOTA-panitumumab (KD = 1.0 ± 0.4 × 10−9 M [24]) or [64Cu]DOTA-panitumumab F(ab′)2 (KD = 0.14 ± 0.05 × 10−9 M [25]).
MicroSPECT/CT and biodistribution studies were performed in NOD/SCID mice with s.c. SK-OV-3 human ovarian cancer xenografts overexpressing HER2 (1.2 × 106 HER2/cell [19]) after i.v. (tail vein) injection of 36–37 MBq (70 μg) of [99mTc]His6-pertuzumab Fab. The specificity of tumor uptake was assessed by comparison to irrelevant [99mTc]Fab 3913. Tumor uptake at 24 h p.i. of [99mTc]His6-pertuzumab Fab is 13.7-fold greater than [99mTc]Fab 3913 (4.1 ± 0.6 %ID/g vs. 0.32 ± 0.02 %ID/g; P < 0.01; Table 2). Further, the T/B and T/M ratios for [99mTc]His6-pertuzumab Fab (3.2 ± 0.2 and 6.9 ± 1.2, respectively) were higher than [99mTc]Fab 3913 (2.2 ± 0.2, and 1.6 ± 0.3, respectively). SK-OV-3 tumors were imaged at 4 h p.i. and with improved tumor visualization at 24 h p.i. of [99mTc]His6-pertuzumab Fab (Fig. 3a,b), while tumors were not visualized at either time point with [99mTc]Fab 3913 (Fig. 3c,d). These results demonstrated that the tumor uptake of [99mTc]His6-pertuzumab Fab was specific and microSPECT/CT allowed visualization of HER2-positive SK-OV-3 tumors in NOD/SCID mice, especially at 24 h p.i. The uptake of [99mTc]His6-pertuzumab Fab at 24 h p.i. in s.c. SK-OV-3 tumors in NOD/SCID mice (4.1 ± 0.6 %ID/g) was lower than we reported at 24 h p.i. of [64Cu]NOTA-pertuzumab F(ab′)2 (1–3 MBq; 50 μg) in NOD/SCID mice (9.8 ± 5.1 %ID/g) in an earlier study [23], which is most likely explained by the more rapid elimination of Fab and its monovalent HER2 binding. In the earlier study, SK-OV-3 tumors were imaged at 24 h p.i. of [64Cu]NOTA-pertuzumab F(ab′)2 by microPET/CT [23]. The impact of bivalent HER2 binding and a longer circulation time for intact IgG forms of mAbs are revealed by the much greater uptake in HER2-overexpressing MDA-MB-361 human BC xenografts (5.4 × 104 HER2/cell) in athymic mice found at 72 h p.i. of [111In]DTPA-pertuzumab (34.6 ± 9.9 %ID/g) [22]. MDA-MB-361 tumors in this study were imaged by microSPECT/CT at 72 h p.i. of [111In]DTPA-pertuzumab.
To further understand the relationship between the tumor uptake of [99mTc]His6-pertuzumab Fab and HER2 expression level, microSPECT/CT and biodistribution studies were performed in NOD/SCID mice with s.c. MDA-MB-361 or MDA-MB-231 human BC xenografts with intermediate or low HER2 expression (5.1 × 105 and 5.4 × 104 HER2/cell, respectively [18]). MDA-MB-231 tumors were not imaged at 24 h p.i. of [99mTc]His6-pertuzumab Fab (Fig. 4a), while MDA-MB-361 tumors were visualized (Fig. 4b). There was lower uptake of [99mTc]His6-pertuzumab Fab in MDA-MB-231 than MDA-MB-361 tumors at 24 h p.i. (0.8 ± 0.1 %ID/g vs. 5.6 ± 1.4 %ID/g, P < 0.05; Table 2). Tumor uptake of [99mTc]His6-pertuzumab Fab at 24 h p.i. in mice with MDA-MB-361 tumors was slightly higher than in mice with SK-OV-3 tumors (5.6 ± 1.4 %ID/g vs. 4.1 ± 0.6 %ID/g, P = 0.37), despite the lower HER2 expression of MDA-MB-361 cells (5.1 × 105 HER2/cell [18]) than SK-OV-3 cells (1.2 × 106 HER2/cell [19]). These results suggest that microSPECT/CT with [99mTc]His6-pertuzumab Fab was not able to discriminate a 2-fold difference in HER2 expression in these tumors but was able to detect the 10–20-fold difference in HER2 expression between MDA-MB-361 or SK-OV-3 tumors and MDA-MB-231 tumors (5.4 × 104 HER2/cell). The inability to discriminate the 2-fold difference in HER2 expression between MDA-MB-361 and SK-OV-3 tumors may be due to other factors such as tumor vascularization, interstitial pressure, and the stromal matrix which presents penetration barriers to radioimmunoconjugates [26]. Nonetheless, our results indicated that tumors with moderate to high HER2 expression, which are represented by MDA-MB-361 or SK-OV-3 xenografts in NOD/SCID mice, could be imaged at 24 h p.i. of [99mTc]His6-pertuzumab Fab, but tumors with low HER2 expression such as MDA-MB-231 could not be imaged. Importantly, in patients with breast cancer (BC), only those who have tumors that express moderate or high levels of HER2 comparable to MDA-MB-361 or SK-OV-3 tumors [18, 19] are eligible for treatment with HER2-targeted therapies [7], and thus imaging with [99mTc]His6-pertuzumab Fab may be useful to identify tumors in patients with these levels of HER2.
PET has been studied to assess tumor HER2 status in patients with BC. In one study, PET visualized lesions at 5–8 days p.i. of [89Zr]DFO-pertuzumab (20–50 mg, 74 MBq) in 6 patients with HER2-positive metastatic BC [11]. In another study, PET with [89Zr]DFO-trastuzumab (10–50 mg, 37 MBq) detected tumors in the liver, lung, bone, and brain in 14 patients with HER2-positive BC, and the optimal time point for tumor uptake was 4–5 d p.i. [27]. Since HER2-positive tumors in mice were imaged by microSPECT/CT at 4 h p.i. of [99mTc]His6-pertuzumab Fab with clear visualization at 24 h p.i. (Figs. 3 and 4), this may allow earlier tumor imaging in patients than by PET using 89Zr-labeled pertuzumab or trastuzumab. Moreover, the high radiochemical purity (92–97 %) of [99mTc]His6-pertuzumab Fab achieved by incubation with [99mTc(CO)3(H2O)3]+ may allow development of a kit formulation to prepare the radiopharmaceutical for patients.
Conclusions
Modification of pertuzumab Fab with His6 peptides enabled labeling with the [99mTc(CO)3(H2O)3]+ complex. [99mTc]His6-pertuzumab Fab retained high affinity specific binding to HER2 on SK-BR-3 cells in vitro and localized specifically in vivo in s.c. SK-OV-3 human ovarian cancer xenografts in NOD/SCID mice. Tumors were visualized by microSPECT/CT at 4 h or 24 h p.i. of [99mTc]His6-pertuzumab Fab. Tumor uptake of [99mTc]His6-pertuzumab Fab and tumor visualization by microSPECT/CT in mice was dependent on HER2 expression level. Only MDA-MB-361 or SK-OV-3 tumors with high or intermediate HER2 expression but not MDA-MB-231 tumors with low HER2 were imaged with [99mTc]His6-pertuzumab Fab. Our results suggest that [99mTc]His6-pertuzumab Fab is a promising agent for imaging tumor HER2 expression which may be helpful for selecting patients for HER2-targeted therapies.
References
Yan M, Schwaederle M, Arguello D, Millis SZ, Gatalica Z, Kurzrock R (2015) HER2 expression status in diverse cancers: review of results from 37,992 patients. Cancer Metastasis Rev 34:157–164
Oh DY, Bang YJ (2020) HER2-targeted therapies - a role beyond breast cancer. Nat Rev Clin Oncol 17:33–48
Slamon DJ, Clark GM, Wong SG, Levin W, Ullrich A, McGuire W (1987) Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 235:177–182
Piccart-Gebhart MJ, Procter M, Leyland-Jones B, Goldhirsch A, Untch M, Smith I, Gianni L, Baselga J, Bell R, Jackisch C, Cameron D, Dowsett M, Barrios CH, Steger G, Huang CS, Andersson M, Inbar M, Lichinitser M, Láng I, Nitz U, Iwata H, Thomssen C, Lohrisch C, Suter TM, Rüschoff J, Suto T, Greatorex V, Ward C, Straehle C, McFadden E, Dolci MS, Gelber RD, Herceptin Adjuvant (HERA) Trial Study Team (2005) Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med 353:1659–1672
Swain SM, Kim SB, Cortés J, Ro J, Semiglazov V, Campone M, Cruelos E, Ferrero JM, Schneeweiss A, Knott A, Clark E, Ross G, Benyunes MC, Baselga J (2013) Pertuzumab, trastuzumab, and docetaxel for HER2-positive metastatic breast cancer (CLEOPATRA study): overall survival results from a randomised, double-blind, placebo-controlled, phase 3 study. Lancet Oncol 14:461–471
Modi S, Saura C, Yamashita T, Park YH, Kim SB, Tamura K, Andre F, Iwata H, Ito Y, Tsurutani J, Sohn J, Denduluri N, Perrin C, Aogi K, Tokunaga E, Im SA, Lee KS, Hurvitz SA, Cortes J, Lee C, Chen S, Zhang L, Shahidi J, Yver A, Krop I, DESTINY-Breast01 Investigators (2020) Trastuzumab deruxtecan in previously treated HER2-positive breast cancer. N Engl J Med 382:610–621
Wolff AC, Hammond MEH, Allison KH, Harvey BE, Mangu PB, Bartlett JMS, Bilous M, Ellis IO, Fitzgibbons P, Hanna W, Jenkins RB, Press MF, Spears PA, Vance GH, Viale G, McShane LM, Dowsett M (2018) Human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. Arch Pathol Lab Med 142:1364–1382
Rossi S, Basso M, Strippoli A, Dadduzio V, Cerchiaro E, Barile R, D'Argento E, Cassano A, Schinzari G, Barone C (2015) Hormone receptor status and HER2 expression in primary breast cancer compared with synchronous axillary metastases or recurrent metastatic disease. Clin Breast Cancer 15:307–312
Vu T, Claret FX (2012) Trastuzumab: updated mechanisms of action and resistance in breast cancer. Front Oncol 2:62
Niikura N, Liu J, Hayashi N, Mittendorf EA, Gong Y, Palla SL, Tokuda Y, Gonzalez-Angulo AM, Hortobagyi GN, Ueno NT (2012) Loss of human epidermal growth factor receptor 2 (HER2) expression in metastatic sites of HER2-overexpressing primary breast tumors. J Clin Oncol 30:593–599
Ulaner GA, Lyashchenko SK, Riedl C et al (2017) First-in-human HER2-argeted imaging using 89Zr-pertuzumab PT/CT: dosimetry and clinical application in patients with breast cancer. J Nucl Med 59:900–906
Gebhart G, Lamberts LE, Wimana Z, Garcia C, Emonts P, Ameye L, Stroobants S, Huizing M, Aftimos P, Tol J, Oyen WJG, Vugts DJ, Hoekstra OS, Schröder CP, Menke-van der Houven van Oordt CW, Guiot T, Brouwers AH, Awada A, de Vries EGE, Flamen P (2016) Molecular imaging as a tool to investigate heterogeneity of advanced HER2-positive breast cancer and to predict patient outcome under trastuzumab emtansine (T-DM1): the ZEPHIR trial. Ann Onco 27:619–624
Ulaner GA, Offin M, Scaltriti M et al (2019) 89Zr-trastuzumab PET/CT for prediction of response to HER2-targeted therapy in patients with HER2 mutant lung cancer: an exploratory phase 2 trial. J Nucl Med 60(Suppl 1):144
Gebhart G, Flamen P, De Vries EG, Jhaveri K, Wimana Z (2016) Imaging diagnostic and therapeutic targets: human epidermal growth factor receptor 2. J Nucl Med 57:81S–88S
Sinclair A, Morrison A, Young C, Pyke L. (2018) The Canadian medical imaging inventory, 2017. Ottawa: CADTH
Ku A, Chan C, Aghevlian S, Cai Z, Cescon D, Bratman SV, Ailles L, Hedley DW, Reilly RM (2019) MicroSPECT/CT imaging of cell-line and patient-derived EGFR-positive tumor xenografts in mice with panitumumab fab modified with hexahistidine peptides to enable labeling with 99mTc(I) tricarbonyl complex. Mol Pharm 16:3559–3568
Waibel R, Alberto R, Willuda J, Finnern R, Schibli R, Stichelberger A, Egli A, Abram U, Mach JP, Plückthun A, Schubiger PA (1999) Stable one-step technetium-99m labeling of his-tagged recombinant proteins with a novel Tc(I)-carbonyl complex. Nat Biotechnol 17:897–901
McLarty K, Cornelissen B, Scollard DA, Done SJ, Chun K, Reilly RM (2009) Associations between the uptake of 111In-DTPA-trastuzumab, HER2 density and response to trastuzumab (Herceptin) in athymic mice bearing subcutaneous human tumour xenografts. Eur J Nucl Med Mol Imaging 36:81–93
Orlova A, Tolmachev V, Pehrson R, Lindborg M, Tran T, Sandström M, Nilsson FY, Wennborg A, Abrahmsén L, Feldwisch J (2007) Synthetic affibody molecules: a novel class of affinity ligands for molecular imaging of HER2-expressing malignant tumors. Cancer Res 67:2178–2186
Gill SC, von Hippel PH (1989) Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem 182:319–326
Chen P, Wang J, Hope K, Jin L, Dick J, Cameron R, Brandwein J, Minden M, Reilly RM (2006) Nuclear localizing sequences (NLS) promote nuclear translocation and enhance the radiotoxicity of the anti-CD33 monoclonal antibody HuM195 labeled with 111In in human myeloid leukemia cells. J Nucl Med 47:827–836
McLarty K, Cornelissen B, Cai Z, Scollard DA, Costantini DL, Done SJ, Reilly RM (2009) Micro-SPECT/CT with 111In-DTPA-pertuzumab sensitively detects trastuzumab-mediated HER2 downregulation and tumor response in athymic mice bearing MDA-MB-361 human breast cancer xenografts. J Nucl Med 50:1340–1348
Lam K, Chan C, Reilly RM (2017) Development and preclinical studies of 64Cu-NOTA-pertuzumab F(ab')2 for imaging changes in tumor HER2 expression associated with response to trastuzumab by PET/CT. MAbs 9:154–164
Aghevlian S, Lu Y, Winnik MA, Hedley DW, Reilly RM (2018) Panitumumab modified with metal-chelating polymers (MCP) complexed to 111In and 177Lu – an EGFR-targeted theranostic for pancreatic cancer. Mol Pharm 15:1150–1159
Boyle AJ, Cao PJ, Hedley DW, Sidhu SS, Winnik MA, Reilly RM (2015) MicroPET/CT imaging of patient-derived pancreatic cancer xenografts implanted subcutaneously or orthotopically in NOD-scid mice using 64Cu-NOTA-panitumumab F(ab')2 fragments. Nucl Med Biol 42:71–77
Jain RK (1990) Physiological barriers to delivery of monoclonal antibodies and other macromolecules in tumors. Cancer Res 50(Suppl 3):814s–819s
Oude Munnink TH, Dijkers E, Lub-de Hooge M, Kosterink J, Brouwers A, de Jong J, van Dongen G, de Vries E (2009) HER-2-PET imaging with 89Zr-trastuzumab in metastatic breast cancer patients. J Clin Oncol 27(Suppl 15):1045
Acknowledgments
The authors thank Deborah Scollard and Teesha Komal at the STTARR Innovation Centre for technical support. The authors also thank Dr. Sachdev Sidhu at the Toronto Recombinant Antibody Centre at the University of Toronto for providing Fab 3913.
Funding
This study was supported by a grant from the Canadian Cancer Society to RMR (Grant #704660). VF and NA received scholarships from the STARS21 strategic training program in radiation research supported by the Terry Fox Foundation. VF is supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) by the Polymer Nanoparticles for Drug Delivery (PoND) Training Program, MDS Nordion Graduate Scholarship in Radiopharmaceutical Sciences (OTSS), and William Knapp Buckley Award. VF and AK received scholarships from the Centre for Pharmaceutical Oncology at the University of Toronto. NA was supported by a fellowship with the Precision Medicine Initiative (PRiME) at the University of Toronto. AK is supported by the Queen Elizabeth II Graduate Scholarship in Science and Technology (QEII-GSST). Pertuzumab was provided by Genentech Inc. (South San Francisco, CA, USA) through a Materials Transfer Agreement with the University of Toronto.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethical Approval
All applicable institutional and/or national guidelines for the care and use of animals were followed.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 199 kb)
Rights and permissions
About this article
Cite this article
Facca, V.J., Al-saden, N., Ku, A. et al. Imaging of HER2-Positive Tumors in NOD/SCID Mice with Pertuzumab Fab-Hexahistidine Peptide Immunoconjugates Labeled with [99mTc]-(I)-Tricarbonyl Complex. Mol Imaging Biol 23, 495–504 (2021). https://doi.org/10.1007/s11307-020-01571-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11307-020-01571-z