Abstract
Purpose
The purposes of this study were to construct immortalized human bone marrow mesenchymal stem cells (UE7T-13) with overexpression of the hepatocyte nuclear factor4α (hHNF4α) and luciferase2-mKate2 dual-fusion reporter gene, further investigate their impact on treating acute liver injury (ALI) in rats, and track their biodistribution and survival by bioluminescence imaging (BLI).
Procedures
The hHNF4α and luciferase2-mKate2 genes were transduced by a lentiviral vector into UE7T-13 cells (named E7-hHNF4α-R cells), and expression was verified by immunofluorescence, RT-PCR, and flow cytometry. E7-hGFP-R cells expressing the luciferase2-mKate2/hGFP gene served as a negative group. A correlation between the bioluminescence signal and cell number was detected by BLI. The ALI rats were established and divided into three groups: PBS, E7-hGFP-R, and E7-hHNF4α-R. After transplantation of 2.0 × 106 cells, BLI was used to dynamically track their biodistribution and survival. The restoration of biological functions was assessed by serum biochemical and histological analyses.
Results
Stable high-level expression of hHNF4α and mKate2 protein was established in the E7-hHNF4α-R cells in vitro. The E7-hHNF4α-R cells strongly expressed hGFP, hHNF4α, and mKate2 proteins, and the hHNF4α gene. hGFP-mKate2 dual-positive cell expression reached approximately 93 %. BLI verified that a linear relationship existed between the cell number and bioluminescence signal (R2 = 0.9991). The cells improved liver function in vivo after transplantation into the ALI rat liver, as evidenced by the fact that AST and ALT temporarily returned to normal levels in the recipient ALI rats. The presence of the transplanted E7-hGFP-R and E7-hHNF4α-R cells in recipient rat livers was confirmed by BLI and immunohistochemistry. However, the cells were cleared by the immune system a short time after transplantation into ALI rats with a normal immune system.
Conclusion
Our data revealed that the E7-hHNF4α-R cells can transiently improve damaged liver function and were rapidly cleared by the immune system. In addition, BLI is a useful tool to track transplanted cell biodistribution and survival.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In recent years, cell transplantation has attracted increasing attention as an important supplement for liver transplantation [1,2,3]. Therefore, finding a promptly available source of hepatocyte-like cells to replace the lost or defective cells affected by end-stage liver diseases has become crucial. One attractive approach is the directed differentiation of mesenchymal stem cells (MSCs) into hepatocyte-like cells (HLCs) by overexpression of liver-enriched transcription factors (LETFs) [4,5,6]. LETFs play an important role in fetal liver maturation, hepatic differentiation, and the process of maintaining cell morphology and function [7]. In particular, hepatocyte nuclear factor (HNF) is a transcription factor that regulates liver-specific gene expression and is essential for the differentiation and maturation process in liver [8]. HNF4α contributes greatly to the liver development and maturation processes [9, 10]. HNF4α regulates the expression of a large number of functional genes that are associated with hepatocyte differentiation, glycolysis, gluconeogenesis, urea generation, liver fatty acid metabolism, bile acid synthesis, apolipoprotein drug metabolism, coagulation factors, and others and plays a critical role in maintaining the unique polarizing structure of liver [11,12,13]. Additionally, HNF4α promotes the hepatocyte benign phenotype and prevents malignant liver cell transformation [14,15,16]. In our previous study [17], we transduced a lentiviral vector containing HNF4α into UE7T-13 immortalized human BM-MSC (named E7-hHNF4α cells). The E7-hHNF4α cells acquired hepatocyte-like cell functions such as indocyanine green (ICG) uptake and release, glycogen storage, urea production, and ALB secretion and expressed some liver-specific genes, including FOXA2, ALB, CYP2B6, and AAT in vitro.
There are numerous questions regarding the viability and biology of transplanted cells in vivo, such as distribution, localization, engraftment, and repopulation. Recent developments in molecular imaging may provide an efficient and noninvasive imaging tool to monitor transplanted cells in vivo [18]. Reporter gene-bioluminescence imaging (BLI) is an optical imaging approach based on light emission and detection by specific cooled charge-coupled device (CCD) cameras and can provide qualitative and quantitative information on cell viability [19]. BLI makes use of the luciferase reporter gene to label the target cells. BLI is commonly used for tracking transplanted cell biodistribution and survival in small living animals using an in vivo imaging system (IVIS) and is appropriate and sensitive for the long-term monitoring of stem cells [20, 21].
Based on our previous studies [17, 22], we constructed E7-hHNF4α-R cells with a stable, high-level expression of the hHNF4α and luciferase2-mKate2 dual-fusion reporter gene. The GFP-mKate2-positive cells were further purified by flow cytometry. Using BLI, we showed that a linear relationship exists in vitro between the labeled cell number and the bioluminescence signal. Acute liver injury (ALI) was established in rats, and the rats were divided into three groups: PBS, E7-hGFP-R, and E7-hHNF4α-R. After transplantation of 2.0 × 106 cells, BLI was used to dynamically track the biodistribution and survival. The restoration of biological functions was assessed by serum biochemical and histological analysis.
Material and Methods
Lentivirus Production and Generation of the MSC-Luc2-mKate2-hHNF4α Cells (E7-hHNF4α-R Cells)
The dual-fusion reporter genes luciferase2 (Luc2, Promega Inc., Madison, WI, USA) and mKate2 (Evrogen Inc., Moscow, Russia) were cloned into the lentiviral vector pLenti6.3/V5-DEST according to a previous report [22]. Based on our previous study [17], the lentiviral particles (EF1α-hHNF4α-hrGFP and EF1α-hrGFP) were produced using the three-plasmid system (Invitrogen). After packaging and concentrating, the lentiviral particles containing the luciferase2-mKate2/hHNF4α-hrGFP gene were used to transfect UE7T-13 cells. After transduction, both the green fluorescent protein (GFP)- and mKate2-positive cells were expanded in vitro. The percentage of the GFP- and mKate2-positive cells was determined, and the cells were further purified by Influx™ FACS (BD Bioscience). We successfully transduced the luciferase2-mKate2 gene and HNF4α gene into UE7T-13 cells, and the percentage of both the GFP- and mKate2-positive cells increased to 90 %. These positive cells (E7-hHNF4α-R cells) were used in further experiments as a stable cell line. The E7-hGFP-R cells containing the luciferase2-mKate2/hGFP gene were used as a negative group.
Cell Culture
The immortalized human bone marrow-derived MSCs (UE7T-13 cells)were kindly provided by Pro. Mori [22, 23] and were cultured with low-glucose DMEM (HyClone) containing 10 % fetal bovine serum (FBS) (HyClone), 100 IU/ml penicillin (HyClone), and 100 mg/ml streptomycin (HyClone).The E7-hHNF4α-R cells and E7-hGFP-R cells were cultured with DMEM-F12 (Invitrogen) containing 10 % FBS, 20 ng/ml oncostatin M (OSM), and 20 ng/ml HGF (PeproTech) in 6-well plates coated with Matrigel (BD Bioscience).
Florescence-Activated Cell Sorting Analysis
Both green fluorescent protein (GFP)- and mKate2-positive cells were identified and further sorted according to the manufacturer’s instructions by fluorescence-activated cell sorting analysis (FACS, BD Biosciences).
RT-PCR Assay
Total RNA was extracted with a Total RNA Extraction kit (Sangon Biotech Co., Ltd). Real-time polymerase chain reaction (RT-PCR) was performed using the SuperScript®III One-Step RT-PCR System (Invitrogen) according to the manufacturer’s instruction. The primer sequences for the target gene HNF4α were as follows: forward, 5′-TTAGCCGGCAGTGCGTGGTG-3′; reverse, 5′-CTGGGAACGCAGCCGCTTGA-3′.The primer sequences for the positive control GAPDH were as follows: forward, 5′-CGGAGTCAACGGATTTGGTCGTAT-3′; reverse, 5′-AGCCTTCTCCATGGTGGT-3′.
Immunofluorescence Analysis
For immunofluorescent staining, the E7-hHNF4α-R and E7-hGFP-R cells were fixed with 4 % paraformaldehyde (PFA) for 20 min, washed three times with phosphate-buffered saline (PBS), and then incubated with PBS containing 0.2 % Triton X-100 (Sigma), and blocked by 3 % bovine serum albumin (BSA, Sigma) in PBS for 60 min at room temperature. Primary antibodies against HNF4α were incubated at 4 °C overnight and then washed three times with PBS followed by incubation with the appropriate secondary antibody conjugated to Alexa Fluor 594 (Invitrogen) in the dark at room temperature for 1 h. Nuclei were stained with 2-(4-amidinophenyl)-6-indolecarbamidine dihydrochloride (DAPI, Sigma).
In Vitro Bioluminescence Imaging
The E7-hHNF4α-R and E7-hGFP-R cells (2.5 × 105, 1 × 105, 5 × 104, 2.5 × 104, 1 × 104, and 5 × 103) were placed on black 48-well plates. D-luciferin substrate (150 μg/ml) was added to each well at room temperature. Cell bioluminescence signals were detected by the IVIS Spectrum Imaging System and analyzed by Carestream molecular imaging software (Caliper Life Sciences, Alameda, CA).
Establishment of the Acute Liver Injury Rat Models
Female Sprague-Dawley (SD) rats (180–220 g) were purchased from the animal center of Sun Yat-sen University (Guangzhou, China). All conditions and experimental procedures involving animals in this study were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Animal Ethics Committee of Sun Yat-sen University. The ALI rat models were induced by a single intraperitoneal injection of D-galactosamine (D-GaIN, Sigma) at 700 mg/kg [24].
Cell Transplantation and In Vivo and Ex Vivo Bioluminescence Imaging Analyses
At 24 h after D-GaIN intraperitoneal injection, the ALI rats were randomly divided into the PBS, E7-hGFP-R, and E7-hHNF4α-R groups (n = 30 for each group). All rats were anesthetized with 10 % chloral hydrate (5 μl/g) by intraperitoneal injection and underwent an abdominal incision. The prepared cells, 2 × 106 in 200 μl of PBS or PBS, were slowly administered via the superior mesenteric vein (SMV). In vivo, bioluminescence imaging was performed before transplantation and at 1, 3, 6, and 7 days after transplantation in the rats of each group. The luciferase substrate (300 mg/kg) was injected into the rats intraperitoneally. Bioluminescence signals were detected with an acquisition time of 30 s.
Biochemical Analysis
The rats were sacrificed at multiple time points (1, 3, 6, and 7 days) after cell transplantation. Blood samples were collected from the inferior vena cava of the rats, and the plasma levels of aspartate aminotransferase (AST) and alanine aminotransferase (ALT) were measured by an auto-analyzer (Hitachi 7600, Japan).
Histology and Immunohistochemistry Analysis
For histopathology evaluation, the liver tissue samples collected at multiple time points (1, 3, 6, and 7 days) were fixed for 24 h with 4 % paraformaldehyde, embedded in paraffin and then cut into 2 μm slices. The morphological changes of the liver were observed by hematoxylin and eosin (H&E) staining.
To detect the liver distribution of luciferase and ALB after transplantation, the liver tissue samples were prepared according to the method described above. After deparaffinization, rehydration, and antigen retrieval, the liver tissue sections were incubated with rabbit polyclonal anti-luciferase antibody (1:200, Abcam) or anti-albumin antibody (1:500, Bethyl) overnight at 4 °C. For primary antibody detection, the liver tissue sections were further incubated with HRP-conjugated goat anti-rabbit antibody (ZSGB-BIO, Beijing, China) and stained using 3,3-diaminobenzidine (DAB, ZSGB-BIO, Beijing, China). After being washed with PBS, the liver tissue sections were stained with hematoxylin, and the liver tissue sections were incubated by substituting PBS for the primary antibody as negative controls.
Statistical Analysis
Data are expresssed as the mean ± standard deviation (SD). Statistical analyses were performed using a statistical software (SPSS, 22.0) and Student’s t test. The difference between means was considered to be statistically significant at P < 0.05 (two-tailed).
Results
Generation of the E7-hHNF4α-R Cells with Stable High-Level Expression of the hHNF4α/luciferase2-mKate2 Gene
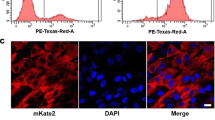
Schematic diagrams of the lentiviral vectors including E7-hHNF4α-R cells and E7-hGFP-R cells are shown in Fig. 1a. A flowchart for the generation of the E7-hHNF4α-R cells is shown in Fig. 1b and includes the following steps: lentivirus transduction, GFP-mKate2-positive cell selection, and E7-hHNF4α-R cell expansion in hepatocyte culture medium. The empty vector controls (E7-hGFP-R cells) were treated exactly as described for the E7-hHNF4α-R cells. Immunofluorescence staining showed that GFP, HNF4α, and mKate2 proteins were expressed in the E7-hHNF4α-R cells (Fig. 2a, b). Approximately, 93 % of the E7-hHNF4α-R cells expressed both GFP and mKate2 proteins as determined by flow cytometry (Fig. 2c). Reverse transcription polymerase chain reaction (RT-PCR) was performed to verify that the HNF4α mRNA was robustly expressed in the E7-hHNF4α-R cells but not in the E7-hGFP-R cells (Fig. 2d). Human primary hepatocytes were used as the positive control.
Schematic diagram of the lentiviral vectors and a flow chart of lentivirus transfection of MSCs. a Empty vector plasmid and HNF4α plasmid. b The flowchart of generation of E7-hHNF4α-R cells: lentivirus transduction, GFP-mKate2 positive cell selection, and E7-hHNF4α-R cell expansion in hepatocyte culture medium.
The successful generation of E7-hHNF4α-R cells. a, b Immunofluorescence staining showed that GFP, HNF4α, and mKate2 proteins were expressed in the E7-hHNF4α-R cells (× 200; scale: 100 μm). c Approximately 93 % of the E7-hHNF4α-R cells expressed both GFP and mKate2 proteins as determined by flow cytometry. d Reverse transcription polymerase chain reaction (RT-PCR) was performed to verify that the HNF4α mRNA was robustly expressed in the E7-hHNF4α-R cells but not in the E7-hGFP-R cells. Human primary hepatocytes were used as the positive control.
In Vitro Bioluminescence Imaging
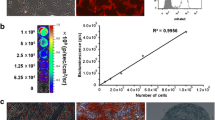
Bioluminescence imaging was used to analyze the correlation between Luc2 reporter gene activity and cell number. As shown in Fig. 3a, b, a linear relationship between the bioluminescence signal and E7-hHNF4α-R cell number was observed (R2 = 0.999). The E7-hGFP-R cells were used as the negative control.
Changes of Liver Function and Liver Pathology
At 48 h after intraperitoneal injection of D-GaIN (700 mg/kg), the plasma levels of AST and ALT were detected to evaluate liver function. As shown in ESM Fig. 1a, the plasma levels of AST and ALT following D-GaIN injection were significantly higher than those in rats without D-GaIN injection. H&E staining analysis was used to evaluate the histological changes of the liver induced by D-GaIN injection. At 48 h after D-GaIN injection, the liver tissues showed hepatic structural disorders and hepatocyte ballooning degeneration in comparison with those without D-GaIN injection (ESM Fig. 1b).
In Vivo and Ex Vivo Bioluminescence Imaging
BLI was used to dynamically detect luciferase reporter gene expression by bioluminescence light intensity, which can provide qualitative and quantitative information on cell viability in vivo. To monitor the distribution and location of the E7-hHNF4α-R cells in tissues and organs, the E7-hHNF4α-R cells labeled with the luciferase reporter gene were carefully transplanted into the ALI rat models via SMV. The negative controls were the E7-hGFP-R cells. We observed the accumulation of the E7-hHNF4α-R cells in rats by detecting bioluminescence light intensity before transplantation and at 1, 3, 6, and 7 days after transplantation. As shown in Fig. 4a, b, the intensity of the bioluminescence signal in the liver region of the rats at 1 day after transplantation was significantly higher than before transplantation (P < 0.05). At 3 and 6 days, the intensity of the bioluminescence signal gradually weakened and declined over time, and it was almost undetectable at 7 days after E7-hHNF4α-R cell transplantation in vivo. To identify the distribution of transplanted cells in specific organs of the ALI rat models, the liver, heart, spleen, lung, and kidney were taken from the body of the rats after transplantation with the E7-hHNF4α-R cells. Ex vivo bioluminescence imaging of the liver, heart, spleen, lung, and kidney was then performed immediately. As shown in Fig. 4c, d, using ex vivo BLI, the E7-hHNF4α-R cells were primarily dispersed in the liver following SMV transplantation. The results with ex vivo BLI further confirmed the results obtained with in vivo BLI. Using in vivo and ex vivo BLI, the results shown in ESM Fig. 2 show that the distribution and changes following E7-hGFP-R cell transplantation were exactly as determined in the E7-hHNF4α-R cells.
In vivo and ex vivo bioluminescence imaging of the E7-hHNF4α-R cells. a At 1, 3, 6, and 7 days after transplantation, the E7-hHNF4α-R cells accumulated in the liver region of the rats by detecting bioluminescence light intensity. b Bioluminescence signal intensity was quantitatively analyzed in vivo liver region of rats with ALI before and after E7-hHNF4α-R cells transplantation (mean ± standard deviation; n = 6; *P < 0.05, comparison with before E7-hHNF4α-R cells transplantation; #P < 0.05, comparison with the liver signal values at the same time point). c In ex vivo BLI, E7-hHNF4α-R cells mainly dispersed distribution in the liver, which further confirmed that the results in vivo BLI. d Bioluminescence signal intensity was quantitatively analyzed in ex vivo of rats with ALI (mean ± standard deviation; n = 6; *P < 0.05, comparison with before E7-hHNF4α-R cells transplantation; #P < 0.05, comparison with the liver signal values at the same time point).
Effect of the E7-hHNF4α-R Cells on the Functional Recovery of Rat Liver with ALI
To further clarify that the E7-hHNF4α-R and E7-hGFP-R cells labeled with the luciferase reporter gene were present in the liver, the liver tissues were collected 1, 3, 6, and 7 days after E7-hHNF4α-R and E7-hGFP-R cell transplantation to detect the anti-firefly luciferase by immunohistochemical staining. As shown in Fig. 5, luciferase expression in the PBS group, E7-hGFP-R cell group, and E7-hHNF4α-R cell group was observed at 1, 3, 6, and 7 days after transplantation. The brown-stained cells represent luciferase-positive cells. Luciferase-positive cells were primarily distributed near the portal vein in the liver and gradually decreased at 1, 3, and 6 days after transplantation. At 7 days, almost no brown luciferase-positive cells were observed.
To observe the effects of the E7-hHNF4α-R cells on the functional recovery of rats with ALI, the changes of plasma AST and ALT levels in the PBS group, E7-hGFP-R group, and E7-hHNF4α-R group were detected at each time point. As shown in Fig. 6a, b, the levels of plasma AST and ALT in the E7-hHNF4α-R group decreased gradually at each time point after transplantation but were significantly lower than that in the PBS group (P < 0.05); however, the difference was not statistically significant compared to that in the E7-hGFP-R group at 1 and 3 days after transplantation. The levels of plasma AST and ALT in the E7-hHNF4α-R group were lower than those in the other two groups at 6 days after transplantation, and the differences were not statistically significant. The levels of plasma AST and ALT in the E7-hGFP-R group decreased gradually at each time point after transplantation, and there was no significant difference compared to those in the other two groups. The levels of plasma AST and ALT in the three groups were close to normal at 7 days after transplantation.
Effect of the E7-hHNF4α-R cells on the functional recovery of rat liver with ALI. a, b The levels of plasma AST and ALT in E7-hHNF4α-R cells group decreased gradually at each time point after transplantation but significantly lower than that in PBS group (P < 0.05), and the difference was not statistically significant than that in E7-hGFP-R cells group at 1 and 3 days after transplantation. The levels of plasma AST and ALT in E7-hHNF4α-R cells group were lower than those in the other two groups at 6 days after transplantation, and the difference was not statistically significant. c Normal rat liver examined by H&E staining. d–f PBS group, E7-hGFP-R group, and E7-hHNF4α-R group of liver tissue showed hepatocytes swelling and vacuolar changes to some extent. In the E7-hHNF4α-R group, the regions of the cells were arranged neatly, and the area of the vacuolar degeneration cells was smaller than those of the PBS group and E7-hGFP-R group by H&E staining at 3 days after transplantation.
Liver injury in the PBS group, E7-hGFP-R group, and E7-hHNF4α-R group was analyzed by H&E staining at 3 days after transplantation. In Fig. 6c, the normal liver tissue was used as the negative control. In Fig. 6d–f, the PBS group, E7-hGFP-R group, and E7-hHNF4α-R group of liver tissues show hepatocyte swelling and vacuolar changes to some extent. In the E7-hHNF4α-R group, the regions of cells were arranged precisely, and the areas of vacuolar degeneration were smaller than those of the PBS group and E7-hGFP-R group.
Discussion
Mesenchymal stem cells (MSCs) are widely used in clinical studies in the treatment of immune diseases such as graft-versus-host disease (GVHD) and Crohn’s disease [25, 26]. In recent years, MSCs have been attracting increasing attention in tissue repair and cell transplantation replacement therapy. Differentiation of MSCs into hepatocyte-like cells has great potential for cell therapy of liver diseases. However, the low efficiency and low reproducibility of MSCs in liver differentiation are still the biggest obstacle to its clinical use [27, 28].
Previous studies have shown that overexpression of LETFs plays an important role in the regulation of MSC differentiation [5, 6, 29]. A variety of LETFs plays an important role in the process of liver development and maturation. Hepatocyte nuclear factor (HNF) belongs to a class of transcription factors that regulate the expression of liver-specific genes, which is essential for the development and maturation of the liver. Among the six members of the HNF family, HNF4α plays a key role in the differentiation and maturation of hepatocytes [30]. Studies have shown that HNF4α begins to be expressed in the early stages of embryonic liver development and is present throughout the entire development of the liver. HNF4α is highly expressed in the differentiated mature liver and is the key transcription protein in regulating hepatocyte differentiation and maintaining hepatocyte biology [9]. HNF4α regulates the expression of a large number of liver function genes, including genes associated with hepatocyte differentiation, glycolysis, gluconeogenesis, urea production, fatty acid metabolism, cholic acid synthesis, drug metabolism apolipoprotein, and coagulation factors [11,12,13]. Hepatocytes lacking HNF4α are small and round, exhibit deep cytoplasmic staining and almost no glycogen, cannot express protein molecules that are associated with cell connections, and produce cells that are unable to form structures such as sinusoids [10]. Studies have shown that HNF4α plays a critical role as a “switch” in bone marrow-derived hematopoietic stem cells that transdifferentiate into hepatocytes [31]. These findings strongly suggest that HNF4α can safely and efficiently promote MSC differentiation into hepatocytes and ensure that the cells function as normal hepatocytes.
In our previous study [17], we successfully transduced HNF4α into hMSCs (UE7T-13 cells). E7-hHNF4α cells carried out partial hepatocyte-like functions such as uptake and release of ICG, glycogen storage, urea secretion, and ALB secretion in vitro and exhibited phenobarbital-induced activation of the CYP3A4 gene, suggesting that E7-hHNF4α cells have potential for drug development.
However, in vitro experimental conditions for stem cell proliferation and differentiation do not reflect the in vivo state. At present, little is known about cell proliferation, migration, differentiation, and outcomes after stem cell transplantation in vivo, and current research remains in the exploratory stage, which is another major obstacle to the clinical application of stem cells. Therefore, it is important to promote MSC clinical applications by conducting in-depth research into cell proliferation, migration, distribution, and differentiation after transplantation of living stem cells.
Bioluminescence imaging (BLI) is a type of optical imaging that provides quantitative information about living cells by detecting the expression of the luciferase reporter gene by using a noninvasive live imaging system (IVIS). Luciferase2-mKate2 [22, 32], which is a dual-reporter gene for firefly luciferase and red fluorescent protein (mKate2) carried by a nonspecific cytomegalovirus enhancer/promoter (CMV), was used in this study. In addition, GFP is a fluorescent reporter gene that we chose for cell labeling, which is primarily used for tracing and sorting E7-hHNF4α cells. Therefore, E7-hHNF4α cells harboring the luciferase2-mKate2 dual-fusion reporter gene can stably and continuously express firefly luciferase and red fluorescent protein. The former can be used for deep tissue and organ imaging of small animals and is more advantageous for in vivo tracing such as dynamic tracing after cell transplantation, while the latter can be used to determine whether the luciferase2-mKate2 dual-fusion reporter gene was expressed and to perform flow cytometry on cells expressing the luciferase2-mKate2 dual-fusion reporter gene. In our study, the E7-hHNF4α-R cells were successfully constructed with a stable and high expression of the HNF4α and luc2-mKate2 genes. In vitro, BLI confirmed that there was a positive correlation between the number of the E7-hHNF4α-R cells and E7-hGFP-R cells and their bioluminescence signals. The preparation allows for the dynamic tracking and quantitative study of living cells after cell transplantation. A previous study with the mouse [22, 32] had explored the quantitative relationship between the bioluminescent signal intensity and the number of cells of immortalized human mesenchymal stem cells (UE7T-13) infected with the luciferase2-mKate2 dual-fusion reporter gene in vitro and in vivo. In our study, in vivo and ex vivo bioluminescence imaging showed that the E7-hHNF4α-R cells were distributed into the liver via the SMV. On the first day after transplantation, the intensity of the bioluminescence signal in the liver region of the rats was significantly higher than before transplantation. The signal intensity gradually decreased after 3 and 6 days and was almost undetectable after 7 days.
On the first and third days after transplantation, the plasma AST and ALT levels in the liver area of the rats were significantly lower than in the PBS group and were lower than in the E7-hGFP-R group, but the difference was not statistically significant. Current research suggests that short-term liver function improvement after embryonic stem cell (ES) and MSC transplantation may be related to paracrine effects of the transplanted cells and immunomodulatory effects [33,34,35,36]. The E7-hHNF4α-R cells in this study are hepatocyte-like cells with partially mature liver cell function, as we described in detail in our previous study [17]. The effectiveness of liver transplantation for the treatment of acute and chronic liver injury in animals after successful induction in vitro to a functional hepatocyte-like cell has also been demonstrated in other studies [37,38,39]. This short-term liver function improvement with stem cell transplantation is an effective method to save the lives of fulminant liver failure (FLF) patients and prolong survival during the period of waiting for liver transplantation [40]. However, the E7-hHNF4α-R cells were rapidly cleared by the immune system (7 days) after being transplanted into ALI rats with a normal immune system and can temporarily improve the damaged liver function. A large number of previous studies have also confirmed that, after transplantation, MSCs are recognized and cleared by the immune system of the recipient and have difficulty surviving in the recipients for long periods of time. One week after stem cell transplantation, it was difficult to trace the exogenous transplanted cells in the liver in vivo. Conversely, in the case of human bone marrow mesenchymal stem cell (hMSC) xenografts for the treatment of mouse/rat liver injury, the immunological rejection and clearance of MSCs after transplantation were more pronounced [32, 34, 41].
There are several limitations to this study. First, the E7-hHNF4α-R cells were studied only in ALI rats with a normal immune system, and further investigations were conducted in an immunodeficient rodent model and chronic liver injury model such as the liver fibrosis model. Second, the long-term fate of the transplanted E7-hHNF4α-R cells in vivo is unclear. Third, it is necessary to further improve the generation efficiency of functional hepatocyte-like cells from MSCs by overexpression of liver-enriched transcription factors.
Conclusion
E7-hHNF4α-R cells were successfully obtained by lentivirus transfection with HNF4α overexpression in human MSCs and with high expression of the HNF4α and luc2-mKate2 dual-fusion reporter genes. Our previous study showed that the E7-hHNF4α cells exhibited partial hepatocyte-like functions and gene expression of mature liver markers. In vitro, BLI confirmed that there was a positive correlation between the number of the E7-hHNF4α-R cells and E7-hGFP-R cells and their bioluminescence signals. In vivo and ex vivo bioluminescence imaging showed that the E7-hHNF4α-R cells were distributed into the liver via the SMV. However, E7-hHNF4α-R cells were rapidly cleared by the immune system (7 days) after being transplanted into ALI rats with a normal immune system and improved the damaged liver function in a short period of time.
References
Wang P, Petrella F, Nicosia L et al (2016) Molecular imaging of stem cell transplantation for liver diseases: monitoring, clinical translation, and theranostics. Stem Cells Int 2016:4058656
Forbes SJ, Gupta S, Dhawan A (2015) Cell therapy for liver disease: from liver transplantation to cell factory. J Hepatol 62:S157–S169
Colmenero J, Sancho-Bru P (2017) Mesenchymal stromal cells for immunomodulatory cell therapy in liver transplantation: one step at a time. J Hepatol 67:7–9
Huang P, Zhang L, Gao Y, He Z, Yao D, Wu Z, Cen J, Chen X, Liu C, Hu Y, Lai D, Hu Z, Chen L, Zhang Y, Cheng X, Ma X, Pan G, Wang X, Hui L (2014) Direct reprogramming of human fibroblasts to functional and expandable hepatocytes. Cell Stem Cell 14:370–384
Chen ML, Lee KD, Huang HC, Tsai YL, Wu YC, Kuo TM, Hu CP, Chang C (2010) HNF-4αlpha determines hepatic differentiation of human mesenchymal stem cells from bone marrow. World J Gastroenterol 16:5092–5103
Cho JW, Lee CY, Ko Y (2012) Therapeutic potential of mesenchymal stem cells overexpressing human forkhead box A2 gene in the regeneration of damaged liver tissues. J Gastroenterol Hepatol 27:1362–1370
Costa RH, Kalinichenko VV, Holterman AX, Wang X (2003) Transcription factors in liver development, differentiation, and regeneration. Hepatology 38:1331–1347
Schrem H, Klempnauer J, Borlak J (2002) Liver-enriched transcription factors in liver function and development. Part I: the hepatocyte nuclear factor network and liver-specific gene expression. Pharmacol Rev 54:129–158
Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ (2001) Hepatocyte nuclear factor 4αlpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol 21:1393–1403
Parviz F, Matullo C, Garrison WD, Savatski L, Adamson JW, Ning G, Kaestner KH, Rossi JM, Zaret KS, Duncan SA (2003) Hepatocyte nuclear factor 4αlpha controls the development of a hepatic epithelium and liver morphogenesis. Nat Genet 34:292–296
Gonzalez FJ (2008) Regulation of hepatocyte nuclear factor 4 alpha-mediated transcription. Drug Metab Pharmacokinet 23:2–7
Chiba H, Gotoh T, Kojima T, Satohisa S, Kikuchi K, Osanai M, Sawada N (2003) Hepatocyte nuclear factor (HNF)-4αlpha triggers formation of functional tight junctions and establishment of polarized epithelial morphology in F9 embryonal carcinoma cells. Exp Cell Res 286:288–297
Satohisa S, Chiba H, Osanai M, Ohno S, Kojima T, Saito T, Sawada N (2005) Behavior of tight-junction, adherens-junction and cell polarity proteins during HNF-4αlpha-induced epithelial polarization. Exp Cell Res 310:66–78
Yin C, Lin Y, Zhang X, Chen YX, Zeng X, Yue HY, Hou JL, Deng X, Zhang JP, Han ZG, Xie WF (2008) Differentiation therapy of hepatocellular carcinoma in mice with recombinant adenovirus carrying hepatocyte nuclear factor-4αlpha gene. Hepatology 48:1528–1539
Lazarevich NL, Cheremnova OA, Varga EV, Ovchinnikov DA, Kudrjavtseva EI, Morozova OV, Fleishman DI, Engelhardt NV, Duncan SA (2004) Progression of HCC in mice is associated with a downregulation in the expression of hepatocyte nuclear factors. Hepatology 39:1038–1047
Ning BF, Ding J, Liu J, Yin C, Xu WP, Cong WM, Zhang Q, Chen F, Han T, Deng X, Wang PQ, Jiang CF, Zhang JP, Zhang X, Wang HY, Xie WF (2014) Hepatocyte nuclear factor 4αlpha-nuclear factor-kappaB feedback circuit modulates liver cancer progression. Hepatology 60:1607–1619
Hu X, Xie P, Li W, Li Z, Shan H (2016) Direct induction of hepatocyte-like cells from immortalized human bone marrow mesenchymal stem cells by overexpression of HNF4αlpha. Biochem Biophys Res Commun 478:791–797
Rizzo S, Petrella F, Politi LS, Wang P (2017) Molecular imaging of stems cells: in vivo tracking and clinical translation. Stem Cells Int 2017:1783841
Mezzanotte L, van ‘t Root M, Karatas H et al (2017) In vivo molecular bioluminescence imaging: new tools and applications. Trends Biotechnol 35:640–652
Chen IY, Greve JM, Gheysens O, Willmann JK, Rodriguez-Porcel M, Chu P, Sheikh AY, Faranesh AZ, Paulmurugan R, Yang PC, Wu JC, Gambhir SS (2009) Comparison of optical bioluminescence reporter gene and superparamagnetic iron oxide MR contrast agent as cell markers for noninvasive imaging of cardiac cell transplantation. Mol Imaging Biol 11:178–187
Parashurama N, Ahn BC, Ziv K, Ito K, Paulmurugan R, Willmann JK, Chung J, Ikeno F, Swanson JC, Merk DR, Lyons JK, Yerushalmi D, Teramoto T, Kosuge H, Dao CN, Ray P, Patel M, Chang YF, Mahmoudi M, Cohen JE, Goldstone AB, Habte F, Bhaumik S, Yaghoubi S, Robbins RC, Dash R, Yang PC, Brinton TJ, Yock PG, McConnell MV, Gambhir SS (2016) Multimodality molecular imaging of cardiac cell transplantation: part I. reporter gene design, characterization, and optical in vivo imaging of bone marrow stromal cells after myocardial infarction. Radiology 280:815–825
Li Z, Hu X, Mao J, Liu X, Zhang L, Liu J, Li D, Shan H (2015) Optimization of mesenchymal stem cells (MSCs) delivery dose and route in mice with acute liver injury by bioluminescence imaging. Mol Imaging Biol 17:185–194
Mori T, Kiyono T, Imabayashi H, Takeda Y, Tsuchiya K, Miyoshi S, Makino H, Matsumoto K, Saito H, Ogawa S, Sakamoto M, Hata JI, Umezawa A (2005) Combination of hTERT and bmi-1, E6, or E7 induces prolongation of the life span of bone marrow stromal cells from an elderly donor without affecting their neurogenic potential. Mol Cell Biol 25:5183–5195
Lam SP, Luk JM, Man K, Ng KTP, Cheung CK, Rose-John S, Lo CM (2010) Activation of interleukin-6-induced glycoprotein 130/signal transducer and activator of transcription 3 pathway in mesenchymal stem cells enhances hepatic differentiation, proliferation, and liver regeneration. Liver Transpl 16:1195–1206
Ciccocioppo R, Corazza GR (2016) Mesenchymal stem cells for fistulising Crohn’s disease. Lancet 388:1251–1252
Lim JY, Ryu DB, Lee SE, Park G, Min CK (2017) Mesenchymal stem cells (MSCs) attenuate cutaneous sclerodermatous graft-versus-host disease (Scl-GVHD) through inhibition of immune cell infiltration in a mouse model. J Invest Dermatol 137:1895–1904
di Bonzo LV, Ferrero I, Cravanzola C, Mareschi K, Rustichell D, Novo E, Sanavio F, Cannito S, Zamara E, Bertero M, Davit A, Francica S, Novelli F, Colombatto S, Fagioli F, Parola M (2008) Human mesenchymal stem cells as a two-edged sword in hepatic regenerative medicine: engraftment and hepatocyte differentiation versus profibrogenic potential. Gut 57:223–231
Fiore EJ, Mazzolini G, Aquino JB (2015) Mesenchymal stem/stromal cells in liver fibrosis: recent findings, old/new caveats and future perspectives. Stem Cell Rev 11:586–597
Ishii K, Yoshida Y, Akechi Y, Sakabe T, Nishio R, Ikeda R, Terabayashi K, Matsumi Y, Gonda K, Okamoto H, Takubo K, Tajima F, Tsuchiya H, Hoshikawa Y, Kurimasa A, Umezawa A, Shiota G (2008) Hepatic differentiation of human bone marrow-derived mesenchymal stem cells by tetracycline-regulated hepatocyte nuclear factor 3beta. Hepatology 48:597–606
Li J, Ning G, Duncan SA (2000) Mammalian hepatocyte differentiation requires the transcription factor HNF-4αlpha. Genes Dev 14:464–474
Khurana S, Jaiswal AK, Mukhopadhyay A (2010) Hepatocyte nuclear factor-4αlpha induces transdifferentiation of hematopoietic cells into hepatocytes. J Biol Chem 285:4725–4731
Liu JJ, Hu XJ, Li ZR, Yan RH, Li D, Wang J, Shan H (2017) In vivo bioluminescence imaging of transplanted mesenchymal stromal cells and their rejection mediated by intrahepatic NK cells. Mol Imaging Biol 19:31–40
Haga H, Yan IK, Takahashi K, Matsuda A, Patel T (2017) Extracellular vesicles from bone marrow-derived mesenchymal stem cells improve survival from lethal hepatic failure in mice. Stem Cells Transl Med 6:1262–1272
Kanazawa H, Fujimoto Y, Teratani T, Iwasaki J, Kasahara N, Negishi K, Tsuruyama T, Uemoto S, Kobayashi E (2011) Bone marrow-derived mesenchymal stem cells ameliorate hepatic ischemia reperfusion injury in a rat model. PLoS One 6:e19195
Higashiyama R, Inagaki Y, Hong YY, Kushida M, Nakao S, Niioka M, Watanabe T, Okano H, Matsuzaki Y, Shiota G, Okazaki I (2007) Bone marrow-derived cells express matrix metalloproteinases and contribute to regression of liver fibrosis in mice. Hepatology 45:213–222
van Poll D, Parekkadan B, Cho CH, Berthiaume F, Nahmias Y, Tilles AW, Yarmush ML (2008) Mesenchymal stem cell-derived molecules directly modulate hepatocellular death and regeneration in vitro and in vivo. Hepatology 47:1634–1643
Christ B, Dollinger MM (2011) The generation of hepatocytes from mesenchymal stem cells and engraftment into the liver. Curr Opin Organ Transplant 16:69–75
Basma H, Soto-Gutierrez A, Yannam GR et al (2009) Differentiation and transplantation of human embryonic stem cell-derived hepatocytes. Gastroenterology 136:990–999
Su B, Liu C, Xiang D, Zhang HB, Yuan SM, Wang MJ, Chen F, Zhu HY, He ZY, Wang X, Hu YP (2011) Xeno-repopulation of fah −/− Nod/Scid mice livers by human hepatocytes. Sci China Life Sci 54:227–234
Herrera MB, Fonsato V, Bruno S, Grange C, Gilbo N, Romagnoli R, Tetta C, Camussi G (2013) Human liver stem cells improve liver injury in a model of fulminant liver failure. Hepatology 57:311–319
Chamberlain G, Fox J, Ashton B, Middleton J (2007) Concise review: mesenchymal stem cells: their phenotype, differentiation capacity, immunological features, and potential for homing. Stem Cells 25:2739–2749
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant Nos. 81201090, 81371554, 81371655, 81071206,81430041,81172193,81201090, U1032002, and 81070349), Guangdong Natural Science Foundation (S2012010008367), and Medical Scientific Research Foundation of Guangdong Province (NO. A2015109).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declared that they have no conflict of interest.
Electronic Supplementary Material
ESM 1
(PDF 407 kb)
Rights and permissions
About this article
Cite this article
Xie, P., Hu, X., Li, D. et al. Bioluminescence Imaging of Transplanted Mesenchymal Stem Cells by Overexpression of Hepatocyte Nuclear Factor4α: Tracking Biodistribution and Survival. Mol Imaging Biol 21, 44–53 (2019). https://doi.org/10.1007/s11307-018-1204-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11307-018-1204-0