Abstract
Purpose
In this study, we compared firefly luciferase (Fluc) reporter gene and superparamagnetic iron oxide (Feridex) as cell markers for longitudinal monitoring of cardiomyoblast graft survival using optical bioluminescence imaging (BLI) and magnetic resonance imaging (MRI), respectively.
Procedures
Rats (n = 31) underwent an intramyocardial injection of cardiomyoblasts (2 × 106) labeled with Fluc, Feridex, or no marker (control) or an injection of Feridex alone (75 μg). Afterward, rats were serially imaged with BLI or MRI and killed at different time points for histological analysis.
Results
BLI revealed a drastically different cell survival kinetics (half-life = 2.65 days over 6 days) than that revealed by MRI (half-life = 16.8 days over 80 days). Injection of Feridex alone led to prolonged tissue retention of Feridex (≥16 days) and persistent MR signal (≥42 days).
Conclusions
Fluc BLI reporter gene imaging is a more accurate gauge of transplanted cell survival as compared to MRI of Feridex-labeled cells.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Transplantation of various cell types (e.g., neonatal cardiomyocytes, bone-marrow-derived mesenchymal stem cells, and skeletal myoblasts) into the ischemic or failing heart has been shown to improve cardiac function in pre-clinical models of myocardial infarction (MI) [1]. The same degree of success, however, has yet to be translated into humans, as recent double-blind, randomized clinical trials using different population of autologous bone-marrow-derived stem cells or circulatory progenitor cells have failed to show significant long-term left ventricular functional improvement in MI patients compared to placebo [2]. Included in the list of possible factors preventing clinical success are poor engraftment, survival, and differentiation of implanted stem cells. However, these variables cannot be feasibly investigated in the clinical setting given the inability to utilize post-mortem histological techniques, which are a mainstay for pre-clinical animal studies [3]. Conceivably, the development of noninvasive imaging techniques to monitor the location, long-term viability, and differentiation status of implanted cells could help facilitate and objectify the assessment of stem cell and other cell therapies in humans [4–6].
Many investigators, including our group, are striving to develop noninvasive imaging techniques to monitor the fate of stem cells after transplantation into living subjects using a variety of dedicated small animal imaging modalities, including micropositron emission tomography (microPET), microsingle photon emission computed tomography (microSPECT), optical bioluminesence imaging (BLI), and magnetic resonance imaging (MRI) [7–9]. The hope is that some of these imaging modalities, in particular MRI, microSPECT, and microPET (which all have counterparts in the clinics), can be translated for use in humans to better evaluate the long-term efficacy of stem cell therapy. While the choice of imaging modality is an important consideration because it principally dictates the imaging sensitivity and spatial resolution, the selection of a cell marker is also critical because it (1) can affect the sensitivity of an imaging assay, (2) may or may not adversely affect cellular function, and (3) can influence how faithfully the imaging signal reflects cellular status (e.g., cell death, proliferation).
Two categories of cell markers have generally been used to label stem cells: reporter genes and chemical-based contrast agents. The former group is exemplified by firefly luciferase (Fluc), herpes simplex virus thymidine kinase, and ferritin for BLI, PET, and MR imaging, respectively [7, 10], whereas the latter group includes superparamagnetic iron oxide (SPIO), 18F-FDG, and 111In-oxine for MRI, PET, and SPECT imaging, respectively [8, 9, 11]. Currently, it is unclear which category of cell marker is more useful for noninvasive imaging of cardiac cell transplantation. The goal of this study was to assess and compare the use of two popular cell markers (one from each category), Fluc reporter gene, and SPIO MR contrast agent (Feridex), with the hope that the findings on the former could be later extrapolated to the more clinically relevant PET reporter genes that we and others have previously validated [7, 12, 13]. Specifically, we aimed (1) to investigate whether these cell markers significantly alter cell morphology and growth in cell culture and (2) to identify the more suitable cell marker for serial monitoring of cell survival following transplantation.
Materials and Methods
Creation of a Stable H9c2 Cardiomyoblast Cell Line Expressing Firefly Luciferase
Using Lipofectamine2000 (Invitrogen, Carlsbad, CA, USA) transfection reagent (1 μg DNA/2 μl Lipofectamine2000), H9c2 rat embryonic cardiomyoblasts (American Type Culture Collection; ATCC, Manassas, VA, USA) were transfected with plasmids (pCMV-Fluc-SV40-neo) expressing Fluc under the regulation of a cytomegalovirus (CMV) promoter and selected for neomysin resistance using Geneticin/G418 Sulfate (500 mg/ml; Invitrogen). After plating at low density and allowing time for individual cells to grow into colonies, single clones of cells were picked, re-plated, and grown to significant amount for in vitro assessment of Fluc expression using a 20/20n luminometer (Turner BioSystems, Sunnyvale, CA, USA) as previously described [14]. The highest Fluc expressor of five clones was passaged repeatedly for 3 months before the cells were used for either in vitro assays or transplantation into rat myocardium.
Labeling of H9c2 Cardiomyoblasts with Superparamagnetic Iron Oxide Nanoparticles
Parental H9c2 rat embryonic cardiomyoblasts with a similar passage number as the stable Fluc-expressing cells were incubated with a mixture of SPIO (Feridex IV, Advanced Magnetics, Cambridge, MA, USA) and Lipofectamine2000 at a ratio of 100 μg Fe/30 μg Lipofectamine2000 suspended in Opti-MEM (Invitrogen). Twenty-four hours later, the cells were harvested by trypsinization (0.05% Trypsin-EDTA, Invitrogen), washed three times with phosphate-buffered saline (PBS; Invitrogen), and counted using an automatic cell counter (Cellometer AutoT4™, Nexcelom Bioscience, Lawrence, MA, USA) before plating for either assessment of cell labeling, measurement of cell growth, or implantation into animals.
Assessment of Cell Labeling and Its Effect on Cell Growth
Fluc-, Feridex-, and non-labeled parental H9c2 cells cultured in supplemented Dulbecco’s Modified Eagle’s Medium (10% fetal bovine serum, 1% penicillin/streptomycin; Invitrogen) were harvested by trypsinization, washed three times with PBS, and re-plated in 35-mm glass bottom dishes (MatTek, Ashland, MA, USA) at a concentration of 104 cells/2 ml of medium for histological confirmation of cell labeling and morphology or in six-well plates at a concentration of 5 × 103 cells/ml of medium/well for assessment of cell growth.
For staining of cell markers, all cells were first fixed in 10% neutral-buffered formalin supplemented with 0.1% Triton X-100 (Sigma-Aldrich, St. Louis, MO, USA) for 5 min and washed with PBS. Fluc-labeled cells were stained for Fluc expression by incubation with goat anti-firefly luciferase, horseradish peroxidase (HRP)-conjugated polyclonal antibody (GeneTex, San Antonio, TX, USA), followed by visualization with DAB substrate (Pierce, Rockford, IL, USA). Feridex-labeled cells were stained for iron uptake using Prussian Blue solution containing equal volumes of 10% HCl (Sigma-Aldrich) and 10% potassium ferricyanide (Mallinckrodt, Hazelwood, MO, USA). Unlabeled cells were stained for either iron or firefly luciferase and used as negative controls. All cells were visualized and imaged under bright field at ×400 magnification.
For assessment of cell growth following plating, labeled and unlabeled cells in triplicate wells were harvested at different time points (e.g., days 0, 1, 2, and 3) by trypsinization, resuspended in PBS, and counted using an automatic cell counter. For analysis of doubling time, a population growth model described by the equation \(N_t = N_0 2^{{t \mathord{\left/ {\vphantom {t {t_d }}} \right. \kern-\nulldelimiterspace} {t_d }}} \) was assumed, where t, N t , N 0, and t d represent time (day), cell number at time t, cell number at time 0 (i.e., 24 h post plating), and doubling time, respectively. Linear regression was performed on a plot of natural log of N t versus time, and the doubling time was extracted from the slope of the linear regression line (i.e., \(t_d = {{\ln \left( 2 \right)} \mathord{\left/ {\vphantom {{\ln \left( 2 \right)} {{\text{slope}}}}} \right. \kern-\nulldelimiterspace} {{\text{slope}}}}\)).
Animal Groups and Study Design
All animal studies were performed according to the protocols approved by the Stanford Institutional Animal Care and Use Committee (IACUC). Four groups of Sprague–Dawley rats (n = 31; 250–300 g; Charles River Laboratories, Wilmington, MA, USA) underwent aseptic left thoracotomy, followed by an intramyocardial injection of cells or free Feridex. Group 1 rats (n = 4) received 2 × 106 Fluc-labeled cells and were imaged daily with BLI for 6 days. Group 2 rats (n = 11) received 2 × 106 Feridex-labeled cells and were either imaged longitudinally with MRI for 80 days (n = 2) or killed on days 2, 9, and 16 for post-mortem histological analysis (n = 3/time point). Group 3 rats (n = 4) received 2 × 106 non-labeled cells and were either imaged on day 1 with BLI (n = 2) or day 2 with MRI (n = 2). Group 4 rats (n = 12) received 75 μg of free Feridex and were either imaged longitudinally with MRI for 42 days (n = 3) or killed on days 2, 9, and 16 for histological analysis (n = 3/time point).
Animal Surgeries for Direct Cell or Label Injections
Rats were induced with isoflurane (3–4%), orotracheally intubated, and ventilated with 2–3% isoflurane in 100% oxygen using Harvard (Cambridge, MA, USA) rodent ventilator. Following a left lateral thoracotomy, animals received an intramyocardial injection of either cells or free Feridex diluted in 40 μl PBS at one site into the anterolateral myocardial wall. After injection, the chest was closed in layers, and pneumothorax was evacuated by suction using a 16-G angiocath. Animals received subcutaneous buprenorphene (0.1 mg/kg) for post-operative analgesia and were recovered in a warmed chamber with 100% oxygen.
Optical Bioluminescence Imaging of Living Rats
Sprague–Dawley rats that received Fluc- or non-labeled H9c2 cells underwent BLI daily for 6 days using the In Vivo Imaging System (IVIS 100; Xenogen, Hopkinton, MA, USA). The imaging protocol consisted of six to eight consecutive 5-min scans after intraperitoneal injection of d-Luciferin (375 mg/kg; Biosynth International, Naperville, IL, USA). Region of interest (ROI) analysis using Living Image 2.50 (Xenogen) was performed for each scan to derive the peak heart signal in terms of maximum radiance (p/s/cm2/sr). The daily ROI-derived signal, averaged over all animals, was used to construct an optical cell signal intensity plot (Fig. 4a). The half-life (\(t_{{1 \mathord{\left/ {\vphantom {1 2}} \right. \kern-\nulldelimiterspace} 2}} \)) of the signal decay was obtained by curve fitting the data points with an exponential function, \(y = ae^{ - bx} \), where a is a constant and \(b = {{\left( {\ln 2} \right)} \mathord{\left/ {\vphantom {{\left( {\ln 2} \right)} {t_{{1 \mathord{\left/ {\vphantom {1 2}} \right. \kern-\nulldelimiterspace} 2}} }}} \right. \kern-\nulldelimiterspace} {t_{{1 \mathord{\left/ {\vphantom {1 2}} \right. \kern-\nulldelimiterspace} 2}} }}\).
Magnetic Resonance Imaging of Living Rats
Cardiac-gated MRI of Sprague–Dawley rats that received either Feridex-labeled H9c2 cells or free Feridex was performed using a 4.7 T small animal MR scanner (Varian, Palo Alto, CA, USA) with a 6-cm inner diameter, quadrature, transmit/receive radio-frequency coil. For each imaging session, a multi-slice gradient-recalled echo (GRE) sequence was used to visualize consecutive short-axis heart slices containing the Feridex-induced signal. The imaging parameters were: Matrix = 128 × 128, TR = 160 ms, TE = 2.5 ms, number of slices = 12, slice thickness = 1 mm, FOV = 6 × 6 cm2, flip angle = 30°. For signal quantification of short-axis images, MRVision (MRVision, Menlo Park, CA, USA) was used to draw a ROI for the Feridex-induced signal, the interventricular septum (normal myocardial tissue), and the background to calculate the average target and septum signals and the standard deviation (SD) of the background noise, respectively. The absolute difference between the average signal intensities of the target and septum, multiplied by the size of the target ROI, was calculated as the area-integrated target signal for each slice. Summing of the area-integrated signal over all 2D slices containing the Feridex-induced signal yielded the final volume-integrated target signal, which was subsequently averaged over all animals and specifically used to construct a MR cell signal intensity plot (Fig. 4b). The half-life (\(t_{{1 \mathord{\left/ {\vphantom {1 2}} \right. \kern-\nulldelimiterspace} 2}} \)) of the signal decay was determined by curve fitting the data points with an exponential function, \(y = ae^{ - bx} \), where a is a constant and \(b = {{\left( {\ln 2} \right)} \mathord{\left/ {\vphantom {{\left( {\ln 2} \right)} {t_{1/2} }}} \right. \kern-\nulldelimiterspace} {t_{1/2} }}\). The contrast-to-noise ratio (CNR) of the target signal was calculated by dividing the volume-integrated signal by both the volume of the Feridex signal (in terms of the number of voxels) and the SD of the background.
Ex Vivo Histology of Hearts Injected with Feridex-Labeled Cells or Free Feridex
On days 2, 9, and 16 after injection of either Feridex-labeled H9c2 cells or free Feridex, rats were killed, and their hearts were excised. The hearts were sliced into three 2–3-mm-thick pieces along the short axis, fixed in 10% neutral-buffered formalin over night, and embedded in paraffin. Ten-micrometer slices were stained for iron using aforementioned Prussian Blue staining protocol, counterstained with hematoxylin–eosin (H&E), and visualized by light microscopy at ×400 magnification.
Statistical Analysis
All data are presented as mean ± standard deviation (SD). The unpaired, two-tailed Student’s t test was used for comparison between two measurements (e.g., mean optical/MR signal of the experimental group versus that of the control group; mean optical/MR signal on a given day versus that on the first day; slopes of two linear curve fits for comparison of doubling times). P values of less than 0.05 were considered statistically significant.
Results
Cell Labeling and Its Effect on Cell Growth in Culture
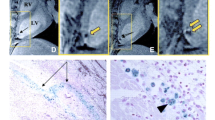
To confirm that cells (H9c2 cardiomyoblasts) could be labeled with either Fluc or Feridex, we performed immunohistochemistry and Prussian Blue staining on Fluc- and Feridex-labeled cells, respectively. Non-labeled cells were used as negative controls. Following incubation with HRP-conjugated antibodies specifically targeting the firefly luciferase enzyme (FL) and the substrate DAB, only Fluc-labeled cells were found to express a high level of FL in the cytoplasm in a peri-nuclear pattern (Fig. 1a). After staining with Prussian Blue, only Feridex-labeled cells were found to have cytosolic accumulation of iron, indicating an effective uptake of Feridex by cells via Lipofectamine2000-assisted transfection (Fig. 1b). Under bright field microscopy, the gross morphology of Fluc-labeled cells was similar to that of non-labeled cells. In contrast, the Feridex-labeled cells were found to contain cytoplasmic vacuoles.
Bright field microscopy and histological examination of cells labeled with either Fluc or Feridex, compared to non-labeled cells. Under ×400 magnification, Fluc- (a, right, Fluc+), Feridex-(b, right, Feridex+), and non-labeled (a, left, Control; b, left, Control) H9c2 cells appear to have similar gross morphology, except for significant cytoplasmic vacuolization in Feridex-labeled cells (red arrow). Immunohistochemistry of Fluc-labeled cells reveals positive cytoplasmic brown DAB stains in an annular peri-nuclear pattern (blue arrow; a, right), indicating the presence of Fluc enzymes. Only negligible cytoplasmic background DAB stains are seen for non-labeled cells (a, left). Prussian Blue iron staining of Feridex-labeled cells shows cytoplasmic deposition of blue crystals, suggesting significant Feridex uptake (blue arrow; b, right), whereas no background stain is seen for non-labeled cells (b, left). Asterisks indicate the location of the nucleus.
To study whether these cell markers could affect cell proliferation in culture, labeled and non-labeled cells in triplicates were plated and allowed to grow over a period of 3 days. Cell counting on day 0 with an automatic cell counter revealed no statistical difference in cell count among all cell groups, confirming comparable plating or cell attachment (Fig. 2). From day 1 to 3, the Fluc-labeled cell group was associated with a significantly lower cell count than non-labeled group (P < 0.05), suggesting a persistent effect of Fluc labeling on cell proliferation. In comparison, the effect of Feridex-labeling appeared to be transient in that the cell count for the Feridex-labeled group was initially statistically significantly lower than that of non-labeled group on days 1 and 2 (P < 0.05) but no longer on day 3. The resulting doubling times obtained from curve fitting using linear regression were found to be 1.41, 1.07, and 1.16 days for the Fluc-, Feridex-, and non-labeled cell groups, respectively. Only the doubling time of Fluc-labeled cells, and not Feridex-labeled cells, was found to be statistically significantly different from that of non-labeled cells (P < 0.05).
Cell growth assay to assess the effect of firefly luciferase or Feridex labeling on doubling time. Fluc- (white solid bars), Feridex- (striped bars), and non-labeled cells (black solid bars) were plated in triplicates in six-well plates for 24 h and allowed to grow for 3 days. Cells were counted using an automatic cell counter on days 0 (24 h post plating), 1, 2, and 3, and the average cell count was plotted against time (day). For any given day, asterisks denote statistical significance (P < 0.05) compared to the control non-labeled group.
Longitudinal Optical Bioluminescence Imaging of Cell Survival
To assess the feasibility of using BLI to monitor the survival of transplanted cells, rats intramyocardially injected with Fluc- or non-labeled cells were imaged longitudinally with BLI. On day 1 following cell implantation, rats injected with labeled cells showed a distinct optical signal in the area of the heart, compared to no distinct signal observed for rats having received non-labeled cells (9,674 ± 1,352 versus 2,850 ± 258 p/s/cm2/sr; P < 0.05; Fig. 3 top panel). The average signal declined over the next 5 days, with a slight increase around day 3 until it reached background levels around day 6: day 1 (9,674 ± 1,352 p/s/cm2/sr), day 2 (8,503 ± 2,703 p/s/cm2/sr), day 3 (8,910 ± 5,820 p/s/cm2/sr), day 4 (7,730 ± 5,806 p/s/cm2/sr), day 5 (3,629 ± 1,107 p/s/cm2/sr), and day 6 (2,658 ± 180 p/s/cm2/sr; Fig. 4a). Exponential curve fitting of the signal decay revealed a half-life of 2.65 days, suggesting that roughly 50% of the cells died within the first 64 h following implantation under the conditions studied.
Longitudinal optical bioluminescence and magnetic resonance imaging of H9c2 cells after transplantation. Optical bioluminescence imaging of a representative rat intramyocardially injected with 2 × 106 Fluc-labeled cells (top panel, right) shows a robust distinct heart signal on day 1 (red arrow), compared to no discernable signal in a representative control rat having received non-labeled cells (top panel, left). The signal increases slightly on day 3 but decreases rapidly to near background levels by day 6. Magnetic resonance imaging of a representative rat injected with 2 × 106 Feridex-labeled cells (bottom panel, right) shows a large hypointense signal (red arrow) in the anterolateral wall of myocardium when viewed in short axis. The size of the signal decreases slightly over time, and the signal persists for at least 80 days post cell injection. No corresponding signal is observed on the short-axis image of a representative control rat having received non-labeled cells (bottom panel, left). Note that only the short-axis 2D slice containing the largest size signal is shown. A, P, R, and L indicate anterior, posterior, right, and left anatomical orientations, respectively.
Decay of mean cell-mediated optical and MR signals. a The mean optical bioluminescence signal (p/s/cm2/sr) of rats intramyocardially injected with 2 × 106 Fluc-labeled cells is plotted against time post injection (days). An exponential fit of the data points shows a half-life of 2.65 days for these cells. The fit is less than ideal at r 2 value of 0.81 because of a slight increase in signal on day 3, which may reflect an actual increase in cell proliferation. b The mean MR signal of rats injected with 2 × 106 Feridex-labeled cells decays monotonically with time. Curve fitting of the data points to an exponential decay shows a high correlation coefficient (r 2 = 0.99) with a half-life of 16.8 days. The horizontal dotted lines indicate background level of optical or MR signal. The error bars represent standard deviation. Asterisks denote statistical significance (P < 0.05) compared to the corresponding optical or MR signal at the first time point (days 1 and 2 for BLI and MRI, respectively).
Longitudinal Magnetic Resonance Imaging of Cell Survival and Feridex Retention
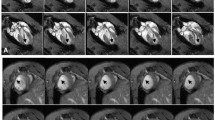
To study whether MRI can be used to serially monitor cell survival following implantation, we injected Feridex- or non-labeled cells into the hearts of living rats and imaged them repetitively for up to 80 days. On day 2, rats injected with Feridex-labeled cells showed a large hypointense signal in the anterolateral myocardial wall, corresponding to the area of cell injection, compared to no hypointense signal observed for rats injected with non-labeled cells (630,873 ± 7,265 versus 2,158 ± 706; P < 0.05; Fig. 3 bottom panel). The average volume-integrated MR signal decreased slowly over 2 months, with imaging signal still seen at the end of the longitudinal study: day 2 (630,873 ± 7,265), day 9 (455,905 ± 82,898), day 16 (332,218 ± 53,646), day 50 (91,533 ± 6,689), and day 80 (24,036 ± 3,724; Fig. 4b). Quantification of the signal decay revealed a half-life of 16.8 days, which is 6.3 times greater than that measured from the optical imaging data. There was a statistically insignificant trend (P > 0.05) for the CNR of the signal to decrease slightly over the study period: day 2 (20.7 ± 2.9), day 9 (20.9 ± 2.1), day 16 (24.3 ± 6.2), day 50 (13.6 ± 2.4), and day 80 (14.7 ± 4.4).
To further investigate whether a slow tissue clearance of Feridex could explain the longer half-life observed for the MR signal, rats that received intramyocardial injections of 75 μg of Feridex only were imaged on days 2, 9, 19, and 42. A hypointense MR signal corresponding to the site of Feridex injection was found to persist for as long as 42 days in all rats, with the imaging signal declining gradually over time, suggesting a slow tissue clearance of Feridex from myocardial tissue even in the absence of any transplanted cells (Fig. 5).
Myocardial clearance of free Feridex following direct injection. Longitudinal MR imaging of a representative rat intramyocardially injected with 75 μg of free Feridex shows a slow clearance of Feridex from the site of injection. The Feridex-induced hypointense signal (red arrows) decreases slowly over time and persists for at least 42 days post injection.
Ex Vivo Histology of Hearts Injected with Feridex-Labeled Cells or Free Feridex
To further analyze the clearance of cellular and free Feridex, rats injected with either Feridex-labeled cells or Feridex alone were killed on days 2, 9, or 16 for histological analysis. On day 2 following injection of Feridex-labeled cells, a large number of iron-positive transplanted cells surrounded by inflammatory cells (either iron positive or iron negative) were visualized near the needle track (Fig. 6, top panel). The total number of cells decreased over the ensuing 2 weeks, during which period the iron stain became increasingly concentrated. Following injection of free Feridex, both iron positive and iron negative inflammatory cells, as well as some non-cell-associated Feridex, were observed along the needle track on day 2. By days 9 and 16, iron stains could still be found either in the fibrotic interstitium or associated with some cell nuclei, after the recession of inflammation. These ex vivo histological results confirmed the slow removal of free Feridex from the myocardium following injection.
Ex vivo histological assessment of Feridex-labeled cells and free Feridex following intramyocardial delivery. Representative double H&E/Prussian Blue staining of rat myocardium at days 2, 9, and 16 following direct injection of either Feridex-labeled cells (top panel) or Feridex alone (bottom panel). Both Feridex-containing cells and free Feridex stain blue (arrows). Asterisks indicate the host myocardium.
Discussion
Recent development of noninvasive molecular imaging techniques has opened the door for studying the fate of stem cells following transplantation into the hearts of living subjects. A critical issue regarding the use of these cell-tracking techniques is the selection of a cell marker, which determines not only the sensitivity of the imaging assay but also the type of biological information that can be inferred from the cell marker-mediated signal (e.g., cell location, death, proliferation, and differentiation). In this study, we compared the use of two popular cell markers, Fluc reporter gene and SPIO MR contrast agent (Feridex), for labeling H9c2 embryonic cardiomyoblasts ex vivo and noninvasively imaging their survival in rats following implantation using BLI and MRI, respectively.
From the cell culture studies, we found that neither the Fluc reporter gene nor Feridex significantly altered cell morphology, with the exception that Feridex when used in combination with Lipofectamine2000 transfection agent-induced cytoplasmic vacuolization, which could be readily observed under light microscopy (Fig. 1b). This observation, though perhaps disconcerting at first sight because of a strong association between cytoplasmic vacuolization and cell stress, is consistent with other reports demonstrating a similar vacuolization under electron microscopy but yet no significant metabolic changes associated with Feridex-labeled cells [15, 16]. Although there was some evidence of Feridex slowing cell proliferation in this study, this effect was found to be short-lived (∼2 days) and could be explained by the dilution of intracellular Feridex to ineffective levels following repeated cell division during growth. The fact that we observed no statistically significant change in the overall doubling time of Feridex-labeled cells compared to non-labeled cells further lends support to the notion that Feridex-labeling does not significantly perturb long-term cellular function. However, it remains to be seen whether such a labeling technique would have a differential effect on other cell types or cellular functions, as one recent report suggested that Feridex-labeling may hinder stem cell differentiation into certain lineages [17].
In contrast to Feridex labeling, stable Fluc labeling using G418 selection appeared to permanently decrease cell proliferation, a phenomenon that was evident throughout the entire assay period (Fig. 2). Such perturbation has also been seen before in a study involving the creation of a stable Fluc-expressing cell line (unpublished data). Cytotoxicity, however, is not always observed for stable transfection/transduction of Fluc, as well as other commonly used reporter genes: As recently detailed by our labs, microarray and proteomic studies on mouse embryonic stem cells stably transduced with a tri-fusion reporter gene (composed of firefly luciferase, monomeric red fluorescent protein, and truncated mutant herpes simplex virus type 1 thymidine kinase) via lentivirus demonstrated minimal changes in cell proliferation, differentiation, and expression of relevant cellular genes [18–20]. Whether the expression of a reporter gene would perturb cell function or phenotype depends on factors such as the site of transgene integration and the number of transgene copies, which could vary depending on the method of transfection/transduction (e.g., 1–3 copies for lentivirus [21] and up to ∼50 copies for plasmid [22]). Thus, for marking cells with reporter genes using stable transfection/transduction of DNA, either a quick method to screen for unperturbed phenotype or an efficient method to ensure safe, site-specific integration of the transgene is needed. The latter approach is in fact under active development and investigation [23].
As for identifying the better cell marker for monitoring the long-term survival of transplanted cells, we performed serial BLI and MRI on rats injected with Fluc- and Feridex-labeled cells, respectively. To our surprise, we found a great discrepancy between the cell survival curves obtained from using the two imaging modalities. The half-life of the BLI-derived cell survival curve was found to be 2.65 days, whereas that derived from MRI was substantially greater at 16.8 days. The shorter half-life associated with BLI could be attributed more to the poorer proliferative capacity of Fluc-labeled cells compared to Feridex- or non-labeled cells, as evident from the in vitro cell proliferation assay, but less so to in vivo silencing of Fluc expression as we have previously studied [24]. This finding is consistent with numerous reports showing that more than 70% of donor cells, regardless of cell type (except embryonic stem cells, which may turn into teratoma), die within the first week of transplantation into the hearts of immunocompetent animals, and it lends support to the notion that BLI in conjunction with Fluc labeling can be used to accurately assess donor cell survival [25–27].
The much prolonged half-life associated with MRI, for the same reason, cannot be explained solely by the better proliferative capacity of Feridex-labeled cells and raises the question as to whether the persistent MR signal could be due to a slow tissue clearance of Feridex upon cell death and lysis. Indeed, our longitudinal MRI of rats intramyocardially injected with 75 μg of free Feridex (∼50% of the maximum amount that can be taken up by 2 million cells ex vivo) showed a persistent MR signal for at least 42 days, thereby supporting this hypothesis. Our MRI results here, however, contrast with a previous report contending that no MR signal could be seen as early as 12 h after intramyocardial injection of free Feridex into the infarcted mouse myocardium [28]. This discrepancy could be explained by a combination of factors, including the different dosage of Feridex administered, animal models tested (i.e., rat versus mouse, non-ischemic heart versus infarcted heart), and MR imaging systems of varying imaging sensitivity used between the two studies. In the present study, we supported our MRI results by performing ex vivo histology at different time points following Feridex injection to assess the tissue clearance of Feridex over time. As expected, we found persistent accumulation of free Feridex in the interstitium for at least 16 days after injection. The fact that some iron-positive endogenous cells were also visualized near the injection site further suggested the involvement of endogenous cells (e.g., macrophage) in phagocytizing and likely removing free Feridex from the myocardium. The change of iron staining pattern over time from diffuse (day 2) to focal (days 9 and 16) for hearts injected with either Feridex-labeled cells or free Feridex further suggested the role of these cells in scavenging and concentrating Feridex, leading to prolonged MR signal that does not accurately reflect the viability of transplanted cells. These findings altogether suggest that MRI, at least in the way utilized in this study, would not be able to distinguish Feridex-labeled cells from free Feridex released upon cell death and possibly engulfed by other endogenous cell types, making Feridex a less suitable marker for tracking long-term cell survival. In contrast, BLI imaging of Fluc-labeled cells does not share the same concern because the Fluc enzyme has a short in vivo half-life (3 h) and should degrade rapidly following cell death and lysis [29]. Thus, the Fluc-mediated signal is specific to only viable transplanted cells. Furthermore, stable integration of Fluc leads to Fluc expression that duplicates with cell division and reflects cell proliferation, whereas chemical-based markers such as Feridex dilute with cell division and would always be limited to short-term imaging of cells that actively proliferate. Moreover, chemical-based markers cannot be used to signify other cellular events relevant to stem cell transplantation such as stem cell differentiation and activation of signal transduction pathways, which can otherwise be imaged using various reporter gene-based systems [30]. For instance, to image stem cell differentiation, we are currently exploring the use of a molecular construct containing a cardiac-specific promoter linked to a reporter gene, such that during stem cell differentiation into cardiomyocytes, the cells containing such a construct can turn on reporter gene expression and be imaged accordingly. In our opinion, the major strength of a cell marker such as Feridex lies in its ability to induce strong hypointense signals that can be readily visualized tomographically using GRE-based MR sequences. Such an attribute makes it an excellent cell marker for use when an application calls for a highly sensitive localization of Feridex-labeled cells and involves minimal cell death (e.g., immediately following implantation, assuming minimal cell death during delivery).
One limitation of this study worth noting concerns MR quantification of cell signals using a GRE pulse sequence, which yields a hypointense signal due to dephasing of spins neighboring Feridex. Unlike optical signals, which are linear with respect to the light source (amount of Fluc/D-luciferin enzymatic reactions), the MR signal intensity obtained with a GRE sequence is non-linear with respect to Feridex concentration and therefore does not lend itself to easy estimation of true Feridex concentration or the number of Feridex-labeled cells. Although the relaxivity R2* is known to correlate linearly with Feridex concentration, and in vivo R2* measurement of transplanted cells has been attempted using a clinical 1.5 T magnet to assess cell survival [31], we have found this approach to be less practical, if not difficult, to achieve at high field (4.7 T) because the MR signal saturates at very short echo times, making curve fitting difficult for extraction of R2*. In this study, we took a semi-quantitative approach to quantifying cell-mediated MR signals by taking the absolute difference between the volume-integrated cell signal and the myocardial background signal. Although such quantification may not allow a direct inference on the amount of Feridex or the number of Feridex-labeled cells present, it captures to a certain degree the rate at which the hypointense signal in a 3D volume decreases slowly with time. Such slow decay could be readily appreciated from the MR images and differs drastically from that obtained from optical imaging. The fact that we found free Feridex to clear slowly from myocardium further supports our claim that Feridex is not a suitable marker for imaging long-term cell survival. Nevertheless, to further address this quantification issue, we are now developing MR sequences, which can visualize stem cells as positive contrast signals that correlate linearly with cell number [32, 33]. With further optimization of the pulse sequences, we should be able to perform more quantitative imaging of Feridex-labeled cells in living animals.
Conclusions
Cell labeling by either stable transfection of the Fluc reporter gene or incubation of Feridex can potentially cause cytotoxicity and should be performed either cautiously or with the appropriate screening techniques to avoid a compromise in the therapeutic efficacy of cell transplantation. Feridex clears slowly from the myocardium following either direct injection or the death of transplanted Feridex-labeled cells and can lead to prolonged, albeit highly sensitive and tomographic, heart signals that are not indicative of cell viability. Long-term evaluation of cell survival can be better performed with Fluc cell labeling, as Fluc reproduces with cell proliferation, and the Fluc enzyme degrades rapidly in the interstitium following cell death. With further optimization of both labeling techniques, it should be feasible in the future to utilize the strengths of each marker by dual labeling cells with Fluc and Feridex so that combined BLI/MRI can be used to image both the location and long-term survival of cells with high sensitivity (Feridex) and accuracy (Fluc). For an eventual clinical translation, a PET reporter gene (e.g., herpes simplex virus type 1 thymidine kinase or dopamine type 2 receptor) [13] would need to be used in place of Fluc to enable combined PET/MRI monitoring of cardiac cell transplantation in humans.
References
Reffelmann T, Kloner RA (2003) Cellular cardiomyoplasty–cardiomyocytes, skeletal myoblasts, or stem cells for regenerating myocardium and treatment of heart failure? Cardiovasc Res 58:358–368
Rosenzweig A (2006) Cardiac cell therapy—mixed results from mixed cells. N Engl J Med 355:1274–1277
Janssens S et al (2006) Autologous bone marrow-derived stem-cell transfer in patients with ST-segment elevation myocardial infarction: double-blind, randomised controlled trial. Lancet 367:113–121
Frangioni JV, Hajjar RJ (2004) In vivo tracking of stem cells for clinical trials in cardiovascular disease. Circulation 110:3378–3383
Bengel FM, Schachinger V, Dimmeler S (2005) Cell-based therapies and imaging in cardiology. Eur J Nucl Med Mol Imaging 32(Suppl 2):S404–S416
Zhou R, Acton PD, Ferrari VA (2006) Imaging stem cells implanted in infarcted myocardium. J Am Coll Cardiol 48:2094–2106
Wu JC et al (2003) Molecular imaging of cardiac cell transplantation in living animals using optical bioluminescence and positron emission tomography. Circulation 108:1302–1305
Zhou R et al (2005) In vivo detection of stem cells grafted in infarcted rat myocardium. J Nucl Med 46:816–822
Kraitchman DL et al (2003) In vivo magnetic resonance imaging of mesenchymal stem cells in myocardial infarction. Circulation 107:2290–2293
Genove G, DeMarco U, Xu H, Goins WF, Ahrens ET (2005) A new transgene reporter for in vivo magnetic resonance imaging. Nat Med 11:450–454
Hofmann M et al (2005) Monitoring of bone marrow cell homing into the infarcted human myocardium. Circulation 111:2198–2202
Ray P et al (2001) Monitoring gene therapy with reporter gene imaging. Semin Nucl Med 31:312–320
Chen IY et al (2004) Micro-positron emission tomography imaging of cardiac gene expression in rats using bicistronic adenoviral vector-mediated gene delivery. Circulation 109:1415–1420
Gheysens O et al (2006) Noninvasive evaluation of immunosuppressive drug efficacy on acute donor cell survival. Mol Imaging Biol 8:163–170
Matuszewski L et al (2005) Cell tagging with clinically approved iron oxides: feasibility and effect of lipofection, particle size, and surface coating on labeling efficiency. Radiology 235:155–161
Arbab AS et al (2003) Characterization of biophysical and metabolic properties of cells labeled with superparamagnetic iron oxide nanoparticles and transfection agent for cellular MR imaging. Radiology 229:838–846
Kostura L, Kraitchman DL, Mackay AM, Pittenger MF, Bulte JW (2004) Feridex labeling of mesenchymal stem cells inhibits chondrogenesis but not adipogenesis or osteogenesis. NMR Biomed 17:513–517
Wu JC et al (2006) Transcriptional profiling of reporter genes used for molecular imaging of embryonic stem cell transplantation. Physiol Genomics 25:29–38
Wu JC et al (2006) Proteomic analysis of reporter genes for molecular imaging of transplanted embryonic stem cells. Proteomics 6:6234–6249
Cao F et al (2006) In vivo visualization of embryonic stem cell survival, proliferation, and migration after cardiac delivery. Circulation 113:1005–1014
Johansen J et al (2002) Evaluation of Tet-on system to avoid transgene down-regulation in ex vivo gene transfer to the CNS. Gene Ther 9:1291–1301
Derouazi M et al (2006) Genetic characterization of CHO production host DG44 and derivative recombinant cell lines. Biochem Biophys Res Commun 340:1069–1077
Olivares EC, Hollis RP, Chalberg TW, Meuse L, Kay MA, Calos MP (2002) Site-specific genomic integration produces therapeutic Factor IX levels in mice. Nat Biotechnol 20:1124–1128
Krishnan M et al (2006) Effects of epigenetic modulation on reporter gene expression: implications for stem cell imaging. Faseb J 20:106–108
Nakamura Y, Yasuda T, Weisel RD, Li RK (2006) Enhanced cell transplantation: preventing apoptosis increases cell survival and ventricular function. Am J Physiol Heart Circ Physiol 291:H939–H947
Muller-Ehmsen J et al (2002) Survival and development of neonatal rat cardiomyocytes transplanted into adult myocardium. J Mol Cell Cardiol 34:107–116
Yau TM, Kim C, Li G, Zhang Y, Weisel RD, Li RK (2005) Maximizing ventricular function with multimodal cell-based gene therapy. Circulation 112:I123–I128
Himes N et al (2004) In vivo MRI of embryonic stem cells in a mouse model of myocardial infarction. Magn Reson Med 52:1214–1219
Naylor LH (1999) Reporter gene technology: the future looks bright. Biochem Pharmacol 58:749–757
Massoud TF, Gambhir SS (2003) Molecular imaging in living subjects: seeing fundamental biological processes in a new light. Genes Dev 17:545–580
Bos C et al (2004) In vivo MR imaging of intravascularly injected magnetically labeled mesenchymal stem cells in rat kidney and liver. Radiology 233:781–789
Cunningham CH, Arai T, Yang PC, McConnell MV, Pauly JM, Conolly SM (2005) Positive contrast magnetic resonance imaging of cells labeled with magnetic nanoparticles. Magn Reson Med 53:999–1005
Foltz WD, Cunningham CH, Mutsaers AJ, Conolly SM, Stewart DJ, Dick AJ (2006) Positive-contrast imaging in the rabbit hind-limb of transplanted cells bearing endocytosed superparamagnetic beads. J Cardiovasc Magn Reson 8:817–823
Acknowledgments
This work was supported in part by NHLBI 5R01HL078632 (S.S.G.), NCI ICMIC P50 CA114747 (S.S.G.), NCI SAIRP (S.S.G.), American Heart Association Pre-doctoral Fellowship (I.Y.C.), Stanford Bio-X Graduate Student Fellowship (I.Y.C), Swiss Foundation of Medical-Biological Grants (JKW), Novartis Research Foundation (JKW), Swiss Society of Radiology (JKW), and AHA Beginning Grant in Aid (J.C.W.).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chen, I.Y., Greve, J.M., Gheysens, O. et al. Comparison of Optical Bioluminescence Reporter Gene and Superparamagnetic Iron Oxide MR Contrast Agent as Cell Markers for Noninvasive Imaging of Cardiac Cell Transplantation. Mol Imaging Biol 11, 178–187 (2009). https://doi.org/10.1007/s11307-008-0182-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11307-008-0182-z