Abstract
Introduction
Obesity occurs partly due to consumption of a high-fat, high-sugar and low fiber diet and is associated with an altered gut microbiome. Prebiotic supplementation can reverse obesity and beneficially alter the gut microbiome, evidenced by previous studies in rodents. However, the role of the small intestinal metabolome in obese and prebiotic supplemented rodents has never been investigated.
Objectives
To investigate and compare the small intestinal metabolome of healthy and obese rats, as well as obese rats supplemented with the prebiotic oligofructose (OFS).
Methods
Untargeted metabolomics was performed on small intestinal contents of healthy chow-fed, high fat diet-induced obese, and obese rats supplemented with oligofructose using UPLC-MS/MS. Quantification of enterohepatic bile acids was performed with UPLC-MS to determine specific effects of obesity and fiber supplementation on the bile acid pool composition.
Results
The small intestinal metabolome of obese rats was distinct from healthy rats. OFS supplementation did not significantly alter the small intestinal metabolome but did alter levels of several metabolites compared to obese rats, including bile acid metabolites, amino acid metabolites, and metabolites related to the gut microbiota. Further, obese rats had lower total bile acids and increased taurine-conjugated bile acid species in enterohepatic circulation; this effect was reversed with OFS supplementation in high fat-feeding.
Conclusion
Obesity is associated with a distinct small intestinal metabolome, and OFS supplementation reverses some metabolite levels that were altered in obese rats. Future research into the effects of specific metabolites identified in this study will provide deeper insight into the mechanism of fiber supplementation on improved body weight.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The obesity pandemic has created a substantial personal and economic burden, costing over $100 billion yearly in the United States alone (Hammond & Levine, 2010). Increased energy consumption of a high-fat, western diet is a salient contributor to development of obesity (Cordain et al., 2005); however, the exact mechanisms remain to be fully elucidated. In humans and animal models, consumption of the western diet or high fat diet (HFD), increases adiposity and worsens glucose and lipid metabolism, all of which are associated with alterations in the gut microbiome (Zhi et al., 2019; Zinöcker & Lindseth, 2018). The gut microbiome is recognized to have a prominent role the development of obesity and type 2 diabetes (T2D) (Fan & Pedersen, 2021). Obesity and metabolic disorders are associated with gut bacterial dysbiosis, characterized by shifts in bacterial composition, richness, and function, making the gut microbiome an attractive target for obesity intervention. The impact of HFD on the gut microbiome is widely reported; however, studies have largely focused on taxonomic shifts, while only more recently research has begun to thoroughly examine bacterial metabolites that represent a main functional output of these shifts. Humans with obesity or diabetes have shifts in the circulating metabolome, with many of these metabolites derived by the gut microbiome (Liu et al., 2017; Menni et al., 2020; Molinaro et al., 2020). More specifically, in rodents, HFD-feeding alters the fecal microbiome along with the serum and fecal metabolome (Daniel et al., 2014; Jo et al., 2021), further indicating that functional microbiome shifts impact systemic metabolite concentrations. Despite this evidence, the connection between gut-derived metabolomic changes and obesity remains poorly investigated.
Given the high bacterial colonization of the colon and ease of collection, studies typically focus on the distal gut microbiota and fecal metabolome. However, the small intestine is the primary site of nutrient digestion, absorption, and gut-brain feedback mechanisms that control food intake and glucose production, all of which are influenced by the small intestinal microbiome (Bauer et al., 2018a, 2018b). Furthermore, while correlations exist between metabolic disorders and circulating gut-derived metabolites, many are likely synthesized in the small intestine and not the colon (Solon-Biet et al., 2019; Wei et al., 2020; Zhao et al., 2020). This is likely because most nutrient absorption and bile acid reabsorption take place in the small intestine, highlighting the importance of their interaction with the microbiome of the small intestine to overall systemic metabolome changes. For example, metabolic disease is associated with changes in circulating amino acid metabolites and secondary bile acids, both of which are likely synthesized or modified by the small intestinal microbiome (Chen et al., 2016; Legry et al., 2017). Despite this, to our knowledge, no studies have examined the difference in small intestine metabolome in obesity. Therefore, research on the small intestinal metabolome is vital to better understand the effect of functional microbiome changes, given that gut bacteria likely produce numerous metabolites that interact with host physiology.
The western diet is notably low in dietary fiber, which contributes to control of body weight, glucose homeostasis, and lipid homeostasis (Parnell & Reimer, 2009; Parnell et al., 2017). The ability of dietary fiber to improve metabolic disease makes this a promising therapeutic supplement to treat obesity. In particular, the extensively researched inulin-type fructan oligofructose (OFS) consistently reduces body weight and adiposity, improves glucose tolerance, and reduces cardiovascular disease risk in diet-induced obese rodents and is a promising therapeutic for humans (de Cossío et al., 2017; Dehghan et al., 2016; Hume et al., 2017; Kumar et al., 2016; Parnell & Reimer, 2009; Zhang et al., 2015). Interestingly, OFS supplementation in rodent models of diet-induced obesity and human studies including individuals with obesity results in shifts of the gut microbiome that are associated with improved metabolic homeostasis (Cani et al., 2007; Drabinska et al., 2018; Klancic et al., 2020). As such, improvements in energy and glucose homeostasis following OFS supplementation are hypothesized to be due to improved nutrient sensing, appetite suppression, and decreased inflammation via beneficial alterations in the gut microbiome and production of short chain fatty acids (Cani et al., 2004, 2006, 2009). Despite the known increases in distal intestinal short chain fatty acid production from OFS treatment that could mediate improvements in metabolic homeostasis (Meyer et al., 2022; Koh et al., 2016), OFS supplementation in HFD-feeding was recently found to improve small intestinal lipid sensing to improve nutrient-induced satiation via alterations in the small intestinal microbiome (unpublished data currently under review). Although these effects could be due to direct interactions of the host with intestinal bacteria, it is more likely a result of changes in downstream metabolites generated by the interaction of ingested nutrients with the bacteria residing in the small intestine (Canfora et al., 2019). As we, and others, have demonstrated that high-fat feeding and OFS shifts the small intestinal microbiome, and gut microbiome-derived metabolites, like short chain fatty acids, have been shown to improve host energy and glucose homeostasis, it is likely that many bacterially derived metabolites are altered in the small intestine that can impact host health; however this has never been investigated.
Therefore, the goal of the present study was to compare the small intestinal metabolome of obese, HFD-fed rats to lean, chow-fed rats. Further, as OFS supplementation in HFD is known to beneficially shift the small intestinal microbiome and impact metabolic homeostasis, we also compared the small intestinal metabolome of obese rats with or without supplementation of 10% OFS with the overarching goals of identifying 1) mechanistic changes in microbial-host metabolism that occur in obesity and that may be responsible for the beneficial effect of OFS supplementation, as well as 2) novel biochemical compounds that can be further investigated as therapeutics for obesity.
2 Materials and methods
2.1 Ethics statement
All rats were housed and maintained in accordance with the University of Arizona Institutional Animal Care and Use Committee (IACUC). All protocols were approved by the IACUC (protocol 17-287).
2.2 Animal model and diets
8–9-week-old male Sprague–Dawley rats were purchased from Charles River Laboratories (Wilmington, MA), cohoused (two per cage), and maintained on a 12-h light/dark cycle with ad libitum access to food. Rats were randomly assigned to receive either chow (ENVIGO 2018 Teklad global 18% protein rodent diets), or high fat diet (HFD; Research Diets D12451) for 3 weeks, then a subset of HFD-fed rats were randomly assigned to receive drinking water supplemented with 10% (w/w) oligofructose (HFD + OFS; BENEO-Orafti P95) for 6 weeks (n = 12/group). Diet macronutrient composition and ingredient lists can be found in Supplementary Table 1. Rats have been previously characterized and utilized for microbiome analysis for another publication (unpublished data currently under review).
2.3 Collection of tissues, plasma, and intestinal contents
At the conclusion of the study, rats were deeply anesthetized and portal blood, small intestinal contents and ileal tissue, colon contents, and liver collected. Approximately 1 mL of portal vein blood was placed in a 1.7 mL Eppendorf tube containing a DPPIV inhibitor cocktail (Millipore Sigma, Burlington, MA). Whole blood was centrifuged, plasma collected, aliquoted, and stored at − 20°C for future analysis. The small intestine and colon were located, isolated, and contents snap frozen in liquid nitrogen and stored at − 80°C for future analysis. Liver, small intestine, and colon tissue samples were collected, snap frozen, and stored at − 80°C for future analyses.
2.4 Metabolite analysis
Luminal small intestinal contents were sent to Metabolon, Inc (Durham, NC) and an untargeted metabolomics analysis conducted as previously described (Evans et al., 2009). Briefly, upon sample receipt, samples were prepared using the MicroLab STAR® Liquid Handling System (Hamilton, Reno, NV). For quality control purposes, several recovery standards were added to samples prior to beginning the extraction process. Samples were deproteinated with methanol. The extract was then aliquoted into 5 fractions: two for analysis by two separate reverse phase (RP) UPLC-MS/MS methods with positive ion mode electrospray ionization (ESI), one for analysis by RP/UPLC-MS/MS with negative ion mode ESI, one for analysis by HILIC/UPLC-MS/MS with negative ion mode ESI, and one sample was reserved. Metabolites were identified using UPLC-MS/MS on an ACQUITY ultra-performance liquid chromatography system (Waters, Milford, MA) and a Q-Exactive high resolution accurate-mass spectrometer (ThermoFisher Scientific, Waltham, MA) interfaced with a heated electrospray ionization source and Orbitrap mass analyzer operated at 35,000 mass resolution. One sample fraction was analyzed in acidic positive ion conditions for hydrophilic compounds. One sample fraction was analyzed in acidic positive ion conditions chromatographically optimized for more hydrophobic compounds. One sample fraction was analyzed using a separate C18 column in basic negative ion optimized conditions. The final sample fraction was eluted from a hydrophilic interaction chromatography UPLC BEH Amide 2.1 × 150 mm, 1.7 µm column (Waters, Wilford, MA) using a gradient of water and acetonitrile with 10 mM ammonium formate and analyzed by negative ionization. The raw data was extracted, peak-identified and quality control processed using Metabolon’s hardware and software. Identification of metabolites was based on retention time/index, mass to charge ratio (m/z), and chromatographic data (including MS/MS spectral data). Area under the curve was used to quantify peaks.
2.5 Bile acid quantification
Bile acids were analyzed at the University of Arizona Cancer Center Analytical Chemistry Shared Resource from small intestine, colon, portal vein plasma, and liver tissue samples via liquid–liquid extraction utilizing ethyl acetate. Chromatographic separation was achieved using a gradient system of acetonitrile and water with 0.1% formic acid on an HSS T3 UPLC column (Waters, Milford, MA). Bile acids were detected using a Xevo G2-S QTof Quadrupole Time-of-Flight Mass Spectrometer (Waters, Milford, MA) operated in negative electrospray ionization mode scanning from 200 to 600 Daltons. Quantification of bile acids and conjugated bile acids was conducted based on the ratio of analyte peak areas to internal standard peak areas in extracted ion chromatograms for the negatively charged parent masses [M-H]−1 of the bile acids, their glycine and taurine conjugates, and internal standards.
2.6 RNA isolation and qRT-PCR
RNA isolation was performed for liver and ileum with the PureLink™ RNA Mini Kit (Ambion, Austin, TX) per the manufacturer’s protocol. NanoDrop™ One Microvolume UV–Vis Spectrophotometer (ThermoFisher Scientific) was used to determine RNA concentration and sample purity. cDNA synthesis was performed on a T100™ Thermal Cycler (BioRad Laboratories, Hercules, CA) with SuperScript™ IV VILO™ Master Mix with ezDNase™ Enzyme (Invitrogen, Carlsbad, CA) per the manufacturer’s protocol. qRT-PCR was performed with CFX96 Touch™ Real-Time PCR Detection System (BioRad Laboratories) with TaqMan™ Gene Expression Assays (ThermoFischer Scientific) for rat Nrb01 (Rn00589173_m1), NR1H4 (Rn00572658_m1), Cyp7a1 (Rn00564065_m1), Cyp8b1 (Rn01445029_s1) and rat 18s ribosomal RNA (Rn03928990_g1).
2.7 Statistical analysis
Statistical analyses and visualization were performed using either GraphPad Prism 8 software (GraphPad Software, San Diego, CA, USA) or Metaboanalyst version 5.0. Sample size of all experiments is n = 12/group, unless otherwise specified. All Metaboanlyst procedures were conducted following log-transformation and Pareto scaling of untargeted metabolomics data. Metabolites visualized by volcano plot were considered differentially regulated based on a false discovery rate (FDR) adjusted p < 0.05 and fold-change threshold > 2. One-way ANOVA for altered metabolites comparing all groups was conducted with Tukey’s honest significant difference (HSD) method post-hoc analysis for multiple comparisons. Hierarchical clustering was performed with the hclust function using Euclidean distance (euclidean) and Ward’s linkage clustering algorithm (ward. D). Principal component analysis (PCA) was performed using the prcomp package and presented with 95% confidence interval. Random forest classification was performed using the randomforest package using 1000 classification trees, each of which was grown by random feature selection from a bootstrap sample at each branch, and pre-specified number of randomly selected variables (the square root of the total number of metabolites (Breiman, 2001)) to identify those that best classify the data into different groups while maintaining the lowest out-of-bag error, and used to determine significant features ranked by the mean decrease in classification accuracy when they are permuted. Body weight, adiposity, and quantified bile acids were compared using one-way ANOVA with Tukey’s post-hoc test for multiple comparisons and visualized with GraphPad Prism 8 software.
3 Results
3.1 Body weight, adiposity, and comparison of the small intestinal metabolome of healthy, obese, and OFS-supplemented rats
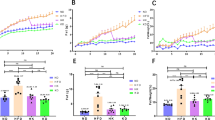
There were no differences in body weight between groups at baseline (312.1 ± 8.02, 315.0 ± 5.0, and 310.8 ± 5.2 for chow, HFD, and HFD + OFS, respectively). Nine weeks of HFD-feeding (Fig. 1A) increased body weight and body fat accumulation compared to chow (Fig. 1B, C); however, rats given OFS supplementation during the last six weeks of HFD (Fig. 1A) had decreased body weight gain beginning at 1 week of supplementation (Fig. 1B) and decreased body fat compared to HFD-induced obese at the conclusion of the study, with no difference from healthy chow (Fig. 1C), in accordance with previous studies (Bomhof et al., 2016; Kumar et al., 2016). Given that gut bacteria participate in metabolism of host- and diet-derived compounds (Agus et al., 2021), we next investigated the metabolome of the small intestinal luminal contents of lean, obese rats, and obese supplemented with OFS. From the Metabolon propriety in-house library, a total of 828 metabolites were detectable in small intestinal contents (Supplementary Table 2). In a comparison of all three dietary groups, 399 out of a total of 828 metabolites were significantly altered (FDR adjusted p < 0.05; Supplementary Table 3). PCA 2D scores plot revealed distinct separation between chow and both HFD and HFD + OFS groups (Fig. 1D), indicating low inter-sample, but high between-group, variability in small intestinal metabolomic signatures in chow vs. HFD and HFD + OFS rats.
High fat-feeding and OFS supplementation in high fat-feeding impacts body weight and adiposity and alters the small intestinal metabolome. A Study design; B body weight; C adiposity; D 2D scores plot of principal component analysis (PCA) with 95% confidence interval of chow (gray), HFD (gold), and HFD + OFS (blue) rats (n = 12/group). Data presented as mean ± SEM where applicable. *p < 0.05, **p < 0.01, ***p < 0.001 determined by one-way ANOVA with Tukey’s multiple comparisons post-hoc test (b and c)
3.2 Untargeted metabolomics analysis of small intestinal metabolites in obese rats compared to lean and OFS-supplemented
Our data indicate a clear distinction in the small intestinal metabolome of obese, HFD-fed rats compared to healthy, chow-fed controls (Fig. 1D). 198 metabolites were significantly downregulated (FDR adjusted p < 0.05) and 158 metabolites were significantly upregulated (FDR adjusted p < 0.05) in obese compared to healthy rats (Fig. 2A). Of these, metabolites involved in vitamin metabolism were significantly downregulated in the small intestine of HFD-induced obese rats, including the carotenoid metabolites carotene diol (1–3) and beta-cryptoxanthin, pyridoxine (Vitamin B6), and tocopherol metabolites, including alpha tocopherol, alpha-tocopherol acetate, delta-tocopherol, gamma-tocotrienol, and alpha-tocotrienol (Fig. 2A). The top 400 metabolites and the top 50 differentially regulated metabolites identified by hierarchical clustering are shown in Fig. 2C. Random Forest (RF) classification comparing the small intestinal metabolome of lean and obese rats provided a predictive accuracy of 100% (data not shown). RF analysis identified metabolites that most strongly contributed to group classification ranked by mean decrease accuracy (MDA), including equol sulfate, 2-acetamidophenol sulfate, soyasaponin I and others (Fig. 2E, Supplementary Table 4). We next evaluated the impact of OFS supplementation during HFD-induced obesity on the small intestinal metabolome. Although PCA analysis did not indicate a significant distinction in the overall small intestinal metabolome (Fig. 1D), N-acetylglucosamine/N-acetylgalactosamine was significantly downregulated, and 4 metabolites were significantly upregulated, including ectoine, 1-methyladenine, 3-ureidopropionate, and pantoate, in rats supplemented with OFS compared to obese rats (Fig. 2B). The top 400 metabolites and the top 50 differentially regulated metabolites identified by hierarchical clustering are shown in heatmaps (Fig. 2D). RF analysis comparing the small intestinal metabolome of OFS supplementation provided a predictive accuracy of 79%, which is significantly better than the 50% accuracy predicted by chance (data not shown). Further, RF analysis identified metabolites that most strongly contributed to group classification ranked by MDA, including dihydroferulic acid sulfate, thiamin, 1-methyladenine, and others (Fig. 2F, Supplementary Table 5).
Untargeted metabolomics reveals differentially abundant small intestinal metabolites in obese HFD-fed rats compared to healthy chow-fed and OFS supplemented rats. Volcano plot of up- and downregulated metabolites in (A) HFD- vs. chow-fed rats and B OFS-supplemented rats (HFD + OFS) vs. HFD-fed rats; vertical lines indicate metabolites increased or decreased by fold-change threshold > 2, and horizontal lines indicate statistical significance FDR-adjusted p < 0.05. Hierarchal clustering analysis of top 400 (left) and top 50 (right) differentially abundant metabolites in (C) HFD- vs. chow-fed and D HFD + OFS vs. HFD-fed rats. Red indicates upregulation of metabolite and blue indicates downregulation of metabolite (see color key). Metabolites identified by random forest analysis and ranked by mean decrease accuracy (MDA) in (E) HFD- vs. chow-fed rats and F HFD + OFS vs. HFD-fed rats. A higher MDA value indicates a greater contribution to group distinction
3.3 Amino acid metabolites in the small intestine
Untargeted metabolomics analysis indicated notable alterations in metabolites involved in metabolism of branched chain amino acids (BCAAs) in obese rats (Fig. 3, Fig. S1, Supplementary Table 6) Specifically, 2-methylbutyrylcarnitine (p = 0.00026) and isobutyrylcarnitine (p = 0.0032) were increased in obese rats compared to chow (Supplementary Table 6). Conversely, alpha-hydroxyisocaproate (p = 0.0021), 2,3-dihydroxy-2-methylbutyrate (p = 0.0001), beta-hydroxyisovalerate (p = 0.012), 2-hydroxy-3-methylvalerate (p = 0.014), N-acetylisoleucine (p = 0.012), and valine betaine (p < 0.0001) were decreased in obese rats (Supplementary Table 6). Of these, isovalerylglycine and ethylmalonate increased with OFS supplementation (Supplementary Table 6). Metabolites involved in histidine metabolism were similarly dysregulated in obese, HFD-fed rats, with significantly increased imidazole propionate (p < 0.0001) and decreased histamine (p = 0.018) levels compared to healthy, chow-fed rats (Supplementary Table 6). Metabolites involved in tryptophan metabolism decreased in obese rats, including N-acetyltryptophan (p = 0.0043), indolelactate (p < 0.0001), indoleacetate (p < 0.0001), and indole-3-carboxylate (p < 0.0001). Of these, indolelactate increased with OFS supplementation (Supplementary Table 6). Finally, the amino acid lysine (p = 0.009) as well as glutarylcarnitine (p = 0.017) increased in obese, HFD-fed rats compared to chow. Fructosyllysine (p < 0.0001), saccharopine (p < 0.0001), N-acetyl-cadaverine (p = 0.0069), and N,N-dimethyl-5-aminovalerate (p = 0.0018) decreased in obese rats (Supplementary Table 6).
Hierarchal clustering analysis of top 50 differentially abundant metabolites of amino acid metabolism analyzed by one-way ANOVA (FDR-adjusted p < 0.05) with Tukey’s honest significant difference post-hoc test (Supplementary Table 6) in healthy chow (gray), obese HFD-fed (gold), and HFD + OFS (blue) rats presented as group average. Red indicates upregulation of metabolite and blue indicates downregulation of metabolite (average per group; see color key)
3.4 Metabolites related to the microbiome of the small intestine
Multiple xenobiotic and gut microbiome-related metabolite species were drastically altered in obese rats compared to healthy chow-fed, with levels of many of these metabolites reversed with OFS supplementation to more closely resemble chow levels (Fig. 4, Fig. S2). Notably, hippurate significantly decreased in obesity and increased with OFS supplementation (p < 0.0001, Fig. 4A). Further, obese rats exhibited decreased lipopolysaccharide components, including 1,1-kestotetraose (p < 0.0001) and 3-deoxyoctulosonate (p = 0.001), compared to lean and OFS supplemented rats (Fig. 4B, C). Finally, ectoine was drastically decreased in obese rats and rescued with OFS supplementation in HFD (p < 0.0001; Fig. 4D).
Metabolites related to the gut microbiota in obese HFD-fed and OFS supplemented rats identified by untargeted metabolomics. All metabolites were considered significantly different via one-way ANOVA of all groups (FDR-adjusted p < 0.05) with Tukey’s honest significant difference post-hoc test (Supplementary Table 3)
3.5 Bile acid metabolites in the small intestine of chow, HFD, and HFD + OFS rats
We observed notable changes in both primary and secondary bile acid species via untargeted metabolomic analysis of the small intestinal contents of obese, HFD-rats compared to healthy chow and OFS supplemented rats (Fig. 5; Fig. S3; Supplementary Table 7). 3-dehydrodeoxycholate (p = 0.0073; Fig. 5B), 6-beta-hydroxylithocholate (p = 0.0021; Fig. 5C), 6-oxolithocholate (p = 0.01; Fig. 5D), alpha-muricholate (p < 0.0001; Fig. 5F, beta-muricholate (p = 0.0017; Fig. 5G), chenodeoxycholate (p < 0.0001; Fig. 5H), cholate (p < 0.0001; Fig. 5I), cholate sulfate (p = 0.0057; Fig. 5J), deoxycholate (p = 0.024; Fig. 5K), glycodeoxycholate (p < 0.0001; Fig. 5L), hyocholate (p = 0.0058; Fig. 5M), hyodeoxycholate (p = 0.0047; Fig. 5N), lithocholate sulfate (p = 0.023; Fig. 5O), ursodeoxycholate (p = 0.0018; Fig. 5Q), and ursodeoxycholate sulfate (p = 0.0056; Fig. 5R) decreased in obese rats, and OFS supplementation significantly increased 3-dehydrodeoxycholate, 6-oxolithocholate, alpha-muricholate, chenodeoxycholate, cholate, cholate sulfate, hyocholate, and ursodeoxycholate (Fig. 5). OFS supplementation increased 3-dehydrocholate (p < 0.0001; Fig. 5A), 7-ketodeoxycholate (p < 0.0001; Fig. 5E), and ursocholate (0.0022; Fig. 5P) compared to both chow and HFD-fed rats.
Bile acid metabolites altered in obese HFD-fed and OFS supplemented rats identified by untargeted metabolomics. All metabolites were considered significantly different via one-way ANOVA of all groups (FDR-adjusted p < 0.05) with Tukey’s honest significant difference post-hoc test (Supplementary Table 7)
3.6 Quantification of bile acids in enterohepatic circulation of chow, HFD, and HFD + OFS rats
We further quantified bile acid levels in the small intestine, hepatic portal vein, liver, and colon to gain a comprehensive understanding of the enterohepatic bile acid pool in obesity (Fig. 6). Obese rats had a modest decrease in total portal vein bile acids compared to chow-fed rats, although this did not reach statistical significance (p = 0.06; Fig. 6B), and an decrease in total portal vein bile acids compared to OFS-supplemented rats (Fig. 6B; p < 0.05). Obese rats had an increased proportion of taurine-conjugated bile acids in the small intestine (Fig. 6E; p < 0.05), liver (Fig. 6G; p < 0.05) compared to chow-fed rats, and an increase in taurine-conjugated bile acids in the small intestine (Fig. 6E; p < 0.01) and portal vein (p < 0.05; Fig. 6F) compared OFS supplemented rats. Further, unconjugated bile acids were lower in obese rats in the small intestine (p < 0.01; Fig. 6E) compared to chow and OFS-supplemented rats, the colon (p < 0.05, Fig. 6H) compared to chow, and portal vein (Fig. 6F; p < 0.01) compared to OFS supplemented rats.
Quantitative analysis of primary and secondary bile acids in enterohepatic circulation of healthy chow fed, HFD-induced obese, or HFD-fed rats with OFS supplementation. Total bile acids in the A small intestine, B portal vein, C liver, and D colon; glycine-, taurine-, and unconjugated bile acids in the E small intestine, F portal vein, G liver, and H colon; composition of the bile acid pool (individual bile acid species calculated as % of total bile acids) in the I small intestine, J portal vein, K liver, and l colon, presented as fold-change from HFD; M simple linear regression of small intestinal bile acids significantly correlated with % body fat (Supplementary Table 8). Data are presented as mean ± SEM. *p < 0.05 vs. HFD; **p < 0.01 vs. HFD; ***p < 0.001 vs. HFD analyzed by one-way ANOVA with Tukey’s multiple comparisons post-hoc test
HFD-induced obesity decreased small intestinal chenodeoxycholic acid (CDCA; p < 0.05) and increased tauroursodeoxycholic acid (TUDCA; p < 0.05) compared to both chow and OFS supplemented rats (Fig. 6I). HFD-fed rats had increased taurocholic acid (TCA) in both the small intestine (Fig. 6I; p < 0.05) and liver (Fig. 6K; p < 0.05) compared to chow and OFS-supplemented rats. Additionally, in the small intestine, HFD-fed obese rats had decreased cholic acid (CA; Fig. 6I; p < 0.05) and ursodeoxycholic acid (UDCA; Fig. 6I; p < 0.01) and increased taurochenodeoxycholic acid (TCDCA; p < 0.01) compared to OFS supplementation (Fig. 6I). HFD-fed obese rats had decreased hepatic glycochenodeoxycholic acid (GDCA) compared to OFS supplemented rats (Fig. 6K; p < 0.05). In the colon, obese rats exhibited decreased UDCA (p < 0.05) and increased glycochenodeoxycholic acid (GCDCA; p < 0.05) compared to lean rats (Fig. 6L); however, there was no effect of OFS supplementation on bile acid levels in the colon (Fig. 6L).
Small intestinal UDCA (p = 0.0435) and lithocholic acid (LCA; p = 0.046) were negatively correlated with body fat percentage, independent of dietary group (Fig. 6M; Supplementary Table 8). Conversely, small intestinal TCDCA (p = 0.0202), TCA (p = 0.0003), and glycocholic acid (GCA; p < 0.0001) were positively correlated with body fat percentage, independent of dietary group (Fig. 6M; Supplementary Table 8).
Obese and OFS supplemented rats had increased hepatic expression of Nr0b2 (small heterodimer partner, SHP) (Fig. S4) compared to healthy chow, with no differences in expression of the farnesoid X receptor (FXR) in the distal small intestine and no changes to expression of hepatic enzymes involved in bile acid synthesis (Fig. S4).
4 Discussion
The small intestinal microbiota impacts host physiology, including energy homeostasis and glucose metabolism (Bauer et al., 2018a, 2018b). HFD-feeding and OFS supplementation are known to impact the gut microbiome composition (Nicolucci et al., 2017; Reimer et al., 2017), and more specifically we have recently found HFD-feeding and OFS alters the small intestinal microbiome (Bauer et al., 2018a) (unpublished data currently in review). While it is possible that gut bacteria can directly influence host epithelial cells due to a thin mucus layer, it is more likely that small intestinal microbiome impacts on the host are mediated by microbially-derived metabolites. An untargeted metabolomic approach revealed that HFD-induced obesity was associated with a unique small intestinal metabolome compared to healthy chow-fed rats, with metabolites from pathways involving vitamin metabolism, fatty acid and steroid biosynthesis, bile acid metabolism, and amino acid metabolism contributing significantly to the observed differences between groups. To our knowledge, this is the first time that small intestine of obese rats has ever been analyzed by untargeted metabolomics. Additionally, given OFS supplementation reverses HFD-induced body weight gain and beneficially alters the gut microbiota (Cluny et al., 2015; Klancic et al., 2020), at least partly due to a reduction in food intake (Cani et al., 2005), we also assessed the impact of OFS supplementation on the small intestinal metabolome to possibly uncover novel mechanisms of action. Many pathways were differentially regulated in the current study, with several pathways already recognized as markers or potential drivers of obesity, including amino acid metabolism. Perhaps most notably were branched chain amino acids (BCAAs), which were altered in the small intestine of obese rats. Circulating BCAAs are increased in individuals with obesity or T2D and are highly associated with obesity and insulin resistance (Newgard et al., 2009; Vanweert et al., 2021). Contrary to previous reports, we found no differences in leucine, isoleucine, or valine in the small intestine of obese rats. However, the current study assessed small intestinal metabolites, whereas most research suggests elevations in circulating BCAAs. In the small intestine, obesity was associated with several decreased intermediates in branched chain amino acid metabolism, including ethylmalonate, 2,3-dihydroxy-2-methylbutyrate, and the N-acyl-alpha amino acid N-acetylisoleucine. This discrepancy may be due to differences in BCAA intake in humans compared to the purified diet used in this study or in the small intestinal microbiome in humans compared to rats. N-acetylleucine and N-acetylisoleucine are proposed biomarkers for T2D (Min et al., 2015; Tsutsui et al., 2011) and warrant further investigation in the context of diet-induced obesity and diabetes. Further, while the metabolic impact of most of these compounds is unknown, OFS supplementation was sufficient to increase small intestinal ethylmalonate, 2,3-dihydroxy-2-methylbutyrate, and N-acetylisoleucine in the context of HFD. Although the metabolic impact of these metabolites is completely unknown, alterations in these metabolites were associated with improvements in OFS, thus these metabolites may be of further interest as to whether they may contribute to the beneficial effects of OFS on adiposity. Additionally, imidazole propionate, a metabolite of histidine metabolism, is increased in serum of individuals with T2D (Molinaro et al., 2020), and impairs insulin signaling in rodents (Koh et al., 2018). In accordance with previous research, we found that obesity was associated with increased imidazole propionate concentrations in the small intestine, indicating that microbially produced metabolites produced in the small intestine contribute to metabolic regulation and glucose tolerance.
Metabolites involved in vitamin metabolism and xenobiotic metabolites were downregulated in the small intestine of HFD-induced obese rats compared to chow-fed, including carotenoids, tocopherol metabolism, pyridoxine, and bioactive compounds like ergothioneine. Interestingly, thiamin was found to contribute to distinction between HFD-fed obese and OFS-supplemented rats. Thiamin is synthesized by several Lactobacillus and Bifidobacterium species, Bacteroides fragilis, Prevotella copri, and others, indicating a potential for gut microbiota modulation by OFS that contributes to production of this vitamin (Yoshii et al., 2019). Further, the vasoprotective and antioxidant metabolites alpha-tocopherol and ergothioneine were recently found to be depleted in plasma of individuals with ischemic heart disease (Fromentin et al., 2022), indicating that small intestinal decreases in these compounds may contribute to systemic depletion and metabolic derangements associated with diet-induced obesity. While several of these metabolites impact host physiology, we hypothesize that this resulted from discrepancies in diet quality, as normal rodent chow diets have increased vitamin content compared to their purified fat-regulated counterparts (González-Blázquez et al., 2020). However, additional research using micronutrient equivalent purified diets on the small intestinal metabolome would provide further insight into the role of the gut microbiome, micronutrients, and food-derived metabolites on the development of obesity and metabolic disease.
Metabolites related to the gut microbiota were specifically upregulated with OFS supplementation compared to HFD-feeding alone. Of these, ectoine, a compatible solute produced by specific bacteria, reduces inflammation and protects gut barrier integrity in rats with experimental colitis (Abdel-Aziz et al., 2013). Thus, differences in ectoine levels may contribute to intestinal inflammation and gut permeability that is aggravated by diet-induced obesity (Cani et al., 2008) and improved with OFS supplementation (Dehghan et al., 2016; Valcheva et al., 2019). Additionally in the current study, HFD-induced obese rats had decreased N-acetylglucosamine/N-acetylgalactosamine. As N-acetylglucosamine is a component of the bacterial and fungal cell wall and plays a role in microbial and host cell signaling (Konopka, 2012), this metabolite may reflect altered small intestinal bacteria with HFD-feeding and should be investigated further. One interesting finding was that obesity was associated with decreased LPS components in the small intestine. This finding contradicts previous data from our lab showing increased abundance of the gram-positive Allobaculum, Bifidobacterium, and Blautia in the small intestine of OFS supplemented HFD-fed rats compared to HFD-fed rats (unpublished data currently under review). Furthermore, circulating LPS levels have been implicated in development of metabolic endotoxemia which contributes to adiposity gain and glucose dysregulation. Hippurate is decreased in obese rats and increased with OFS supplementation. Hippurate is derived from microbial benzoate production that is positively associated with gut microbial richness and metabolic health (Brial et al., 2021; Pallister et al., 2017), and therefore levels of this metabolite may reflect diet-induced microbiota modification. Administration of subcutaneous hippurate has also been shown to improve glucose tolerance and increase insulin secretion in HFD-fed mice (Brial et al., 2021); however, the mechanism remains to be determined. Further, hippurate is produced in the liver and kidney by glycine conjugation of benzoic acid (Amsel & Levy, 1969). Despite this, we observe diet-induced changes in hippurate levels in the small intestine, indicating a possibility that unique bacterial species can produce hippurate. However, the impact of gut-derived hippurate versus endogenous production remains to be determined. Taken together, these data indicate altered microbial metabolism of dietary compounds that likely reflects small intestinal microbiota alterations in obesity and prebiotic supplementation and may contribute to mucosal inflammation and host metabolic health. It should be noted that, while we expect major differences in xenobiotics between lean and obese rats given the differences in the dietary compositions, the only difference between the obese, HFD-fed rats and HFD + OFS rats is supplementation of OFS in the drinking water.
The gut microbiota greatly influences the composition of the enterohepatic bile acid pool via bile salt deconjugation and biotransformation of primary bile acids into secondary bile acids (Wahlström et al., 2016). In line with this, the present study uncovered that pathways involved in bile acid synthesis heavily contributed to the distinction of small intestinal metabolomes in obesity, prompting a further quantitative investigation of the enterohepatic bile acid pool. Total bile acids in the portal circulation were decreased in HFD-indued obesity, in contrast with previous studies showing that individuals with obesity and T2D have increased circulating bile acid levels (Vincent et al., 2013). However, portal vein bile acid levels more accurately reflect bile acid absorption than circulating plasma, and therefore this data suggests increased bile acid reabsorption, possibly due to a decrease in conjugated bile acids (Hofmann, 1963). Obesity was also associated with increased hepatic and small intestinal taurine-conjugated bile acids. Circulating taurine-conjugated bile acids are increased in individuals with T2D and positively correlated with fasting blood glucose, HbA1C, and HOMA-IR (Wewalka et al., 2014). Specifically, TCDCA, TCA, and TUDCA were elevated in the small intestine of obese rats compared to both chow and OFS supplemented rats, and both small intestinal TCDCA and TCA were positively associated with body fat percentage in all groups. TCDCA is an FXR agonist elevated with HFD-feeding (Zhang et al., 2020), which is dependent on the small intestinal microbiota and impairs small intestinal nutrient sensing (Waise et al., 2021). Given that HFD-feeding also impairs nutrient sensing, elevation of TCDCA in HFD-feeding may, at least partially, mediate this effect and warrants further investigation. Additionally, we found that HFD-feeding was associated with increased hepatic expression of SHP, a nuclear transcription factor that is upregulated with FXR activation. FXR influences glucose and energy metabolism, and whole body FXR-deficient mice are protected from diet-induced obesity (Prawitt et al., 2011), indicating that bile acid-mediated FXR agonism may contribute to the disruption of energy and glucose metabolism associated with HFD-induced obesity. Because we observed no differences in hepatic bile acid synthesis enzymes, we hypothesize that the alterations in bile acid levels between groups are not due to changes in synthesis, but rather changes in bile acid conjugation and/or excretion. However, a more comprehensive study investigating hepatic enzymatic activity and bile acid transport would help clarify this point. Taken together, these data suggest that bile acid metabolism is dysregulated in HFD-induced obesity and specific bile acid species likely contribute to the metabolic dysregulation associated with obesity. Further, bile acid alterations during obesity can be effectively reversed with OFS supplementation, implicating this metabolic pathway in the beneficial effects of OFS on energy and glucose homeostasis. Given that improvements in OFS are associated with reductions in food intake (Cani et al., 2005), it is possible this is due to changes in bile acid signaling that reduces overall caloric intake to decrease body weight and adiposity (Castellanos-Jankiewicz et al., 2021). For example, increased TGR5 signaling, and reduced intestinal FXR signaling, increase GLP-1 release (Thomas et al., 2009; Trabelsi et al., 2015), known to reduce food intake (Wachsmuth et al., 2022). Interestingly, OFS is associated with increased GLP-1 levels (Cani et al., 2005), therefore it is possible that altered intestinal bile acid signaling during OFS increases gut peptide release to promote satiation and satiety. This requires further investigation. Overall, HFD-induced obesity results in broad changes in the small intestinal metabolome, highlighting the potential importance of the interaction between the diet and small intestinal bacteria on host metabolism.
Data availability
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request.
References
Agus, A., Clément, K., & Sokol, H. (2021). Gut microbiota-derived metabolites as central regulators in metabolic disorders. Gut, 70, 1174–1182.
Amsel, L. P., & Levy, G. (1969). Drug biotransformation interactions in man. II. A pharmacokinetic study of the simultaneous conjugation of benzoic and salicylic acids with glycine. Journal of Pharmaceutical Sciences, 58, 321–326.
Abdel-Aziz, H., Wadie, W., Abdallah, D. M., Lentzen, G., & Khayyal, M. T. (2013). Novel effects of ectoine, a bacteria-derived natural tetrahydropyrimidine, in experimental colitis. Phytomedicine, 20, 585–591.
Bauer, P. V., Duca, F. A., Waise, T. M. Z., Dranse, H. J., Rasmussen, B. A., Puri, A., Rasti, M., O’Brien, C. A., & Lam, T. K. T. (2018a). Lactobacillus gasseri in the upper small intestine impacts an ACSL3-dependent fatty acid-sensing pathway regulating whole-body glucose homeostasis. Cell Metabolism, 27(572–587), e6.
Bauer, P. V., Duca, F. A., Waise, T. M. Z., Rasmussen, B. A., Abraham, M. A., Dranse, H. J., Puri, A., O’Brien, C. A., & Lam, T. K. T. (2018b). Metformin alters upper small intestinal microbiota that impact a glucose-SGLT1-sensing glucoregulatory pathway. Cell Metabolism, 27, 101-117.e5.
Bomhof, M. R., Paul, H. A., Geuking, M. B., Eller, L. K., & Reimer, R. A. (2016). Improvement in adiposity with oligofructose is modified by antibiotics in obese rats. The FASEB Journal, 30, 2720–2732.
Breiman, L. (2001). Random Forests. Machine Learning, 45, 5–32.
Brial, F., Chilloux, J., Nielsen, T., Vieira-Silva, S., Falony, G., Andrikopoulos, P., Olanipekun, M., Hoyles, L., Djouadi, F., Neves, A. L., Rodriguez-Martinez, A., Mouawad, G. I., Pons, N., Forslund, S., Le-Chatelier, E., Le Lay, A., Nicholson, J., Hansen, T., Hyötyläinen, T., … Dumas, M. E. (2021). Human and preclinical studies of the host-gut microbiome co-metabolite hippurate as a marker and mediator of metabolic health. Gut. https://doi.org/10.1136/gutjnl-2020-323314
Canfora, E. E., Meex, R. C. R., Venema, K., & Blaak, E. E. (2019). Gut microbial metabolites in obesity, NAFLD and T2DM. Nature Reviews Endocrinology, 15, 261–273.
Cani, P. D., Bibiloni, R., Knauf, C., Waget, A., Neyrinck, A. M., Delzenne, N. M., & Burcelin, R. (2008). Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes, 57, 1470–1481.
Cani, P. D., Dewever, C., & Delzenne, N. M. (2004). Inulin-type fructans modulate gastrointestinal peptides involved in appetite regulation (glucagon-like peptide-1 and ghrelin) in rats. British Journal of Nutrition, 92, 521–526.
Cani, P. D., Knauf, C., Iglesias, M. A., Drucker, D. J., Delzenne, N. M., & Burcelin, R. (2006). Improvement of glucose tolerance and hepatic insulin sensitivity by oligofructose requires a functional glucagon-like peptide 1 receptor. Diabetes, 55, 1484–1490.
Cani, P. D., Neyrinck, A. M., Maton, N., & Delzenne, N. M. (2005). Oligofructose promotes satiety in rats fed a high-fat diet: Involvement of glucagon-like Peptide-1. Obesity Research, 13, 1000–1007.
Cani, P. D., Neyrinck, A. M., Fava, F., Knauf, C., Burcelin, R. G., Tuohy, K. M., Gibson, G. R., & Delzenne, N. M. (2007). Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia, 50, 2374–2383.
Cani, P. D., Possemiers, S., Van de Wiele, T., Guiot, Y., Everard, A., Rottier, O., Geurts, L., Naslain, D., Neyrinck, A., Lambert, D. M., Muccioli, G. G., & Delzenne, N. M. (2009). Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut, 58, 1091–1103.
Castellanos-Jankiewicz, A., Guzmán-Quevedo, O., Fénelon, V. S., Zizzari, P., Quarta, C., Bellocchio, L., Tailleux, A., Charton, J., Fernandois, D., Henricsson, M., Piveteau, C., Simon, V., Allard, C., Quemener, S., Guinot, V., Hennuyer, N., Perino, A., Duveau, A., Maitre, M., … Cota, D. (2021). Hypothalamic bile acid-TGR5 signaling protects from obesity. Cell Metabolism, 33, 1483-1492.e10.
Chen, T., Zheng, X., Ma, X., Bao, Y., Ni, Y., Hu, C., Rajani, C., Huang, F., Zhao, A., Jia, W., & Jia, W. (2016). Tryptophan predicts the risk for future type 2 diabetes. PLoS ONE, 11, e0162192.
Cluny, N. L., Eller, L. K., Keenan, C. M., Reimer, R. A., & Sharkey, K. A. (2015). Interactive effects of oligofructose and obesity predisposition on gut hormones and microbiota in diet-induced obese rats. Obesity (silver Spring), 23, 769–778.
Cordain, L., Eaton, S. B., Sebastian, A., Mann, N., Lindeberg, S., Watkins, B. A., O’Keefe, J. H., & Brand-Miller, J. (2005). Origins and evolution of the Western diet: Health implications for the 21st century. American Journal of Clinical Nutrition, 81, 341–354.
de Cossío, L. F., Fourrier, C., Sauvant, J., Everard, A., Capuron, L., Cani, P. D., Layé, S., & Castanon, N. (2017). Impact of prebiotics on metabolic and behavioral alterations in a mouse model of metabolic syndrome. Brain, Behavior, and Immunity, 64, 33–49.
Daniel, H., Gholami, A. M., Berry, D., Desmarchelier, C., Hahne, H., Loh, G., Mondot, S., Lepage, P., Rothballer, M., Walker, A., Böhm, C., Wenning, M., Wagner, M., Blaut, M., Schmitt-Kopplin, P., Kuster, B., Haller, D., & Clavel, T. (2014). High-fat diet alters gut microbiota physiology in mice. ISME Journal, 8, 295–308.
Dehghan, P., Farhangi, M. A., Tavakoli, F., Aliasgarzadeh, A., & Akbari, A. M. (2016). Impact of prebiotic supplementation on T-cell subsets and their related cytokines, anthropometric features and blood pressure in patients with type 2 diabetes mellitus: A randomized placebo-controlled Trial. Complementary Therapies in Medicine, 24, 96–102.
Drabinska, N., Jarocka-Cyrta, E., Markiewicz, L. H., & Krupa-Kozak, U. (2018). The effect of oligofructose-enriched inulin on faecal bacterial counts and microbiota-associated characteristics in celiac disease children following a gluten-free Diet: Results of a randomized placebo-controlled trial. Nutrients. https://doi.org/10.3390/nu10020201
Evans, A. M., DeHaven, C. D., Barrett, T., Mitchell, M., & Milgram, E. (2009). Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Analytical Chemistry, 81, 6656–6667.
Fromentin, S., Forslund, S. K., Chechi, K., Aron-Wisnewsky, J., Chakaroun, R., Nielsen, T., Tremaroli, V., Ji, B., Prifti, E., Myridakis, A., Chilloux, J., Andrikopoulos, P., Fan, Y., Olanipekun, M. T., Alves, R., Adiouch, S., Bar, N., Talmor-Barkan, Y., Belda, E., … Pedersen, O. (2022). Microbiome and metabolome features of the cardiometabolic disease spectrum. Nature Medicine, 28, 303–314.
Fan, Y., & Pedersen, O. (2021). Gut microbiota in human metabolic health and disease. Nature Reviews Microbiology, 19, 55–71.
González-Blázquez, R., Alcalá, M., Fernández-Alfonso, M. S., Villa-Valverde, P., Viana, M., Gil-Ortega, M., & Somoza, B. (2020). Relevance of control diet choice in metabolic studies: Impact in glucose homeostasis and vascular function. Science and Reports, 10, 2902.
Hammond, R. A., & Levine, R. (2010). The economic impact of obesity in the United States. Diabetes Metab Syndr Obes, 3, 285–295.
Hofmann, A. F. (1963). The function of bile salts in fat absorption. The solvent properties of dilute micellar solutions of conjugated bile salts. The Biochemical Journal, 89, 57–68.
Hume, M. P., Nicolucci, A. C., & Reimer, R. A. (2017). Prebiotic supplementation improves appetite control in children with overweight and obesity: A randomized controlled trial. American Journal of Clinical Nutrition, 105, 790–799.
Jo, J. K., Seo, S. H., Park, S. E., Kim, H. W., Kim, E. J., Kim, J. S., Pyo, J. Y., Cho, K. M., Kwon, S. J., Park, D. H., & Son, H. S. (2021). Gut microbiome and metabolome profiles associated with high-fat diet in mice. Metabolites. https://doi.org/10.3390/metabo11080482
Klancic, T., Laforest-Lapointe, I., Choo, A., Nettleton, J. E., Chleilat, F., Noye Tuplin, E. W., Alukic, E., Cho, N. A., Nicolucci, A. C., Arrieta, M. C., & Reimer, R. A. (2020). Prebiotic oligofructose prevents antibiotic-induced obesity risk and improves metabolic and gut microbiota profiles in rat dams and offspring. Molecular Nutrition & Food Research, 64, e2000288.
Koh, A., De Vadder, F., Kovatcheva-Datchary, P., & Bäckhed, F. (2016). From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell, 165, 1332–1345.
Koh, A., Molinaro, A., Ståhlman, M., Khan, M. T., Schmidt, C., Mannerås-Holm, L., Wu, H., Carreras, A., Jeong, H., Olofsson, L. E., Bergh, P. O., Gerdes, V., Hartstra, A., de Brauw, M., Perkins, R., Nieuwdorp, M., Bergström, G., & Bäckhed, F. (2018). Microbially Produced Imidazole Propionate Impairs Insulin Signaling through mTORC1. Cell, 175, 947-961.e17.
Konopka, J.B. (2012) N-acetylglucosamine (GlcNAc) functions in cell signaling. Scientifica (Cairo) 2012.
Kumar, S. A., Ward, L. C., & Brown, L. (2016). Inulin oligofructose attenuates metabolic syndrome in high-carbohydrate, high-fat diet-fed rats. British Journal of Nutrition, 116, 1502–1511.
Legry, V., Francque, S., Haas, J. T., Verrijken, A., Caron, S., Chávez-Talavera, O., Vallez, E., Vonghia, L., Dirinck, E., Verhaegen, A., Kouach, M., Lestavel, S., Lefebvre, P., Van Gaal, L., Tailleux, A., Paumelle, R., & Staels, B. (2017). Bile Acid alterations are associated with insulin resistance, but not with NASH, in obese subjects. The Journal of Clinical Endocrinology & Metabolism, 102, 3783–3794.
Liu, R., Hong, J., Xu, X., Feng, Q., Zhang, D., Gu, Y., Shi, J., Zhao, S., Liu, W., Wang, X., Xia, H., Liu, Z., Cui, B., Liang, P., Xi, L., Jin, J., Ying, X., Wang, X., Zhao, X., … Wang, W. (2017). Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nature Medicine, 23, 859–868.
Menni, C., Zhu, J., Le Roy, C. I., Mompeo, O., Young, K., Rebholz, C. M., Selvin, E., North, K. E., Mohney, R. P., Bell, J. T., Boerwinkle, E., Spector, T. D., Mangino, M., Yu, B., & Valdes, A. M. (2020). Serum metabolites reflecting gut microbiome alpha diversity predict type 2 diabetes. Gut Microbes, 11, 1632–1642.
Meyer, R. K., Lane, A. I., Weninger, S. N., Martinez, T. M., Kangath, A., Laubitz, D., & Duca, F. A. (2022). Oligofructose restores postprandial short-chain fatty acid levels during high-fat feeding. Obesity (Silver Spring), 30, 1442–1452.
Min, J. Z., Tomiyasu, Y., Morotomi, T., Jiang, Y. Z., Li, G., Shi, Q., Yu, H. F., Inoue, K., Todoroki, K., & Toyo’oka, T. (2015). First observation of N-acetyl leucine and N-acetyl isoleucine in diabetic patient hair and quantitative analysis by UPLC-ESI-MS/MS. Clinica Chimica Acta, 444, 143–148.
Molinaro, A., Bel Lassen, P., Henricsson, M., Wu, H., Adriouch, S., Belda, E., Chakaroun, R., Nielsen, T., Bergh, P. O., Rouault, C., André, S., Marquet, F., Andreelli, F., Salem, J. E., Assmann, K., Bastard, J. P., Forslund, S., Le Chatelier, E., Falony, G., … Bäckhed, F. (2020). Imidazole propionate is increased in diabetes and associated with dietary patterns and altered microbial ecology. Nature Communications, 11, 5881.
Newgard, C. B., An, J., Bain, J. R., Muehlbauer, M. J., Stevens, R. D., Lien, L. F., Haqq, A. M., Shah, S. H., Arlotto, M., Slentz, C. A., Rochon, J., Gallup, D., Ilkayeva, O., Wenner, B. R., Yancy, W. S., Jr., Eisenson, H., Musante, G., Surwit, R. S., Millington, D. S., … Svetkey, L. P. (2009). A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metabolism, 9, 311–326.
Nicolucci, A. C., Hume, M. P., Martínez, I., Mayengbam, S., Walter, J., & Reimer, R. A. (2017). Prebiotics reduce body fat and alter intestinal microbiota in children who are overweight or with obesity. Gastroenterology, 153, 711–722.
Pallister, T., Jackson, M. A., Martin, T. C., Zierer, J., Jennings, A., Mohney, R. P., MacGregor, A., Steves, C. J., Cassidy, A., Spector, T. D., & Menni, C. (2017). Hippurate as a metabolomic marker of gut microbiome diversity: Modulation by diet and relationship to metabolic syndrome. Science and Reports, 7, 13670.
Parnell, J. A., & Reimer, R. A. (2009). Weight loss during oligofructose supplementation is associated with decreased ghrelin and increased peptide YY in overweight and obese adults. American Journal of Clinical Nutrition, 89, 1751–1759.
Parnell, J. A., Klancic, T., & Reimer, R. A. (2017). Oligofructose decreases serum lipopolysaccharide and plasminogen activator inhibitor-1 in adults with overweight/obesity. Obesity (silver Spring), 25, 510–513.
Prawitt, J., Abdelkarim, M., Stroeve, J. H., Popescu, I., Duez, H., Velagapudi, V. R., Dumont, J., Bouchaert, E., van Dijk, T. H., Lucas, A., Dorchies, E., Daoudi, M., Lestavel, S., Gonzalez, F. J., Oresic, M., Cariou, B., Kuipers, F., Caron, S., & Staels, B. (2011). Farnesoid X receptor deficiency improves glucose homeostasis in mouse models of obesity. Diabetes, 60, 1861–1871.
Reimer, R. A., Willis, H. J., Tunnicliffe, J. M., Park, H., Madsen, K. L., & Soto-Vaca, A. (2017). Inulin-type fructans and whey protein both modulate appetite but only fructans alter gut microbiota in adults with overweight/obesity: A randomized controlled trial. Molecular Nutrition & Food Research, 61(11), 1700484.
Solon-Biet, S. M., Cogger, V. C., Pulpitel, T., Wahl, D., Clark, X., Bagley, E., Gregoriou, G. C., Senior, A. M., Wang, Q. P., Brandon, A. E., Perks, R., O’Sullivan, J., Koay, Y. C., Bell-Anderson, K., Kebede, M., Yau, B., Atkinson, C., Svineng, G., Dodgson, T., … Simpson, S. J. (2019). Branched chain amino acids impact health and lifespan indirectly via amino acid balance and appetite control. Nature Metabolism, 1, 532–545.
Thomas, C., Gioiello, A., Noriega, L., Strehle, A., Oury, J., Rizzo, G., Macchiarulo, A., Yamamoto, H., Mataki, C., Pruzanski, M., Pellicciari, R., Auwerx, J., & Schoonjans, K. (2009). TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metabolism, 10, 167–177.
Trabelsi, M. S., Daoudi, M., Prawitt, J., Ducastel, S., Touche, V., Sayin, S. I., Perino, A., Brighton, C. A., Sebti, Y., Kluza, J., Briand, O., Dehondt, H., Vallez, E., Dorchies, E., Baud, G., Spinelli, V., Hennuyer, N., Caron, S., Bantubungi, K., … Lestavel, S. (2015). Farnesoid X receptor inhibits glucagon-like peptide-1 production by enteroendocrine L cells. Nature Communications, 6, 7629.
Tsutsui, H., Maeda, T., Min, J. Z., Inagaki, S., Higashi, T., Kagawa, Y., & Toyo’oka, T. (2011). Biomarker discovery in biological specimens (plasma, hair, liver and kidney) of diabetic mice based upon metabolite profiling using ultra-performance liquid chromatography with electrospray ionization time-of-flight mass spectrometry. Clinica Chimica Acta, 412, 861–872.
Valcheva, R., Koleva, P., Martinez, I., Walter, J., Ganzle, M. G., & Dieleman, L. A. (2019). Inulin-type fructans improve active ulcerative colitis associated with microbiota changes and increased short-chain fatty acids levels. Gut Microbes, 10, 334–357.
Vanweert, F., de Ligt, M., Hoeks, J., Hesselink, M. K. C., Schrauwen, P., & Phielix, E. (2021). Elevated plasma branched-chain amino acid levels correlate with type 2 diabetes-related metabolic disturbances. Journal of Clinical Endocrinology and Metabolism, 106, e1827–e1836.
Vincent, R. P., Omar, S., Ghozlan, S., Taylor, D. R., Cross, G., Sherwood, R. A., Fandriks, L., Olbers, T., Werling, M., Alaghband-Zadeh, J., & le Roux, C. W. (2013). Higher circulating bile acid concentrations in obese patients with type 2 diabetes. Annals of Clinical Biochemistry, 50, 360–364.
Wachsmuth, H. R., Weninger, S. N., & Duca, F. A. (2022). Role of the gut-brain axis in energy and glucose metabolism. Experimental & Molecular Medicine, 54, 377–392.
Wahlström, A., Sayin, S. I., Marschall, H. U., & Bäckhed, F. (2016). Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metabolism, 24, 41–50.
Waise, T. M. Z., Lim, Y. M., Danaei, Z., Zhang, S. Y., & Lam, T. K. T. (2021). Small intestinal taurochenodeoxycholic acid-FXR axis alters local nutrient-sensing glucoregulatory pathways in rats. Mol Metab, 44, 101132.
Wei, M., Huang, F., Zhao, L., Zhang, Y., Yang, W., Wang, S., Li, M., Han, X., Ge, K., Qu, C., Rajani, C., Xie, G., Zheng, X., Zhao, A., Bian, Z., & Jia, W. (2020). A dysregulated bile acid-gut microbiota axis contributes to obesity susceptibility. eBioMedicine, 55, 102766.
Wewalka, M., Patti, M. E., Barbato, C., Houten, S. M., & Goldfine, A. B. (2014). Fasting serum taurine-conjugated bile acids are elevated in type 2 diabetes and do not change with intensification of insulin. Journal of Clinical Endocrinology and Metabolism, 99, 1442–1451.
Yoshii, K., Hosomi, K., Sawane, K., & Kunisawa, J. (2019). Metabolism of dietary and microbial vitamin B family in the regulation of host immunity. Frontiers in Nutrition, 6, 48.
Zhang, C., Yin, A., Li, H., Wang, R., Wu, G., Shen, J., Zhang, M., Wang, L., Hou, Y., Ouyang, H., Zhang, Y., Zheng, Y., Wang, J., Lv, X., Wang, Y., Zhang, F., Zeng, B., Li, W., Yan, F., … Zhao, L. (2015). Dietary modulation of gut microbiota contributes to alleviation of both genetic and simple obesity in children. eBioMedicine, 2, 968–984.
Zhang, S. Y., Li, R. J. W., Lim, Y. M., Batchuluun, B., Liu, H., Waise, T. M. Z., & Lam, T. K. T. (2020). FXR in the dorsal vagal complex is sufficient and necessary for upper small intestinal microbiome-mediated changes of TCDCA to alter insulin action in rats. Gut, 70(9), 1675–1683.
Zhao, H., Zhang, F., Sun, D., Wang, X., Zhang, X., Zhang, J., Yan, F., Huang, C., Xie, H., Lin, C., Liu, Y., Fan, M., Yan, W., Chen, Y., Lian, K., Li, Y., Zhang, L., Wang, S., & Tao, L. (2020). Branched-chain amino acids exacerbate obesity-related hepatic glucose and lipid metabolic disorders via attenuating Akt2 signaling. Diabetes, 69, 1164–1177.
Zhi, C., Huang, J., Wang, J., Cao, H., Bai, Y., Guo, J., & Su, Z. (2019). Connection between gut microbiome and the development of obesity. European Journal of Clinical Microbiology and Infectious Diseases, 38, 1987–1998.
Zinöcker, M. K., & Lindseth, I. A. (2018). The western diet-microbiome-host interaction and its role in metabolic disease. Nutrients. https://doi.org/10.3390/nu10030365
Funding
This study was funded by the Arizona Biomedical Research Commission New Investigator Award (ADHS18-198857), National Institute of Food and Agriculture (67017-29252), and NIDDK (R01ES033993).
Author information
Authors and Affiliations
Contributions
FAD conceived and designed research. FAD, RKM, and MAB conducted experiments. RKM and MAB analyzed data. FAD and RKM wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Meyer, R.K., Bime, M.A. & Duca, F.A. Small intestinal metabolomics analysis reveals differentially regulated metabolite profiles in obese rats and with prebiotic supplementation. Metabolomics 18, 60 (2022). https://doi.org/10.1007/s11306-022-01920-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-022-01920-9