Abstract
Introduction
Methods for the automated and accurate identification of metabolites in 1D 1H-NMR samples are crucial, but this is still an unsolved problem. Most available tools are mainly focused on metabolite quantification, thus limiting the number of metabolites that can be identified. Also, most only use reference spectra obtained under the same specific conditions of the target sample, limiting the use of available knowledge.
Objectives
The main goal of this work was to develop novel methods to perform metabolite annotation from 1D 1H-NMR peaks with enhanced reliability, to aid the users in metabolite identification. An essential step was to construct a vast and up-do-date library of reference 1D 1H-NMR peak lists collected under distinct experimental conditions.
Methods
Three different algorithms were evaluated for their capacity to correctly annotate metabolites present in both synthetic and real samples and compared to publicly available tools. The best proposed method was evaluated in a plethora of scenarios, including missing references, missing peaks and peak shifts, to assess its annotation accuracy, precision and recall.
Results
We gathered 1816 peak lists for 1387 different metabolites from several sources across different conditions for our reference library. A new method, NMRFinder, is proposed and allows matching 1D 1H-NMR samples with all the reference peak lists in the library, regardless of acquisition conditions. Metabolites are scored according to the number of peaks matching the samples, how unique their peaks are in the library and how close the spectrum acquisition conditions are in relation to those of the samples. Results show a true positive rate of 0.984 when analysing computationally created samples, while 71.8% of the metabolites were annotated when analysing samples from previously identified public datasets.
Conclusion
NMRFinder performs metabolite annotation reliably and outperforms previous methods, being of great value in helping the user to ultimately identify metabolites. It is implemented in the R package specmine.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Metabolomics is a recent omics technology with a wide range of applications, spanning from the study of human diseases, such as finding diagnostic and prognostic biomarkers, and predict treatment response, to nutrition and drug discovery (Alonso et al. 2015; Villas-Boas et al. 2007).
Several techniques are used in metabolomics, being Nuclear Magnetic Resonance (NMR) one of the most important. In NMR spectroscopy, the spectra obtained display chemical shifts on the x-axis and the intensity of those shifts on the y-axis. Some atomic nuclei (1H, 13C, 15 N, e.g.) possess a magnetic moment (i.e., nuclear spin), which gives rise, upon a magnetic field, to different energy levels and resonance frequencies. In order to correctly characterise and specify the location of NMR signals, usually their location in a spectrum is reported relative to a reference signal from a standard compound added to the sample, such as trimethylsilylpropanoic acid (TSP) or tetramethylsilane (TMS). Since the separation (or dispersion) of NMR signals is magnetic field dependent, an unambiguous location unit must be provided for the resonances of a set of nuclei. Thus, the frequency differences are corrected by dividing each resonance value by the NMR’s spectrometer frequency (400, 500, 600 MHz, e.g.). Since the resulting number is very small, it is multiplied by 106, resulting in a locator number named the chemical shift (δ), with parts-per-million (ppm) as unit.
The most used atomic nucleus for the determination of the chemical shifts is hydrogen (1H-NMR), by virtue of being the most abundant in biological samples. Although equipment costs are high, NMR is a non-destructive technique and highly reproducible. It has been widely applied to obtain metabolic profiles of complex biological mixtures. In fact, each metabolite has a characteristic pattern of chemical shifts, i.e., chemical signature, due to its composition of the nuclei in question and placement of atoms in the molecule (Alonso et al. 2015).
The identification of the metabolites present in an NMR sample can be achieved by comparing the sample spectrum to a set of reference spectra. Each of these reference spectra, ideally obtained under the same experimental conditions of the sample’s spectrum, represents a metabolite. When some (ideally all) peaks of a reference spectrum match peaks of the sample, that metabolite can be considered as identified with high confidence. However, this is not a trivial task, especially if performed by hand. Among other difficulties, some of the peaks of a reference metabolite can be shared with others, leading to false positives, and there can be a scarcity of metabolites with reference spectra acquired under the same exact conditions as those of the samples. In this sense, automated methods capable of performing such a task accurately are crucial.
Publicly available software tools that identify metabolites from 1D 1H-NMR data are mainly focused on performing metabolite quantification. To perform this task, these tools end up limiting the number of metabolites that can be identified. While some rely on the user to give a small list of metabolites whose quantification is intended, like Batman (Hao et al. 2012), others only contain NMR spectra obtained under one frequency, like ASICS (Lefort et al. 2019). Tools like Bayesil (Ravanbakhsh et al. 2015) are strict in the conditions in which the samples’ spectra must be collected so that the identification can be reliable and were only conceptualised to work with mammalian serum, plasma or CSF. Other interesting tools like MIDTool (Filntisi et al. 2017) do not seem to take into consideration the conditions under which the spectra were acquired to filter the reference library according to the data to analyse. Finally, some of these tools do not give the users much liberty in choosing the best parameters for their specific case, and most only receive as input raw spectra, not allowing the users to provide lists of peaks, with the exception of MetaboHunter (Tulpan et al. 2011).
In this scenario, three new algorithms were designed, implemented and tested for their capacity to correctly annotate the metabolites present in 1D 1H-NMR synthetic samples, and also compared to publicly available tools. From these results, we selected the best performing algorithm, which we name NMRFinder, as the one that most reliably performs metabolite annotation, showing better performance than the publicly available methods tested. NMRFinder allows matching the samples with all the reference peak lists in the library, regardless of whether the conditions under which these references were acquired are the same or not to those of the samples. This allows scoring the putative metabolites in the library according to several criteria: how many peaks match the samples, how unique their peaks are in the library and how close the spectrum acquisition conditions are to those of the samples. This way, the number of metabolites that could be left undetected decreases, without increasing the number of false positives.
Our method was tested by generating synthetic data simulating the effects that several situations have on the capacity to correctly perform metabolite annotation: number of different metabolites present in the samples, references missing for metabolites, missing peaks and peak shifts. Our method has shown a remarkable robustness even in the most extreme scenarios, and compared favourably to previously available methods. Our method was further validated with experimental data from several studies available in the MetaboLights (Haug et al. 2020) database. Around 72% of the metabolites reported by the authors were annotated by our method, with an additional annotation of on average 311 metabolites, which are present in the respective organisms’ metabolome.
A vast, robust and up-do-date reference library has also been proven to be very important for correct metabolite identification and annotation. As such, we collected reference peak lists from several sources to populate our library. We were able to gather a total of 1816 references for 1387 different metabolites across different acquisition conditions. We have more than 500 peak lists acquired with 400, 500 or 600 MHz NMR spectrometers, while solvents like water, chloroform (CDCl3), and DMSO each have more than 100 references.
NMRFinder, together with the constructed library and the other algorithms developed in this work, were implemented in the latest version of the R package specmine, available through CRAN. This allows full reproducibility of our results, and also that the community can freely apply these new methods to annotate their data and thus aid in metabolite identification.
2 Materials and methods
2.1 Matching algorithms
Three different algorithms were tested to compare the capacity for correctly annotating the metabolites present in 1D 1H-NMR samples, so that an annotation method could be proposed. All these algorithms are based on evaluating how the peaks from a sample match the peaks of the different reference compounds in a library, by solely comparing the chemical shift values of those peaks. A peak from a sample is considered to match a peak from a reference when the difference between the chemical shifts of those two peaks is less or equal to a tolerance value. For example, while the peaks 0.1 and 0.1 will always be considered a match, the peaks 0.1 and 0.12 are only considered a match if a peak tolerance of at least 0.02 is allowed. One peak in a sample is always allowed to match peaks from more than one reference metabolite. With this in mind, we next explain how the three different algorithms use these matches to score the presence of a reference metabolite in a sample.
2.1.1 Hypergeometric tests
We started by evaluating the use of hypergeometric tests as a way to annotate the metabolites present in a sample. These tests are often used to test which sub-populations are over- or under-represented in a population, by calculating the probability of having k successes caused by chance. In our case, we used the hypergeometric distribution to calculate the probability of a group of k peaks matching to a certain reference peak list being caused by chance (Eq. 1):
where N is the population size (i.e., number of different peaks in the library), K the number of success states in the population (i.e., peaks in the reference), n the number of draws in a trial (i.e., peaks in a sample), and k the number of observed successes (peaks in a sample that match the reference).
Thus, a P(X = k) value below a certain defined threshold denotes that the metabolite corresponding to the reference peak list in question is considered as being present in the sample. When repeating this process for all compounds in the reference library, these values are adjusted using the False Discovery Rate (FDR) approach, with the Benjamini & Hochberg method (Benjamini and Hochberg 1995).
The P(X = k) values are then transformed into a scale from zero to one, allowing to calculate an hypergeometric test score (Eq. 2) for each compound (HTS). This allows to assign normalised scores which may be integrated with other evidences of the presence of that same compound. All compounds with P(X = k) above a certain α are scored 0 in this score. The remaining get a score that approaches 1 as the P(X = k) value approaches 0.
2.1.2 Hypergeometric tests with uniqueness scores
More compounds present in a sample most certainly result in more different peaks detected. This can cause an increase in the number of peaks that belong to more than one different reference. This high peak overlap can hinder an algorithm’s capacity to accurately identify or annotate the compounds present, possibly increasing false positives. Thus, we decided to test if the algorithm could perform better when taking into consideration in the final score how much the peaks in each reference peak list overlap with other references. Towards this end, we decided to implement a uniqueness score.
For each peak p, we first define the uniqueness rate of that peak (Eq. 3) by dividing 1 by the number of references that peak belongs to (np), based on the reference library used.
The uniqueness score of a reference compound c is then calculated as the mean of the uniqueness rate of all peaks in its peak list (Eq. 4):
where K is the number of different peaks in the reference, and pi the i-th peak in the reference of compound c. The greater the uniqueness value, the less overlap it has with other peak lists. In other words, a uniqueness score of 1 corresponds to a compound whose peaks in its peak list do not overlap with any other peak list in the library.
A combined matching score of a compound can be calculated by averaging the hypergeometric test score and the uniqueness score (Eq. 5).
The closer to 1 the final score is, the more reliable is the identification of the metabolite in question.
2.1.3 Matched ratio scores with uniqueness scores
We also decided to explore how well the identification is accomplished by combining the uniqueness scores with a score given by the ratio of peaks matched to the peak list of the reference compound, instead of using the hypergeometric tests. This third algorithm thus combines the uniqueness score (Eq. 4) with the matched ratio score (MRS), which gives the ratio between the number of peaks from the peak list of a reference compound c that matched with the peaks in the sample (nmatched(c)) and the total number of different peaks in that reference peak list (NR) (Eq. 6).
The combined score of a reference compound is, in this case, obtained by calculating the average between the matched ratio score and the Uniqueness Score (Eq. 7).
2.2 NMRFinder
After evaluating the alternatives above, we reached NMRFinder as our final method, where a sample is matched with each reference compound in the library at a time, regardless of whether the conditions under which the references were acquired are equal or not to those of the samples. The matching score used is the combination of matched ratio scores with uniqueness scores from the previous section, as it was the best performing of the three alternatives.
After scoring a match between a reference compound and a sample, the compound is further scored regarding the conditions under which the reference spectrum was acquired as compared to the acquisition parameters of the sample. To do so, the user needs to define a score between 0 and 1 for every possible value of the parameters NMR spectrometer frequency (e.g. 400, 500 MHz) and solvent (e.g. water, DMSO, acetone) in the library. Higher values denote more “similarity” with the samples’ conditions. Default values are provided in our implementation. pH and temperature used for acquirement of reference spectra were not considered, as most of the references gathered from the various databases lacked information regarding these conditions and, when present, most were acquired under the same conditions (see Sect. 3.1).
The users also have the possibility to score each reference compound according to its presence or absence in the organism(s) or group(s) of organisms under study. This aims to ensure that metabolites not reported to be present in an organism/group of interest are less likely to be identified. The presence of a metabolite in an organism/group is assessed using the information in the KEGG database (Kanehisa and Goto 2000). Considering that this database is likely incomplete regarding the metabolites present, the user has also the possibility to define a score for those metabolites not present in the database and those not present in the specified organism(s)/group(s). The scores given to each organism/group should vary from 0 to 1.
The final score of a compound is the average of the matching score (MS) and the user given scores for frequency (freq_score—FS), solvent (solv_score—SS) and organism (org_score—OS) (Eq. 8).
There are two main reasons supporting the introduction of the condition scores: (i) allow matching of the samples with all the references in a library, regardless of the conditions, so that the uniqueness score can better rate the uniqueness of a reference peak list; and (ii) the closer the conditions are to those of the samples, the better the final score is, giving more confidence to the metabolite annotated.
When multiple peak lists acquired under different conditions are available for a single metabolite, all of them are used in the annotation process, even if one of the peak lists has the same exact conditions as those of the sample(s). This allows to increase the chances of the metabolite not being incorrectly missed, but the user must have in mind that a single metabolite might be annotated more than once for the same data.
Finally, NMRFinder, being a method that only annotates the metabolites that might be present in the sample(s), will not report as unknown peaks that do not match any reference in the library metabolite.
2.3 Code implementation
The proposed 1D 1H-NMR annotation methods are implemented in the R package specmine (Costa et al. 2016), available in CRAN. The results are returned to the user in the form of a data frame. For each reference peak list, the user is given the library’s name and identifiers of the corresponding metabolite, the final score and the score of each component used (matching score, frequency score, solvent score, organism score), the number of peaks matched and an identifier allowing to access more detailed results, which are stored in a list. The detailed results for each reference comprise the peaks of the reference that matched the sample, the peaks of the sample that matched the reference, and all the reference’s peaks.
The user is able to set the condition scores and the value of tolerance in ppm to consider when matching two peaks, eventually associating it to the NMR spectrometer’s digital resolution used to acquire the spectrum. The information regarding the presence of metabolites in the organisms or groups of organisms is taken from the KEGG database through their KEGGREST R package (Tenenbaum 2016).
2.4 Methodology for assessing the performance of the algorithms
To assess the performance and limitations of the three algorithms for scoring reference metabolites described in the previous section, we ran a series of computational experiments to evaluate how correctly and precisely the algorithms could annotate the set of metabolites in different samples.
In the first set of experiments, so that the composition of the samples would be entirely known, and thus correctly evaluate the algorithms’ performance, we created synthetic datasets from reference peak lists. In these preliminary experiments, we restricted our library to peak lists whose spectra were acquired at a frequency of 500 MHz and using water as solvent, and only those known to be part of the human metabolism according to the KEGG database. We thus obtained a restricted library whose references had the same conditions of acquisition.
From this restricted library, 19 datasets were created, each containing samples with a given number of compounds, whose reference peak lists were used to create the respective sample peaks. The number of compounds varied from 10 in the first dataset to 190 in the last (increasing with a step of 10 compounds). Each dataset contains 50 different replicates (samples), where the number of compounds is the same, but the metabolite composition differs, by being randomly chosen in each case. When joining the reference peak lists for the different compounds together in a sample, peaks with a ppm difference lower than 0.001 ppm were considered the same peak. The chemical shifts of these peaks were thus merged, and respective intensities summed. As the methods tested only aim at identifying or annotating the metabolites by solely analysing the chemical shifts, no changes in peak lists’ intensity were made to take into consideration metabolites concentrations in the simulation of synthetic mixtures. The synthetic datasets are available in Online Resource 1.
Our three algorithms were then tested for metabolite annotation, using only the restricted library created. We also used these synthetic datasets to evaluate other tools publicly available and compare to the results of our algorithms. We only chose those tools that are able to perform metabolite identification or annotation for 1D 1H-NMR data, and that can work with different acquisition conditions and take them into consideration when performing the identification or annotation. The only tool that filled all of these requirements, and that allowed a list of peaks as input, was MetaboHunter (Tulpan et al. 2011).
MetaboHunter is a web tool that performs metabolite annotation from spectral data points or peak lists. It provides three different algorithms: MH1: Highest number of matched peaks, MH2: Highest number of matched peaks with shift tolerance, MH3: Greedy selection of metabolites with disjoint peaks. Because the MH1 algorithm did not allow peak tolerances, we did not use this method in our comparisons. MH2 allows peak tolerances when calculating the significance score of the metabolites. MH3 is a greedy selection algorithm, developed to identify as little false positives as possible. After matching all references to the sample, it iteratively chooses the highest scored metabolite, i.e., the one with the highest number of peaks and whose peaks do not yet belong to previously selected metabolites.
When running MetaboHunter, we filtered its reference library to make the annotation as similar as possible to ours except for the scoring. We were able to choose the peak lists with the following options: ‘Solvent’ (Water) and ‘Frequency’ (500 MHz). Because we filtered our library to only use peak lists of metabolites known to be present in human metabolism (using KEGG database), and MetaboHunter only groups the peak lists according to groups of species and not also by species, we could not filter the library to not contain metabolites not normally present in humans. Instead, the samples were matched using the group where Homo sapiens belongs to, by setting the type of metabolite to Mammalian. The tool further requires choosing between peak lists from the MMCD (Madison Metabolomics Consortium Database) (Cui et al. 2008) or the HMDB (Human Metabolome Database) (Wishart et al. 2018) databases. We chose the HMDB database as all metabolites in the reference library for our experiments were extracted from the HMDB database. We also implemented the algorithms MH2 and MH3 to use them with our restricted library of reference peak lists. This allowed us to better compare the algorithms, as the differences observed in the results could be caused by the differing libraries and not by the algorithms.
Next, we used the full library of collected reference peak lists to further evaluate NMRFinder with the synthetic datasets.
We started by testing the effect of removing references that were used to construct the simulated datasets. For each sample, a certain percentage of the references used to construct that said sample were randomly removed from the library prior to metabolite annotation. After each sample was analysed, references were added back to the library and the same process repeated to the next sample. Eleven different levels of “missing” references (from 0 to 100%, increasing with a step of 10%) were thus defined for each sample in the dataset including samples with 190 compounds. Of note, 100% of missing peak lists means that all references that were used to construct a sample were not present in the library used for annotation of that sample, i.e., references not used to construct a sample were still present in the library.
We then evaluated the effect that noise in chemical shifts has in NMRFinder’s metabolite annotation of the dataset of samples with 190 compounds each. To mimic the absence of peaks, we randomly removed from each sample a certain percentage (from 0 to 60%, increasing with a step of 10%) of peaks prior to annotation. Separately, peak shifts were also tested, by adding six different levels of peak shifts (from 0 to 5) to the samples at a time prior to annotation. For example, while in level 1 all peaks are randomly shifted with 0 or 0.01 ppm with equal probabilities, in level 5 all peaks are randomly shifted with 0, 0.01, 0.02, 0.03, 0.04 or 0.05 ppm. Level 0 corresponds to no addition of peak shifts.
In a second set of experiments, we used real experimental data, gathered from the MetaboLights database (Haug et al. 2020). We evaluated if the best performing algorithm, NMRFinder, could annotate the metabolites reported by the authors and others not reported, by adding the condition and organism specific scores.
To evaluate and compare different algorithms we used receiver operating characteristic (ROC) curves. ROC curves are created by plotting the true positive rate (TPR), also known as sensitivity, over the false positive rate (FPR), also known as fall-out, through different levels of a decision. ROC curves thus help evaluating the performance of an algorithm by giving a visual representation of the cost/benefit. The closer the curve is to the yy axis, the better the algorithm. The area under the ROC curve (AUC) is used to numerically represent this, i.e., the optimal AUC is 1. With this in mind, we evaluated the performance of the algorithms by looking into their ROC curves when varying the threshold stating which the metabolites are considered identified.
3 Results
3.1 Reference library
Reference peak lists for metabolites were collected from databases such as the Human Metabolome Database (HMDB) (Wishart et al. 2018), Biological Magnetic Resonance Data Bank (BMRDB) (Ulrich et al. 2007) and Spectral Database for Organic Compounds (SDBS) (National Institute of Advanced Industrial Science and Technology 2020) to populate our library. Some in-house peak lists were also produced and inserted into the library. We were able to gather a total of 1816 peak lists for 1387 different metabolites across different acquisition conditions. Each list is composed by the peaks’ chemical shift values and corresponding intensities. This library is provided with the R package specmine, where the proposed 1D 1H-NMR annotation methods are implemented.
From these lists, over 700 compounds were acquired with a frequency of 400 MHz, 566 with 500 MHz and 532 with 600 MHz (Figure S1(a) from Online Resource 2). We were only able to gather 1 peak list acquired with 700 MHz. Regarding the solvents, water and deuterated chloroform (CDCl3) make up around 80% of the library (Figure S1(b) from Online Resource 2), with 51% using water and around 29% using CDCl3. Out of the 1816 references, 1015 were acquired with a pH between 7 and 8, while 753 lacked information. Also, 1160 peak lists were acquired with 25ºC, but 647 have no information on temperature of acquisition.
We evaluated the number of metabolites which have a peak at each ppm position (Figure S3 from Online Resource 2). It is possible to realise that the spectral region between 1 and 4 ppm contains the highest concentration of overlaps. The peak with the most overlap is 1.34 ppm, present in 112 reference peak lists. Outside this region, only 33 other peaks are present in more than 30 reference peak lists. While the average overlap is 4.933, the median is 2.
There are 155 reference peak lists that totally overlap with 1 or more other compounds in the library that have the same or more number of peaks. From these, 84 only have one peak and 13 have two peaks. There are 10 compounds (5 pairs) that have the same exact peaks: fumaric acid and maleic acid, L-lactic acid and D-lactic acid, FAD and FADH, D-lysine and L-lysine, and D-tagatose and L-sorbose. These compounds are, e.g., epimers (tagatose and sorbose) or enantiomers (L- and D- lactic acids, lysine), and are not discriminated. Each pair of compounds will always be inevitably annotated together.
Nevertheless, 3589 (out of 9081) peaks are only present in one reference list, which corresponds to 676 references that may be unequivocally annotated based on that peak. Still, only 27 references out of these 676 contain only peaks that do not overlap with any other peak list.
3.2 Evaluation of the metabolite annotation algorithms
3.2.1 Comparison of the matching algorithms
Metabolite annotation was conducted on all synthetic datasets for all the different matching algorithms evaluated. Due to how datasets were created, we allowed a peak tolerance of 0.001 ppm. At this stage, we only used as our reference library the peak lists whose acquisition conditions were the same as those used to create the datasets. As such, our library was composed by 198 references in total.
We also evaluated the evolution of the false and true positive rates with an increasing number of compounds in samples after calculating the best threshold score for each dataset of compounds, for each algorithm. The best score corresponds to the one whose difference between the TPR and FPR is the largest. The results were summarised in Fig. 1; Table 1. Detailed results are given in Online Resource 2 (Figures S4 to S8).
Evolution of the area under the ROC curve (AUC), false (FPR) and true (TPR) positive rates through the increased number of compounds that make up the samples in the datasets. Metric values for each dataset are the mean of each sample’s metrics. Hyper hypergeometric tests; Hyper_uniq hypergeometric tests with uniqueness scores; Match_uniq matched ratio scores with uniqueness scores; MH2_ourLibrary MetaboHunter’s MH2 algorithm implemented with our reference library; MH3_ourLibrary MetaboHunter’s MH3 algorithm implemented with our reference library. The dashed grey line represents the value 0.6
Overall, the highest and most constant TPR values along the datasets are given by the algorithm that uses hypergeometric tests (Hyper), with a mean TPR of 0.709 across all datasets. However, the FPR is high in this algorithm, with values very close to 0.5 in most datasets. A lower rate of FPs would be more advised. When the uniqueness scores are used with the hypergeometric tests (Hyper_uniq algorithm), the AUC increases (Table 1). The average AUC across all datasets is 0.713 and the average FPR decreases to 0.272. This denotes that the uniqueness scores help to avoid false positives. However, the TPR decreases to 0.611, yet less than the decrease in FPR.
The algorithm using the matched ratio and uniqueness scores (Match_uniq) has the second best mean TPR, near the best one, 0.697. Moreover, the FPR is substantially lower when compared to the two algorithms mentioned previously, reaching a mean of 0.145. Indeed, the mean AUC value is clearly the best out of all algorithms tested, 0.835. This means that it is the algorithm that best balances the rate of metabolites correctly identified and those incorrectly identified.
As regards the MetaboHunter, the MH2 implemented with our reference library (MH2_ourLibrary) is the algorithm that has the closest performance to that of Match_uniq. In fact, the mean FPR across all datasets for algorithm MH2_ourLibrary is the same as that of Match_uniq. However, the average TPR is slightly smaller, leading to a smaller AUC.
The algorithm that clearly performs worst regarding TPR and AUC is the MH3 implemented with our reference library (MH3_ourLibrary). This denotes that the scoring is too restrictive and does not allow the annotation of a reasonable number of metabolites for posterior analysis of the samples in a biologically meaningful way. As mentioned here before, and also by the authors (Tulpan et al. 2011), this method was created to minimise FPs, which is supported by our results (the FPR is below 1%).
Finally, while MetaboHunter’s MH2 performs very closely to Match_uniq when using our library of reference peak lists, such performance decreases significantly when running the MH2 algorithm through MetaboHunter’s webtool (results present in Online Resource 2). Although MetaboHunter’s library used in this case was composed by 273 metabolites and ours was by only 198 metabolites, MetabHunter’s library comprises all mammalian metabolites and not just those from humans. Also, 35 out of the 198 metabolites in our library are not present in MetaboHunter’s library. With this, approximately 82% of the library used in this case (only references for 500 MHz, water and human metabolism) overlaps with the MetaboHunter’s library. It is thus understandable why the MetaboHunter results are better when using our library (Figures S9 to S12 from Online Resource 2). As we propose our method to have a new algorithm and a library as vast and complete as possible, our method outperforms MH2 slightly as regards to the algorithm, but considerably when it comes to the reference library.
3.2.2 Effect of missing references
As most metabolites do not have reference spectra for all the different conditions under which the NMR samples may be collected, we next evaluated the effect of not having all references for those metabolites that are present in our samples. We defined 11 different levels of “missing” references (from 0 to 100%), as explained in detail in Sect. 2.4. We used all reference peak lists in our library, regardless of acquisition conditions, to assess if the metabolites with no reference and that have an alternative peak list in other conditions could still be identified. This was performed using the best algorithm NMRFinder, using the matched ratio and uniqueness scores (Match_uniq) combined with the condition scores (see Sect. 2.2). Several different combinations of frequency and solvent scores were tested (Online Resource 2).
Figure 2 shows the results for three situations: (1) using only as references those with the exact same condition as those of the samples (Specific References), (2) using all references available in the library but giving a score of 1 to all conditions equal to those of the samples and a score of 0 to all others (Specific Scores), (3) using all references available in the library and the best combination of condition scores that are not the Specific Scores (Best Non-Specific Scores). The best combination was the one that had a higher difference between TPR and FPR. Detailed results are given in Online Resource 2.
Comparison of the TPR and FPR values between using Specific References (only the references with the exact same frequency and solvent conditions as those of the samples were used); Specific Scores (it uses all references in the library, and all reference conditions equal to those of the samples have a score of 1 as those that that are not equal have a score of 0); and Best Non-Specific Scores (it uses all references in the library and the best combination of condition scores that is not the specific score). Each panel represents the results for a certain percentage of “missing” peak lists (from 0 to 100%). Only the specific peak lists regarding the compounds that make up the respective samples were considered for removal
Throughout all cases of missing references, using only the peak lists that were acquired under the same conditions as those of the samples (Specific References) leads to the worst results. This difference is more significant when there are fewer missing peak lists. Thus, allowing the comparison of the samples to all references in the library improves the correct annotation of metabolites, as it allows peak lists acquired under similar conditions, but not the same, to be correctly considered a match and to not leave a metabolite undetected. Also, the calculation of the uniqueness scores leads to a more precise score of the matches.
For samples with 0 and 10% missing references, the TPs are, on average, equal between the Specific Scores and Best Non-Specific Scores, while the number of FPs increases slightly. Thus, when the number of missing references is low, using the Specific Scores is better than giving some liberty in the condition scores (Best Non-Specific Scores). This low number of missing peak lists may allow the Specific Scores to be restrictive enough to lower the detection of FPs, but without losing the capacity of annotating the metabolites that are actually present.
In the samples with 30% or more missing references, it is possible to observe a slight increase in the identification capacity from the Specific Scores to the Best Non-Specific Scores. As expected, the increase of missing references decreases the capacity to detect the present metabolites. However, the Best Non-Specific Scores improves, even though slightly, the results. Also, it is only with the samples with 60% or more of missing peak lists that it is not possible to obtain more than 50% of the metabolites present with the Best Non-Specific Scores. The Specific Scores can only identify more than 50% of the present metabolites for samples with 40% or less of missing references.
As having a library with reference peak lists for all existing metabolites in all the different conditions of acquisition is a very difficult task, the use of condition scores shows to improve the annotation. However, this can only be of help if the metabolites present have alternative peak lists in other conditions. Regarding this case, only 91 out of the 198 references used have alternative peak lists.
3.2.3 Effect of missing peaks and peak shifts
We next evaluated how much the annotation capacity of our algorithm is affected by the presence of missing peaks and peak shifts in the samples. These problems can happen very frequently in NMR samples due to physical or chemical variations during sample preparation or even spectra acquisition. To address these, we modified the samples created originally so that they would contain such spectral variations and evaluated the effect on metabolite identification using our best algorithm NMRFinder (see Sect. 2.2). We used the Specific Scores defined in the previous case, which led to the best results. We only used the dataset of samples with 190 compounds.
3.2.3.1 Effect of missing peaks
To mimic the absence of peaks, we randomly removed from each sample a certain percentage (from 0 to 60%) of peaks prior to annotation. The results are provided in Table 2. As expected, the increase of missing peaks causes a decrease in TPR, while the FPR does not show a clear trend. Nevertheless, the TPR never falls below 0.9, showing that the algorithm has the capacity to overcome missing peaks without much effect on the annotation.
3.2.3.2 Effect of peak shifts
To mimic peak shifts, these were added to each sample prior to identification. Six different levels of peak shifts were tested (see Sect. 2.4). In this case, we allowed a peak tolerance of 0.03 ppm when matching peaks. This value was set so that the existence of peak shifts is considered, but without being too loose that could potentially cause identification of too many FPs. The results were summarised in Table 3.
As expected, the increase in the level of peak shifts causes a decrease in TPR, while the FPR does not show a clear tendency. Peaks shifts, however, seem to affect more the capacity to correctly annotate the metabolites present than the absence of peaks. Indeed, the TPR falls below 0.9 at level 3 or above, which corresponds to the tolerance given (0.03 ppm). Still, the TPR never falls below 0.88, accompanied by a very low value of incorrectly detected metabolites.
3.3 Metabolite annotation with real datasets
We next tested our algorithm by performing annotation on publicly available datasets, stored in the MetaboLights database (Haug et al. 2020). From the NMR studies available, 29 were used. The studies had to satisfy two requirements to be considered in this analysis: (a) the format of the data files had to follow that of Bruker or Varian processed files, or be a list of identified peaks; (b) have available the list of identified metabolites by the authors (at least 10). Some of the studies separated the samples into different groups prior to identification. For these cases, we also separated the samples accordingly, totalling 35 different datasets. A full list of the datasets used, as well as the description of the processing steps performed on the data prior to metabolite annotation, are given in Online Resource 2.
The NMRFinder (see Sect. 2.2) was used, with a peak tolerance of 0.03 ppm. The scoring was similar to the one used in the previous sections (Specific Scores). In those situations where the organism studied was not present in KEGG, the smallest group of organisms in which that organism is inserted was used for organism scoring. The threshold score for annotation was set based on the best threshold obtained for the dataset including samples with 190 compounds in the case in Sect. 3.2.1. This best threshold, as explained in Online Resource 2, is the threshold in the ROC curves that corresponds to the one whose difference between TPR and FPR is larger. The value obtained was 0.578. However, for the annotation of the real datasets, we decided to be a little less strict and used the value 0.5. The full list of metabolites annotated by NMRFinder for each dataset is presented in Online Resource 3.
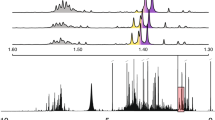
When reported metabolites were annotated by our method, we searched if the reference that allowed such identification had the exact same acquisition conditions as those under which the samples were obtained (Identified with specific references), or if it had not (Identified with alternative references). When a reported metabolite was not annotated, we also searched, in spite of that, if it had a peak list acquired under the same exact conditions (Not identified although with specific references) or a peak list acquired under other conditions (Not identified although with alternative references). In some cases, the reported metabolites were not present in our library (No references). These results were summarised in Fig. 3.
Identification outcome of our method regarding the metabolites reported by the authors. Metabolites were classified into 5 different categories (see main text): No reference; Not identified although with alternative references; Not identified although with specific references; Identified with alternative references; and Identified with specific references. The number of metabolites identified by the studies’ authors is present at the bottom of each bar. The datasets marked with the red rectangle were acquired with a frequency of 700 MHz for 1D 1H-NMR spectra
Regarding the datasets acquired with a frequency below 700 MHz for 1D 1H-NMR spectra, our method annotated on average 71.8% of the metabolites reported by the authors. Furthermore, only an average of 40.8% of the reported metabolites would be annotated if only the metabolites with specific references were considered. Nevertheless, no reported metabolite with a reference acquired under the same conditions as those of the samples was left undetected. Although we only have one metabolite in our library with a peak list acquired with a frequency of 700 MHz, we were still able to annotate a mean of 55.1% of the metabolites reported by the authors. This was only possible through the references acquired under alternative conditions. These results show the relevance of using peak lists acquired under different conditions in the annotation of metabolites.
It is important to notice that our method’s inability to annotate some of the reported metabolites may be due to the processing of the raw spectra not being the exact same as that performed by the authors. In fact, some studies did not provide enough information on the data processing. Also, some studies were not completely clear about the conditions under which the samples’ data were acquired. In both cases, such information could improve the understanding of the outcomes reported regarding the ability of the methods in annotating metabolites in chemically complex samples, for instance.
We next evaluated the metabolites that our method annotated that were not reported by the authors (Figure S13 from Online Resource 2). These metabolites were separated into 4 different categories, regarding if they were annotated with specific references or alternative ones, and if they are present or not in the organism according to KEGG. While the mean number of metabolites reported by the authors is 39, we were able to additionally annotate, on average, 286 new metabolites. Some were annotated through peak lists acquired under the same conditions as those of the samples, but mostly by references with alternative conditions regarding frequencies and solvents. Furthermore, an average of 96.6% of these metabolites are present in the respective organism/ group, according to KEGG. This shows that the additionally annotated metabolites have a good level of reliability, although further validation is always necessary to consider a metabolite annotated by our method as identified.
4 Conclusion
Most public tools that identify or annotate metabolites from 1D 1H-NMR data are mainly focused on performing metabolite quantification. To perform such a task, these tools end up limiting the number of metabolites that can be identified and quantified. While some rely on the user to give a small list of metabolites present whose quantification is intended, others only contain spectra obtained under one frequency, or are even very strict in the conditions in which the samples’ spectra must be collected so that the identification can be reliable. Also, some tools do not give the users much liberty in choosing the best parameters for their specific case, and most only receive as input raw spectra.
To address these issues, we constructed a new method that annotates the metabolites that may be present from reference peak lists with great reliability. Thus, the user has total liberty to decide, in the end, which metabolites should be considered identified, with NMRFinder serving as a useful and reliable starting point for the user to identify metabolites. This method was added to the R package specmine (Costa et al. 2016), together with other methods, allowing the user to choose and test which is more suitable. We not only provide the metabolite annotated and respective final score, but also the individual scores and the peaks that matched between the respective references and the samples.
Data availability
The proposed 1H-NMR annotation methods are present in the R package specmine (Costa et al., 2016), available in CRAN.
References
Alonso, A., Marsal, S., & Julià, A. (2015). Analytical methods in untargeted metabolomics: State of the art in 2015. Frontiers in BioEngineering and BioTechnology, 3, 23.
Benjamini, Y., & Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological), 57(1), 289–300.
Costa, C., Maraschin, M., & Rocha, M. (2016). An R package for the integrated analysis of metabolomics and spectral data. Computer Methods and Programs in Biomedicine, 129, 117–124.
Cui, Q., Lewis, I. A., Hegeman, A. D., Anderson, M. E., Li, J., Schulte, C. F., et al. (2008). Metabolite identification via the Madison metabolomics consortium database. Nature Biotechnology, 26(2), 162–164.
Filntisi, A., Fotakis, C., Asvestas, P., Matsopoulos, G. K., Zoumpoulakis, P., & Cavouras, D. (2017). Automated metabolite identification from biological fluid 1H NMR spectra. Metabolomics, 13, 146.
Hao, J., Astle, W., De Iorio, M., & Ebbels, T. M. (2012). BATMAN—An R package for the automated quantification of metabolites from nuclear magnetic resonance spectra using a Bayesian model. Bioinformatics, 28(15), 2088–2090.
Haug, K., Cochrane, K., Nainala, V. C., Williams, M., Chang, J., Jayaseelan, K. V., & O’Donovan, C. (2020). MetaboLights: A resource evolving in response to the needs of its scientific community. Nucleic Acids Research, 48(D1), D440–D444.
Kanehisa, M., & Goto, S. (2000). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Research, 28(1), 27–30.
Lefort, G., Liaubet, L., Canlet, C., Tardivel, P., Père, M. C., Quesnel, H., et al. (2019). ASICS: An R package for a whole analysis workflow of 1D 1H-NMR spectra. Bioinformatics, 35(21), 4356–4363.
National Institute of Advanced Industrial Science and Technology (2020) SDBS—Spectral database for organic compounds. Retrieved from https://sdbs.db.aist.go.jp.
Ravanbakhsh, S., Liu, P., Bjordahl, T. C., Mandal, R., Grant, J. R., Wilson, M., et al. (2015). Accurate, fully-automated NMR spectral profiling for metabolomics. PLoS ONE, 10(5), e0124219.
Tenenbaum D. (2016) KEGGREST: Client-side REST access to KEGG. R package version 1(1).
Tulpan, D., Léger, S., Belliveau, L., Culf, A., & Čuperlović-Culf, M. (2011). MetaboHunter: An automatic approach for identification of metabolites from 1H-NMR spectra of complex mixtures. BMC Bioinformatics, 12(1), 400.
Ulrich, E. L., Akutsu, H., Doreleijers, J. F., Harano, Y., Ioannidis, Y. E., Lin, J., et al. (2007). BioMagResBank. Nucleic Acids Research, 36(suppl_1), D402–D408.
Villas-Boas, S. G., Nielsen, J., Smedsgaard, J., Hansen, M. A., & Roessner-Tunali, U. (2007). Metabolome analysis: An introduction (Vol. 24). Hoboken: Wiley.
Wishart, D. S., Feunang, Y. D., Marcu, A., Guo, A. C., Liang, K., Vázquez-Fresno, R., et al. (2018). HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Research, 46(D1), D608–D617.
Funding
This study was funded by the PhD scholarship with reference SFRH/BD/138951/2018, awarded by the Portuguese Foundation for Science and Technology (FCT).
Author information
Authors and Affiliations
Contributions
SC and DC collected data and implemented the new peak lists library. SC and MR designed the algorithms. SC implemented the algorithms. SC, MM and MR designed the experiments and analysed the results. SC and MR wrote the manuscript draft. All authors read, reviewed and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human and/or animal participants
This article does not contain any studies with human and/or animal participants performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cardoso, S., Cabral, D., Maraschin, M. et al. NMRFinder: a novel method for 1D 1H-NMR metabolite annotation. Metabolomics 17, 21 (2021). https://doi.org/10.1007/s11306-021-01772-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-021-01772-9