Abstract
Introduction
Untargeted metabolomics workflows include numerous points where variance and systematic errors can be introduced. Due to the diversity of the lipidome, manual peak picking and quantitation using molecule specific internal standards is unrealistic, and therefore quality peak picking algorithms and further feature processing and normalization algorithms are important. Subsequent normalization, data filtering, statistical analysis, and biological interpretation are simplified when quality data acquisition and feature processing are employed.
Objectives
Metrics for QC are important throughout the workflow. The robust workflow presented here provides techniques to ensure that QC checks are implemented throughout sample preparation, data acquisition, pre-processing, and analysis.
Methods
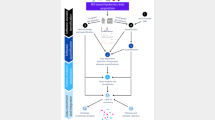
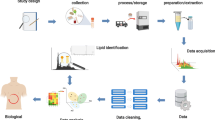
The untargeted lipidomics workflow includes sample standardization prior to acquisition, blocks of QC standards and blanks run at systematic intervals between randomized blocks of experimental data, blank feature filtering (BFF) to remove features not originating from the sample, and QC analysis of data acquisition and processing.
Results
The workflow was successfully applied to mouse liver samples, which were investigated to discern lipidomic changes throughout the development of nonalcoholic fatty liver disease (NAFLD). The workflow, including a novel filtering method, BFF, allows improved confidence in results and conclusions for lipidomic applications.
Conclusion
Using a mouse model developed for the study of the transition of NAFLD from an early stage known as simple steatosis, to the later stage, nonalcoholic steatohepatitis, in combination with our novel workflow, we have identified phosphatidylcholines, phosphatidylethanolamines, and triacylglycerols that may contribute to disease onset and/or progression.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Untargeted metabolomics aims to measure the widest array of metabolites possible. Well-defined workflows maintain quality and reproducibility of metabolomics data, (Kirwan et al. 2014; Vorkas et al. 2015) but are often overlooked by researchers. Unfortunately, standard protocols do not exist for the evaluation of untargeted metabolomics despite advocacy from the Metabolomics Standards Initiative (Sumner et al. 2007). Metabolite standards are generally used to equilibrate sample variation, but single internal standards are insufficient in untargeted metabolomics to provide adequate downstream normalization, as noted by Speed et al. (De Livera et al. 2012). This workflow (Fig. 1) combines novel and adapted methods for quality control (QC) and evaluation of lipidomics data to enable reasonable biological conclusions.
Specifically, elements of experimental design, including randomization during preparation and analysis, simplify data preprocessing (Dunn et al. 2011). One challenge of metabolomics is ensuring differences in signal between samples are due to independent variable(s), despite everything a sample endures from exposure (e.g. diet), preparation (e.g. storage, extraction, drying, reconstitution), data acquisition (e.g., ion suppression, column performance), and data processing (e.g. peak picking, normalization, scaling, statistics). Historically, homogenized tissue is subjected to analysis, followed by post-acquisition normalization to total protein, total ion signal (TIC), or a housekeeping metabolite (Silva et al. 2014; Worley and Powers 2013). Highly concentrated samples cause inconsistent ion suppression throughout chromatography, while less concentrated samples may have lower signal-to-noise, preventing proper filtration of background ions. The usual correction includes pre-processing data normalization, but small differences at each step become confounded, leading to under or over-correction post acquisition (Redestig et al. 2009). Therefore, meticulously adjusting solvent volume based on weight or protein prior to extraction negates excess normalization. Injection of equal concentrations (w/v) reduces the variables contributing to error in raw signal comparisons.
Internal and injection standards indicate sample preparation quality, ion suppression, and source cleanliness, which combined with randomization within sequence setup, ensures trends are not artificial. Deliberate integration of blanks and standards allows for quality checking and eases further data processing.
Further, data quality validation is imperative prior to further analysis, enabling confidence in results including inspection of: TIC variation, retention time (RT) drift, contamination, and injection standard peak area (PA). Feature detection and injection quality were assessed with metrics such as Bland–Altman (BA) plots (Broadhurst and Kell 2006), Standardized Euclidean distance plots (Qi and Voit 2016), distribution plots and the variation in RT and signal for the same features across samples within a condition.
Chromatogram and RT inspection indicates column integrity throughout the sequence and between batches. Contamination inspection ensures minimal interference of common contaminants including polyethylene glycol and polysiloxane (Dunn et al. 2011). Injection standard inspection, completed during data acquisition, maintains sample integrity for re-injection. Unsupervised multivariate statistics such as PCA identifies extreme outlying samples, indicating separation of blanks, QCs, samples and technical replicates. We analyzed QC parameters after each stage of filtering and processing to ensure data were of sufficient quality to proceed. All of these steps provided a quality comparison of healthy and diseased livers to improve the biological interpretation concerning lipid changes and disease manifestations.
Pre-processing aligned the features across the data set. Features were defined by a specific m/z and RT representing a single ion or group of co-eluting isomeric ions. Untargeted collection resulted in features from which those of biological origin must be distinguished from noise. Therefore, we introduced Blank Feature Filtering (BFF), where features with high signal in the extraction blank compared to the sample were removed, eliminating features with low S/N. This workflow improves confidence in results and evaluates quality in untargeted metabolomics experiments.
Nonalcoholic fatty liver disease (NAFLD) affects nearly one-third of Americans and the number is expected to double by the year 2025 (Browning et al. 2004). Closely associated with insulin resistance, obesity and type 2 diabetes mellitus (Koliaki and Roden 2013), NAFLD is comprised of a few stages. Triglyceride accumulation alone or with mild liver inflammation is diagnosed as simple steatosis (SS) (Pellicoro et al. 2014). The SS stage of NAFLD is reversible with diet and exercise, but rarely is the disease caught early due to asymptomatic nature and the need for a liver biopsy for diagnosis. Chronic disease is often associated with hepatocyte injury, chronic liver inflammation and fibrosis (i.e., formation of scar tissue), known as nonalcoholic steatohepatitis (NASH) (Chalasani et al. 2012; Patterson et al. 2016). The presence of NASH carries a significant risk of end-stage liver disease (cirrhosis), and even hepatocellular carcinoma (Bril and Cusi 2016).
Mouse models of NAFLD by genetic knockout or diet/drug treatment (Nagarajan 2012) have been used to shed light upon the progression of SS to NASH and fibrosis. This work used a diet treatment to probe the progression of SS to NASH. Previous work (Patterson et al. 2016) has demonstrated the large change in diacylglycerides, ceramides, and acylcarnitines in disease onset and progression. Untargeted lipidomics provides an opportunity to assess how lipids differ in disease progression for a mouse model. These changes may shed light on disease mechanisms and targets for treatment.
2 Materials and methods
2.1 Animals and diets
Animal studies were approved by the Institutional Animal Care and Use Committee at the University of Florida, and were the same mice used in a previous Patterson et al. publication (Patterson et al. 2016). Mice (C57BL/6) purchased from Jackson Laboratories (Bar Harbor, ME) at 6–8 weeks were fed either a control diet: (CTRL; 10% fat calories; Research Diets, Inc., New Brunswich, NJ. # D09100304) or a high trans-fat high fructose diet (TFD; 40% fat calories; Research Diets, Inc.# D09100301) for 8 or 24 weeks. At 8 weeks of TFD feeding, mice had already developed SS, whereas 24 weeks of the TFD resulted in development of the histological features of NASH, which are consistent with previous literature reports (Clapper et al. 2013; Trevaskis et al. 2012).
2.2 Storage and handling of samples
Mouse livers were flash frozen in liquid nitrogen immediately after collection of whole blood from the aorta. Frozen livers were individually cryogenically ground using a liquid nitrogen-insulated mortar and ceramic pestle into a fine powder for weighing. Upon sample preparation, liver masses were recorded for all experiments. Stock powder and freshly weighed tissue were kept in liquid nitrogen until quenched with solvent during sample preparation.
2.3 Solvents and standards
Folch extraction solvents included methanol (Fisher Scientific, San Jose, CA) containing 1 mM butylhydroxytoluene (BHT) (Sigma Aldrich, St. Louis, MO), chloroform, and water (both Fisher Scientific). Isopropanol used for reconstitution was purchased from Fisher Scientific (San Jose, CA).
Internal standards used in the untargeted liver lipid analysis covered a range of lipid classes and structures. Generally, non-endogenous chain lengths were used (Castro-Perez et al. 2010) including PC 19:0/19:0, DG 14:0/14:0, TG 15:0/15:0/15:0 (1 ppm final concentration injected), SM d18:1/17:0, 13C2-cholesterol, PE 15:0/15:0 (0.15 ppm), and LPC 19:0, PG 14:0/14:0, PS 14:0/14:0, Cer d18:1/17:0 (50 ppb). The differing concentrations coincided with concentration variations among lipid classes within the liver. These standards, except for TG and 13C2-cholesterol were purchased from Avanti Polar Lipids Inc (Alabaster, AL). TG 15:0/15:0/15:0 was purchased from Sigma (St. Louis, MO), and 13C2-cholesterol was purchased from Cambridge Isotope Laboratories (Andover, MA). A stock mix dissolved in Folch extraction solvent [chloroform:methanol (2:1, v:v)] was prepared to spike into the liver prior to homogenization, and simultaneously with extraction solvent.
2.4 Homogenization and lipid extraction
Cryoground mouse livers stored at − 80 °C were transferred to liquid nitrogen before weighing. All liver samples were randomized prior to weighing in triplicate, and randomized again prior to subsequent sample preparation, with each replicate treated as an individual sample in the randomization process. Masses were estimated at 15 mg (actual average 19.4 ± 0.3 mg) and recorded to calculate volume of internal standard-spiked (IS-spiked) solvent used for homogenization. Homogenization solvent volume was equal to 20 times the mg of liver weighed (e.g., 15.0 mg tissue = 300 mL IS-spiked solvent), allowing the ratio of liver to solvent to remain consistent. Folch solvent was added to the liver with homogenization beads (zirconium oxide 1.0 mm diameter, Next Advance, Averill Park, NY) and homogenized for 120 s (Bead Ruptor4, OMNI International, Kennesaw, GA). Following incubation on ice (20 min) with occasional vortexing, water was added at ¼ the volume used for Folch solvent addition (e.g., 75 µL water for 15.0 mg tissue). Further incubation on ice (10 min) followed, with occasional vortexing and centrifugation (5 min, 6 °C, 20,000 rcf). The lower phase (170 µL) was transferred to a centrifuge tube and the remaining phases were re-extracted with chloroform:methanol (2:1, v/v) at a volume of ½ of the IS-spiked solvent (e.g., 150 µL for 15.0 mg tissue). During incubation on ice (10 min), samples were occasionally vortexed and centrifuged. More organic phase (75 µL) was removed and combined with the clean organic phase, dried under nitrogen gas (30 °C), and reconstituted in 500 µL isopropanol. Reconstituted samples were vortexed, centrifuged (3 min), and stored at 4 °C (20 min) to ensure clean injections. LC vials had SM d18:1/06:0, the injection standard (5 µL of 100 ppm) added and dried under nitrogen. SM d18:1/06:0 was chosen as an injection standard rather than an extraction standard because the short 06:0 fatty acid chain causes elution before most endogenous SMs, so SM d18:1/06:0 cannot be used for normalization purposes. An aliquot from each reconstituted sample (25 µL) was added to each corresponding LC vial followed by 975 µL isopropanol. A 10-fold dilution was necessary to ensure prevention of signal saturation.
Each group was independently pooled to provide QCs. Controls, including 8 and 24 week samples were pooled to form QCA. QCB was a pooled sample of the 8 week TFD samples. QCC pooled the 24 week TFD samples. Finally, a total pooled sample was made from all groups, labeled QCD. A dilution series of 1:1, 1:2, 1:4, and 1:8 ratios of the QCD (Vorkas et al. 2015) was constructed.
Extraction blanks (referred to as blanks throughout) were subjected to the same conditions and solvents as the samples, but were not exposed to liver extracts. This accounts for signals arising from solvents and tubes as well as mobile phase and column background.
2.5 Liquid chromatography—mass spectrometry methods
A Dionex 3000 Ultimate (Thermo, San Jose, CA) autosampler (4 °C) stored samples for injection onto a Waters Acquity C18 BEH column (50 mm × 2.1 mm, 1.7 µm pore size) (Milford, MA) at 50 °C with a 3 µL injection using a quaternary HPLC pump. Mobile phase A contained acetonitrile and water (90:10, v:v). Mobile phase B included isopropanol, acetonitrile, and water (90:8:1, v:v:v). Both added 10 mM ammonium formate and 0.1% formic acid. The 23 min gradient began at 20% B and rose to 98% B at 17 min, remained isocratic for 1 min before returning to starting conditions over another minute and re-equilibrating for 4 min.
A Thermo Q-Exactive collected full scans in polarity switching mode with the following source parameters: spray voltage 3.5 kV, capillary temperature 300 °C, sheath gas 30 arb, auxiliary gas 5 arb, probe heater temp 350 °C, s-lens RF level 35.0. One spectrum was collected per mode per second with 35,000 resolving power, 3 × 106 AGC, and m/z 200–1100 scan range. Data files were converted to .mzXML using Proteowizard’s MSConvert. Although positive and negative mode data were collected, the data for this workflow processed the positive mode data only—the mode in which the chromatography conditions were optimized.
2.6 Sequence setup
Systematic standards allowed monitoring of instrument performance over time. QC standards and a blank were injected every ten samples including blank, pooled QC, neat QC. Following this systematic block of QCs, a randomized set of ten samples were selected, followed by another blank, QC block, and set of randomized samples. Technical replicates of each sample were treated as individual replicates for randomization purposes. Randomization before data acquisition accounted for variability occurring with injection sequence, apart from sample preparation order variation.
2.7 Quality control analysis of the data
In data quality evaluation, the injection standard SM d18:1/06:0 was processed in each injection using Thermo’s Xcalibur and Microsoft Excel, and inspected. A PCA scores plot was created to determine injection and data quality by evaluating pooled QC clusters and disease state separation. Internal standard PAs were processed and inspected over the course of the sequence.
2.8 Data pre-processing and filtration
Files collected in polarity-switching mode were sliced using Thermo’s proprietary slice processing method. Thermo proprietary .RAW files were converted to .mzXML format using Proteowizard’s MSConvert (proteowizard.sourceforge.net) (Chambers et al. 2012) with the following parameters: absolute 0.001 most intense threshold and peak picking MS level 1–1.
These .mzXML file types were processed by open-source MZmine (version 2.15) (Pluskal et al. 2010) for peak picking, alignment, and gap-filling. LipidMatch, an R-based lipid identification software was used for lipid identifications (Koelmel et al. 2017). MZmine parameters can be found in Supplementary Table 1.
The extraction blanks allowed identification of background elements not of biological origin. Here the feature signals in the blanks were compared to the signals in the samples for each feature individually, in a novel method called BFF, where a threshold is calculated below which signals are excluded from further analysis. The BFF threshold (BFFthreshold) was computed as shown in Eq. 1. Equation 2 indicates the requirements for a signal to remain based on the average value for each group of samples (Saverage). If the average value for a particular feature in the blanks group was zero (not present in blanks), a threshold for the chromatographic background was used for filtering, in this case, 5000.
The feature remains if this statement is true:
A caveat exists in that background signals from extraction and chromatography may behave differently in the solvent compared to the biological matrix, thus inappropriately influencing BFF. This difference has not been fully evaluated.
For this experiment, 1000 features from the QC injections were visually inspected by a single individual (REP) and separated into true and false features according to: peak shape, signal intensity, and ability to consistently integrate using the same parameters. The performance of BFF was compared to feature classification based on expert judgement (REP) (Table 1). Two alternative filtering methods were evaluated. The first was based on a simple threshold performed feature-wise, where all features with signals below 30,000 (or 300,000) or missing signal in 50% of the samples in all groups were removed from the data. The simple threshold method did not include blank samples in the mzMine analysis. The second method included a series of statistical summaries and tests for each feature based on BA plot (Bland and Altman 1986) outliers and RT distributions. BA outliers were obtained from a simple linear regression fitted to BA plots, where features more than 3 standard deviations from the fitted line were flagged. The line itself is a diagnostic as the slope is expected to be zero. If the difference in the RT between the 95th and 5th percentile was greater than 0.2 min or, in features where the difference between the RT maximum and median was greater than 0.1 min, or in features where the difference between the RT minimum and median was greater than 0.1 min, the feature was flagged. Each of the three filtration methods were compared to the expert judgement to estimate false positive and false negative rates.
2.9 Normalization
Samples were calibrated to have equal concentration prior to injection, removing most of the expected artifacts from technical variation, as discussed in the sample preparation section. Data was normalized to the PA median value of each sample.
2.10 Differentially expressed features and related statistics
ANOVA identified differences between the four treatment groups, and significantly different features were identified. Furthermore, the compounds remaining after BFF were identified via LipidMatch. First, features were sorted by tentative LipidMatch annotations based on MS/MS. All post-BFF features (963) were analyzed with ANOVA, hierarchical clustering, and PCA. Annotated features were further examined. Compounds identified with fatty acyl constituents and headgroup, or headgroup and sum composition, were matched to those identified as significant in ANOVA analysis. Sum composition indicates the total number of carbons and double bonds in the fatty acyl moiety, but not the individual tails. MetaboAnalyst 2.0 visualized the data using heat maps and PCA.
3 Results and discussion
3.1 Quality control analysis of the results
The importance of consistent and reliable sample preparation in metabolomics cannot be overstated. The workflow described, as applied to mouse liver lipidomics, allows multiple quality checks ensuring proper data preparation, acquisition and analysis. Confirmation of the same ratio of weight to solvent allows the injections to be compared effectively. QC during data acquisition included inspection of chromatography, mass accuracy, and injection standard PA. Chromatography inspection of endogenous lipids in the same pooled sample demonstrated column integrity (Supplemental Fig. 1). Profile chromatograms demonstrated consistency in RT and signal (Fig. 2). Internal standard mass accuracy were all < 2.5 ppm error. The PCA scores plot including pooled QCs (Supplemental Fig. 2) evaluated quality preparation and injection, and demonstrated QCs clustering and disease state separation.
A plot of injection standard (SM d18:1/17:0) signal over the sequence (RSD of 7.7%) (Supplemental Fig. 3) including blanks and the dilution series (last five injections), provides a tool indicating needs for reinjection if injection standard signal is low. Signal increase corresponded to pooled groups, which always followed a blank, indicating the blank injection had an effect on signal intensity and should be investigated further. The lack of a negative slope indicated maintenance of source cleanliness, whereas tight PAs throughout the sequence indicate consistent injections. Signal at zero from the blanks indicated negligible carry over. The last few data points are representative of the dilution series injected at the end of the sequence used to find biologically relevant species (Vorkas et al. 2015).
Similar graphs inspected the extraction standards (Supplemental Fig. 4). Note larger variability compared to injection standard, but similar trend in consistency. RSDs for the standards shown: 7.7% (SM), 20.5% (Cer), 21.9% (PC), and 20.8% (LPC). Samples 9a and 7b (red) had higher signal in the extraction standards, but not the injection standard. This indicated that 9a and 7b had slightly more internal standard added but the injection volume remained precise. Monitoring these samples after BFF, such as in Supplemental Fig. 5 indicated that they were not different from other samples within the group.
Peak picking using MZmine selected 7812 features, and ~ 1000 were evaluated by an expert. Table 1 demonstrates false positive values among three filtering methods in comparison to expert judgment. The BFF method demonstrated balanced, low false positives (3%) and negatives (3%), while threshold filtering performed particularly poorly, with 49% false positives and 6% false negatives for 300,000 threshold filter. Filtering data by BA plot outlier removal, or removing features with large deviations in RT between samples, did not perform as well as BBF when compared to expert judgement. Therefore, BFF was adopted to filter the features from all treatment groups. The Venn diagram shown (Supplemental Fig. 6) compares peaks picked between a dataset processed with and without blanks using BFF or 30 k filtering, respectively. Furthermore, it shows false positive rates of the features compared to expert judgement where BFF had only a 2% false positive rate.
To demonstrate filtration efficacy, Fig. 3 shows PCA before and after BFF, with better separation after applying BFF, which identified a substantial number of false features. This is a direct result of the liberal values set at peak picking, as more conservative MZMine values revealed more false negatives. We propose a liberal peak picking algorithm combined with background filtering to minimize false negatives and false positives. After BFF the number of features was 963 (a reduction in 87% from 7812). BFF allows for feature identification during peak calling by providing automatic filtering of anomalous results with a low false positive and false negative rate, as noted in Table 1. Low intensity peaks representing trace molecular species would not be detected using stringent peak processing parameters, but are retained using BFF. In order to deploy BFF properly, many blanks are necessary within the sequence. This data set had one blank for every ten samples. We do not recommend using BFF when the number of blanks is low relative to the number of samples.
PCA on the treatment groups alone before (a) and after BFF (b). BFF removes a substantial number of peaks with signals that are not consistently above background measured in blanks that are sampled between each batch of samples. Separation of the disease from the control is improved after the removal of noise
Sample quality was further evaluated by examining differences between biological (A) and technical replicates (B) in the BA plots in Supplemental Fig. 7. The technical replicates are less variable than the biological replicates. All of these methods are implemented in SECIMTools (https://github.com/secimTools/SECIMTools, Kirpich submitted).
3.2 Univariate analysis
ANOVA indicated that 370 lipids were significantly different between sample groups (p < 0.0001) in positive mode after BFF. Fifty-five annotated features had a p-value less than 0.0001. The primary goal was to understand the changes associated with disease progression, comparing 8 week TFD (8T) to 24 week TFD (24T), while also noting changes associated with age instead of diet.
3.3 Combined statistical analyses: post ANOVA data visualization
Heat maps were generated using Euclidean distance similarity measure and Ward’s linkage clustering from the post-ANOVA compounds (p < 0.0001), shown in Fig. 4, where diseased (8T and 24T) cluster together. The controls were more diffuse, however more variation exists between the disease states than between the age differences. The red or blue colors indicate up or down regulation between the groups in context with the controls. Met 2831, identified as PE (18:0_22:6) is the only identified lipid that decreases with disease stage compared to healthy controls. Compounds with confident annotations that notably increase with disease onset and possibly progression include: features 3194, 4318, 6994, 4576, 2857, 2389, and 2415, all corresponding to TG species with 58 or 56 carbons, ranging from 3 to 6 double bonds. Table 2 demonstrates those compounds from the post-BFF HCA with LipidMatch annotations. Several values indicate the presence of isomeric TGs at different RTs.
Two-way Hierarchical clustering analysis (HCA) with samples on the X axis and lipids on the Y axis. The samples are sorted based on intensity and clear separation of the disease from the control, and between the two disease time points is apparent. Cells are colored based on the relative intensity of the peak. The 50 top peaks from ANOVA (P < 0.0001) are depicted
Removal of unidentified features prior to analysis can be controversial, as some seek to minimize false negatives and reason that false features are unlikely to be significant in analysis. Where a statistical test of difference in abundance is desired, the inclusion of false peaks will increase the multiple testing penalty substantially. Here instead of correcting for ~ 800 peaks a correction for ~ 8000 peaks would be required, setting the bar for identifying compounds substantially higher. While limiting the scope to only identified features may fail to identify novel trends and changes in the data, that issue is beyond the scope of this paper. Of the 963 positive mode unannotated features, some significant compounds are not identified. A few correlate cholesteryl ester (CE) fragments and RT, but would have to be confirmed with targeted studies.
PCA by class explored differences between disease states within lipid class. Annotated features after filtering were subjected to PCA by class. The figures shown (Supplemental Fig. 8) demonstrate that PEs are separated by component 1 with the most variance accounted for (37%) compared to the other classes, but the PCs, neutral lipids (DGs and TGs combined) and TGs alone, separate the control diets from the trans-fat diets well. Arguably, PC 2 separates the two disease states best when using PE data only.
The general trends indicate PEs and PCs are downregulated in NASH mice, and TGs are upregulated, compared to SS and controls. TGs that increase with disease progression have lower numbers of unsaturation, averaging about 1 per fatty acid chain. More information about these fatty acid constituents could reveal more pathway knowledge regarding saturated TGs, monounsaturated TGs, and polyunsaturated TGs in disease progression.
3.4 Biological interpretation
Pathways for lipids are not as well-known as those for metabolites, and typically emphasize head group or fatty acid moiety. The pathways scrutinized surround mitochondrial metabolism, focusing on lipid classification, and the fatty acid chain. Our observations support disease progression theories within the context of the TCA cycle or Kennedy pathway, but are not limited to these biochemical mechanisms. As a summary of lipid metabolism in relation to the TCA cycle, TGs are converted into fatty acids (pathway also goes in reverse), and proceed through beta-oxidation to feed the TCA cycle (Garrett and Grisham 2010). An increase of TGs with induction of the TCA cycle indicates dysfunction of beta oxidation, because the TGs would normally decrease with induction of the TCA cycle (Satapati et al. 2012). Higher amounts of TGs may result in decreased PC and PE levels, if fatty acid moieties are contained in the TGs instead. This idea is supported because glycerol phospholipids (GPLs) are built from intermediates of TG synthesis.(Cheng et al. 2011) One theory for progression of SS to NASH surrounds the relationship of insulin resistance, lipid oxidation, hepatic tri-carboxylic acid (TCA) flux, and production of reactive oxygen species via mitochondrial respiratory dysfunction (Patterson et al. 2016; Satapati et al. 2012; Sunny et al. 2011, 2016).
Certain synthetic pathways are known, such as the production of PEs and PCs via the Kennedy pathway (Gibellini and Smith 2010). In this pathway, fatty acid chains for phospholipids are retrieved from DGs. It is generally unknown what effect different fatty acid chains (e.g., 18:2 vs. 16:1) have on biological processes. One exception is arachidonic acid (20:4), which has been implicated in anti-inflammation pathways, (Castro-Perez et al. 2010) and in cirrhosis (Gorden et al. 2011). PC 18:0_20:4 has increased concentrations in the TFD model compared to the age matched controls, but as the disease progresses, the level decreases. PE 16:0_20:4 decreases with disease progression, and when compared to age-matched controls.
4 Conclusion
The workflow presented provides a platform for quality data acquisition, analysis, and interpretation allowing checks at every decision point. Adjusting extraction solvent based on tissue weight and randomization of samples throughout sample preparation and acquisition reduce batch-to-batch variability and the need for extensive normalization post-acquisition. QC analysis during acquisition and thorough feature filtering ensure noise is reduced without affecting the opportunity to understand true variance. BFF ensures reduction of variables from sample preparation or instrumental causes, while reducing features to a reliable, robust, and reproducible list.
Evidence supports both a decrease in PEs and PCs and an increase of certain TG species with disease progression. Unfortunately many TGs co-elute, adding uncertainty about fatty acid contributions. Nevertheless, certain trends are evident: TGs of interest contain at least three degrees of unsaturation, while most of the PCs and PEs consist of one saturated chain. This observed trend is supported in literature (Arendt et al. 2013; Jacobs et al. 2013; Li et al. 2006): one suggests the fatty acid chains are used in TGs instead of PCs and PEs, therefore reducing GPLs and increasing TGs. No one fatty acid consistently dominates PC and PE results, which include 16:0, 18:0, 18:1, 18:2, 20:4, and 22:6. The most promising PE and PC features according to multiple tests and validation include: PE 16:0_22:6, PE 18:0_22:6, TG 58:5, TG 58:6, PE 16:0_20:4, PC 16:0_20:4, PC 40:6, TG 56:5, TG 54:2, TG 56:4, and TG 56:5. These results, combined with the presence of increased DGs and ceramides in targeted analyses (Patterson et al. 2016), supports a reorganization of fatty acids from GPLs into TGs, DGs, and CERs as the disease progresses.
The limited number of DGs and sphingolipids present in the results may be attributed to relatively low concentrations when compared to PCs, PEs, and TGs. Therefore, targeted extraction methods might reveal more about lower abundant lipid classes such as CEs and sphingolipids. Additionally, further research of the TCA cycle intermediates and other metabolites (choline, ethanolamine, ADP, ATP, etc.) may enable a better understanding of the transitions between lipid classes.
Gorden et al. targeted a wide range of lipid classes in NAFLD plasma and liver by using the best extraction and analysis techniques for each lipid class (Gorden et al. 2015). Their work showed a large contribution of PC and PE classes in distinguishing between NASH and SS. Linear Discriminant Analysis by class demonstrated a greater influence of sphingolipids and glycerophospholipids in distinguishing disease state than eicosanoids and neutral lipids. Our work corroborates the effect of PC and PEs on disease state differences; however, we found more of an influence of TGs on disease progression and onset.
Abbreviations
- BFF:
-
Blank feature filtering
- Cer:
-
Ceramide
- DG:
-
Diacylglyceride
- LC:
-
Liquid chromatography
- MS:
-
Mass spectrometry
- NAFLD:
-
Nonalcoholic fatty liver disease
- NASH:
-
Nonalcoholic steatohepatitis
- QC:
-
Quality control
- RT:
-
Retention time
- SS:
-
Simple steatosis
- TG:
-
Triacylglyceride
References
Arendt, B. M., Ma, D. W., Simons, B., Noureldin, S. A., Therapondos, G., Guindi, M., et al. (2013). Nonalcoholic fatty liver disease is associated with lower hepatic and erythrocyte ratios of phosphatidylcholine to phosphatidylethanolamine. Applied Physiology, Nutrition, and Metabolism, 38(3), 334–340. doi:10.1139/apnm-2012-0261.
Bril, F., & Cusi, K. (2016). Nonalcoholic fatty liver disease: The new complication of type 2 diabetes mellitus. Endocrinology and Metabolism Clinics of North America, 45(4), 765–781. doi:10.1016/j.ecl.2016.06.005.
Broadhurst, D. I., & Kell, D. B. (2006). Statistical strategies for avoiding false discoveries in metabolomics and related experiments. Metabolomics, 2(4), 171–196. doi:10.1007/s11306-006-0037-z.
Browning, J. D., Szczepaniak, L. S., Dobbins, R., Nuremberg, P., Horton, J. D., Cohen, J. C., et al. (2004). Prevalence of hepatic steatosis in an urban population in the United States: Impact of ethnicity. Hepatology, 40(6), 1387–1395.
Castro-Perez, J. M., Kamphorst, J., Degroot, J., Lafeber, F., Goshawk, J., Yu, K., et al. (2010). Comprehensive LC–MS E lipidomic analysis using a shotgun approach and its application to biomarker detection and identification in osteoarthritis patients research articles. Journal of Proteome Research, 9, 2377–2389.
Chalasani, N., Younossi, Z., Lavine, J. E., Diehl, A. M., Brunt, E. M., Cusi, K., et al. (2012). The diagnosis and management of non-alcoholic fatty liver disease: Practice guideline by the American Gastroenterological Association, American Association for the Study of Liver Diseases, and American College of Gastroenterology. Gastroenterology, 142(7), 1592–1609. doi:10.1053/j.gastro.2012.04.001.
Chambers, M. C., Maclean, B., Burke, R., Amodei, D., Ruderman, D. L., Neumann, S., et al. (2012). A cross-platform toolkit for mass spectrometry and proteomics. Nature Biotechnology, 30(10), 918–920. doi:10.1038/nbt.2377.
Cheng, D., Jenner, A. M., Shui, G., Cheong, W. F., Mitchell, T. W., Nealon, J. R., et al. (2011). Lipid pathway alterations in Parkinson’s disease primary visual cortex. PLoS ONE, 6(2), e17299. doi:10.1371/journal.pone.0017299.
Clapper, J. R., Hendricks, M. D., Gu, G., Wittmer, C., Dolman, C. S., Herich, J., et al. (2013). Diet-induced mouse model of fatty liver disease and nonalcoholic steatohepatitis reflecting clinical disease progression and methods of assessment. American Journal of Physiology Gastrointestinal and Liver Physiology, 305(7), G483–G495. doi:10.1152/ajpgi.00079.2013.
De Livera, A. M., Dias, D. A., De Souza, D., Rupasinghe, T., Tull, D. L., Roessner, U., et al. (2012). Normalising and integrating metabolomics data. Analytical Chemistry, 84, 10768–10776
Dunn, W. B., Broadhurst, D., Begley, P., Zelena, E., Francis-McIntyre, S., Anderson, N., et al. (2011). Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nature Protocols, 6(7), 1060–1083. doi:10.1038/nprot.2011.335.
Garrett, R. H., & Grisham, C. M. (2010). The tricarboxylic acid cycle. In L. Lockwood, S. Kiselica, A. Summers, & L. Weber (Eds.), Biochemistry (4th ed.). Boston: Cengage Learning.
Gibellini, F., & Smith, T. K. (2010). The Kennedy pathway-de novo synthesis of phosphatidylethanolamine and phosphatidylcholine. IUBMB Life, 62(6), 414–428. doi:10.1002/iub.337.
Gorden, D. L., Ivanova, P. T., Myers, D. S., McIntyre, J. O., VanSaun, M. N., Wright, J. K., et al. (2011). Increased diacylglycerols characterize hepatic lipid changes in progression of human nonalcoholic fatty liver disease; comparison to a murine model. PLoS ONE, 6(8), 1–10. doi:10.1371/journal.pone.0022775.
Gorden, D. L., Myers, D. S., Ivanova, P. T., Fahy, E., Maurya, M. R., Gupta, S., et al. (2015). Biomarkers of NAFLD progression: A lipidomics approach to an epidemic. Journal of Lipid Research, 56(3), 722–736. doi:10.1194/jlr.P056002.
Jacobs, R. L., van der Veen, J. N., & Vance, D. E. (2013). Finding the balance: The role of S-adenosylmethionine and phosphatidylcholine metabolism in development of nonalcoholic fatty liver disease. Hepatology (Baltimore, Md.), 58, 1207–1209. doi:10.1002/hep.26499.
Kirwan, J. A., Weber, R. J. M., Broadhurst, D. I., & Viant, M. R. (2014). Direct infusion mass spectrometry metabolomics dataset: A benchmark for data processing and quality control. Scientific Data, 1, 140012. doi:10.1038/sdata.2014.12.
Koelmel, J. P., Kroeger, N. M., Ulmer, C. Z., Bowden, J. A., Patterson, R. E., Cochran, J. A., et al. (2017). LipidMatch: An automated workflow for rule-based lipid identification using untargeted high-resolution tandem mass spectrometry data. BMC Bioinformatics, 18, 331. doi:10.1186/s12859-017-1744-3.
Koliaki, C., & Roden, M. (2013). Hepatic energy metabolism in human diabetes mellitus, obesity and non-alcoholic fatty liver disease. Molecular and Cellular Endocrinology, 379(1–2), 35–42. doi:10.1016/j.mce.2013.06.002.
Li, Z., Agellon, L. B., Allen, T. M., Umeda, M., Jewell, L., Mason, A., & Vance, D. E. (2006). The ratio of phosphatidylcholine to phosphatidylethanolamine influences membrane integrity and steatohepatitis. Cell Metabolism, 3(5), 321–331. doi:10.1016/j.cmet.2006.03.007.
Martin Bland, J., & Altman, D. (1986). Statistical methods for assessing agreement between two methods of clinical measurement. Lancet, 327(8476), 307–310. doi:10.1016/S0140-6736(86)90837-8.
Nagarajan, P. (2012). Genetically modified mouse models for the study of nonalcoholic fatty liver disease. World Journal of Gastroenterology, 18(11), 1141. doi:10.3748/wjg.v18.i11.1141.
Patterson, R. E., Kalavalapalli, S., Williams, C. M., Nautiyal, M., Mathew, J. T., Martinez, J., et al. (2016). Lipotoxicity in steatohepatitis occurs despite an increase in tricarboxylic acid cycle activity. American Journal of Physiology—Endocrinology and Metabolism, 310(7), E484–E494. doi:10.1152/ajpendo.00492.2015.
Pellicoro, A., Ramachandran, P., Iredale, J. P., & Fallowfield, J. A. (2014). Liver fibrosis and repair: Immune regulation of wound healing in a solid organ. Nature Reviews Immunology, 14(3), 181–194. doi:10.1038/nri3623.
Pluskal, T., Castillo, S., Villar-Briones, A., & Orešič, M. (2010). MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics, 11(1), 395. doi:10.1186/1471-2105-11-395.
Qi, Z., & Voit, E. O. (2016). Strategies for comparing metabolic profiles: Implications for the inference of biochemical mechanisms from metabolomics data. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 99, 1–32. doi:10.1109/TCBB.2016.2586065.
Redestig, H., Fukushima, A., Stenlund, H., Moritz, T., Arita, M., Saito, K., & Kusano, M. (2009). Compensation for systematic cross-contribution improves normalization of mass spectrometry based metabolomics data. Analytical Chemistry, 81(19), 7974–7980. doi:10.1021/ac901143w.
Satapati, S., Sunny, N. E., Kucejova, B., Fu, X., He, T. T., Méndez-Lucas, A., et al. (2012). Elevated TCA cycle function in the pathology of diet-induced hepatic insulin resistance and fatty liver. Journal of Lipid Research, 53(6), 1080–1092. doi:10.1194/jlr.M023382.
Silva, L. P., Lorenzi, P. L., Purwaha, P., Yong, V., Hawke, D. H., & Weinstein, J. N. (2014). Measurement of DNA concentration as a normalization strategy for metabolomic data from adherent cell lines. Analytical Chemistry, 85(20), 9536–9542. doi:10.1021/ac401559v.
Sumner, L. W., Amberg, A., Barrett, D., Beale, M. H., Beger, R., Daykin, C. a., et al. (2007). Proposed minimum reporting standards for chemical analysis: Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics, 3(3), 211–221. doi:10.1007/s11306-007-0082-2.
Sunny, N. E., Bril, F., & Cusi, K. (2016). Mitochondrial adaptation in nonalcoholic fatty liver disease: Novel mechanisms and treatment strategies. Trends in Endocrinology & Metabolism, 28, 1–11. doi:10.1016/j.tem.2016.11.006.
Sunny, N. E., Parks, E. J., Browning, J. D., & Burgess, S. C. (2011). Excessive hepatic mitochondrial TCA cycle and gluconeogenesis in humans with nonalcoholic fatty liver disease. Cell Metabolism, 14(6), 804–810. doi:10.1016/j.cmet.2011.11.004.
Trevaskis, J. L., Griffin, P. S., Wittmer, C., Neuschwander-Tetri, B. A., Brunt, E. M., Dolman, C. S., et al. (2012). Glucagon-like peptide-1 receptor agonism improves metabolic, biochemical, and histopathological indices of nonalcoholic steatohepatitis in mice. American Journal of Physiology Gastrointestinal and Liver Physiology, 302(38), G762–G772. doi:10.1152/ajpgi.00476.2011.
Vorkas, P. A., Isaac, G., Anwar, M. A., Davies, A. H., Want, E. J., Nicholson, J. K., & Holmes, E. (2015). Untargeted UPLC-MS profiling pipeline to expand tissue metabolome coverage: Application to cardiovascular disease. Analytical Chemistry, 87(8), 4184–4193. doi:10.1021/ac503775m.
Worley, B., & Powers, R. (2013). Multivariate analysis in metabolomics. Current Metabolomics, 1, 92–107.
Acknowledgements
The authors would like to thank Miguel Ibarra, Oleksandr Moskalenko, and Justin Richardson for their computer programming expertise. We would also like to thank funding sources including the Southeastern Center for Integrated Metabolomics (NIH grant U24 DK097209), the UF Clinical Translational Science Institute (CTSI NIH Grant UL1 TR000064), and the Eastman Chemical Company Analytical Summer Fellowship.
Funding
This study was funded by NIH grant U24 DK097209, UF Clinical Translational Science Institute (CTSI NIH Grant UL1 TR000064).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Approval
All applicable international, national, and institutional guidelines for the care and use of animals were followed. This article does not contain any studies with human participants performed by any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Patterson, R.E., Kirpich, A.S., Koelmel, J.P. et al. Improved experimental data processing for UHPLC–HRMS/MS lipidomics applied to nonalcoholic fatty liver disease. Metabolomics 13, 142 (2017). https://doi.org/10.1007/s11306-017-1280-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-017-1280-1