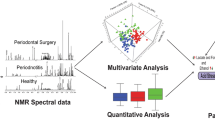
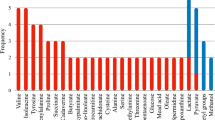
Abstract
Introduction
Periodontitis is a chronic, non-reversible inflammatory disease of the oral cavity leading to destruction of periodontal tissues. Thus, the estimation of bacterial metabolite, tissue damage and secretory metabolites of the triggered inflammatory cells likely to yield results. It may be of value for understanding the pathophysiology of the disease by metabolic profiling of saliva samples using high-resolution NMR spectroscopy.
Objective
The study will evaluate the difference in salivary metabolites in healthy and periodontal condition along with fetching of possible biomarkers in case of chronic periodontitis.
Methods
1H- NMR spectroscopy has been employed in 114 saliva samples in search of distinctive differences and spectral data were further subjected to multivariate analysis.
Result
One-hundred metabolites were characterised and assigned in the 1H NMR spectra of saliva. The statistical analysis of control (Healthy subjects) and diseased (Periodontal subjects) using PLS-DA model resulted in R2 of 0.84 and Q2 of 0.79. There was an elevation in the concentration of statistically discriminant metabolites. The twenty newly identified metabolites in saliva indicates bacterial population shift along with change in homeostasis. These disturbs the biofilm, a real protector against any possible bio-damage on tooth surface. These newly identified metabolites could define better geographically diversified periodontal condition.
Conclusion
Analysis clearly differentiates healthy subjects from the diseased ones. Few newly identified metabolites along with the pool of metabolites may serve as biomarkers for distinguishing the severity and complexity of periodontitis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Periodontitis, a disease of oral origin is a result of disturbance in homeostasis with respect to bacterial population at microenvironment level characterized by inflammation followed by infection in gum or gingiva. This leads to destruction of periodontium in a stepwise manner resulting in irreversible tooth loss and therefore can be proclaimed as a multifactorial disease (Wolf 2006). Generally more than 700 bacterial species are supposed to reside in sub gingival plaque which forms biofilm in a sequential manner as gram negative, gram positive, aerobic and anaerobic bacteria etc. on tooth surface (Brogden and Guthmiller 2002; Estrela et al. 2010; Haffajee and Socransky 2006). Different types of bacterial population that comes at the beginning on the tooth surface are known as early colonizers. They are being bridged by Fusobacterium nucleatum to the late colonizing bacteria (Merritt et al. 2009). Sometimes these populations do not interact with each other. Periodontitis is an infective condition attributable to certain pathogens, namely, aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis, Bacteroides forsythus, Prevotella intermedia, Campylobacter rectus, Treponema denticola, Fusobacterium nucleatum etc. (Dumitrescu 2010; Perez-Chaparro et al. 2014). Crevicular fluid often contains inflammatory mediators and the oral pathogens associated with periodontitis. The mechanism underlying this destructive process involves both direct tissue damage from the bacterial plaque products and indirect damage through bacterial induction of the host inflammatory and immune responses. The shift of symbiosis to dysbiosis of microbial population is a real culprit behind this disease as dominance of anaerobic bacteria such as proteobacteria, spirochaetes etc. over facultative bacteria (early colonizers) such as Actinomyces and Streptococci species. The loss of alveolar bone, the major supporting structure of tooth is the outcome of pathological manifestation during the infection (Page and Kornman 1997). Although periodontal bacteria are the causative agent in periodontitis, subsequent progression and disease severity are determined by host immune response in which many cell types notably polymorphonuclear leukocytes, macrophages, lymphocytes and fibroblasts are involved (Meyle 1993; Offenbacher 1996). In India, it is highly recommended to enhance the oral health literacy in order to shift the paradigm from normal to professional health care. It is generally the problem of more rural population as compared to urban where dentists are within the reach. We should organize the oral health programme in order to understand the signs and symptoms of oral health infection for better quality of life. Saliva has a cleansing effect on the teeth. Normally, 700–800 ml of saliva is secreted per day (Eastoe 1988). Saliva samples are readily available, its collection is also non-invasive and it could be developed as diagnostic tool to identify the diseases.
Metabolomics is the study of low molecular weight compounds by revealing their chemical property known as metabolite profiling in biological samples. Metabolites are indicators that act as a biomarker during transition from healthy to diseased state. Salivary metabolomics is all about the study of saliva for important biomarkers in easy and non-invasive method in order to establish saliva as a diagnostic tool for many diseases in very early phase. Understanding the metabolite chemistry of saliva will help researcher to know many biomarkers to be revealed in form of endogenous metabolites including amino acids, peptides, nucleic acids, organic acids and thiols etc. for disease indicator (Arakaki et al. 2008; Park et al. 2012). Therefore, more markers of disease are eagerly needed, particularly, for the detection of disease; highly sensitive and specific biomarkers as primary indicators are relatively more useful (Wang et al. 2011; Yu et al. 2006). Metabolomics offers potential advantages that classically diagnostic approaches do not, based on the following discovery of a suite of clinically relevant biomarker that is simultaneously affected by the disease. Earlier 75 salivary metabolites were identified via NMR (Dame et al. 2015) and some of the metabolites such as propionate, N-acetyl signal, pyruvate, succinate, trimethylamine and phenylalanine were found to be distinctive in case of generalized chronic periodontitis as compare to healthy subjects (Aimetti et al. 2012). Since saliva is in direct contact with the periodontitis pathology, we have therefore attempted to evaluate the differences in metabolite indices in saliva (Dame et al. 2015) specimens in Indian context from control subjects using high resolution NMR spectroscopic studies. It will be important in identifying specific metabolites, which may act as an indicator of the disease in its chronic form.
2 Materials and methods
2.1 Subjects and sample collection
One hundred fourteen samples were collected of Healthy (Control) (n = 52), chronic periodontitis (Case) (n = 62) subjects having a pocket size of >5.0 mm (clinical attachment loss) with no other and oral comorbidities (Supplementary Table 1) along with the age and gender where flap surgery would be required. Subjects were seated comfortably and then asked to collect unstimulated saliva into a sterile container (containing 2.0 mg of Sodium Azide) for a period of 10 min in morning before brushing (All Volunteers/patients requested to refrain from oral activities for a period of 2 h prior to sample collection) and snap frozen immediately in liquid nitrogen. The selected patients were fully informed and their consents were taken well before the investigations. The study was ethically approved by the committee (Ethical approval No. - B-07: CBMR/EMP/IEC/3/26-02-2011).
2.2 Sample preparation and acquisition
Frozen samples were thawed at room temperature and centrifuged at 5000 g for a period of 30 min at 4 °C. Supernatant was taken for further experimental procedure and 300 µl of supernatant mixed with 300 µl of Sodium phosphate buffer in 5 mm NMR tube (Wilmad Glass USA) and vortexed for 30 s for sample homogeneity (Aimetti et al. 2012). NMR experiments were then performed using a Bruker Biospin Avance III 800 MHz NMR (Bruker, GmBH) spectrometer equipped with a 5 mm Triple resonance inverse (TCI)1H/13C/15N cryoprobe with a Z-shielded gradient and standard vertical bore, operating at a proton frequency of 800.21 MHz (18.8 T).
2.3 NMR experimental conditions
The 1H NMR spectra of saliva samples were acquired with water presaturation at 300 K with 128 scans and 4 dummy scans. The spectra were acquired using 1D NOESY with water presaturation and Carr-Purcell-Meiboom-Gill (CPMG) pulse sequence (for suppressing short T2 resonances arising due to macromolecules) on each sample. The following experimental parameters were used: spectral width of 12,019 Hz, time domain data points of 64 K, relaxation delay 4.0 s, 128 scans with 4 dummy scans, with a total recording time of approximately 15 min. CPMG pulse sequence with water suppression [PRESET −90°−(δ−180°−δ) n−Aq] with an echo time of 300µs was performed. For Saliva samples, the spectra were recorded using water suppression pulse sequence 1D NOESY gradient-preset with water irradiation during a relaxation delay of 4 s and mixing time of 10 ms. other parameters were: spectral width 18,028 Hz, time domain data points 64 K, acquisition time 1.81 s, number of scans 128 with 4 dummy scans. This results in a total acquisition time of 12 min 54 s per sample. All spectra were processed by applying a line broadening of 0.3 Hz to the FID prior to Fourier Transformation using the software TOPSPIN (version 3.1). The spectra were manually phase and baseline corrected using automatic baseline correction. To confirm the assignments, two-dimensional 1H–1H homo nuclear correlation spectroscopy (COSY) and 1H–13C hetero nuclear single quantum correlation spectroscopy (HSQC) were performed using Bruker’s standard pulse program library. The parameters used for COSY were: 2048 data points were collected in the t2 domain over the spectral width of 11,194 Hz, 256 t1 increments were collected with 88 transients, relaxation delay of 2 Sec, acquisition time of 91 ms and pre-saturated water resonance during the relaxation delay. The resulting data were zero-filled to 1024 data points, and were weighted with sine bell window functions in both the dimensions prior to Fourier transformation. The parameters used for 1H–13C HSQC were: 2048 data points were collected in t2 dimension over the spectral width of 11,194 Hz, 256 t1 increments were collected with 88 transients, relaxation delay of 2.0 s, acquisition time of 91 ms and a 90° pulse of 10.98 μs. The phase sensitive data were obtained by the echo-antiecho-time proportional phase increments method. The resulting data were zero-filled to 1024 data points and were weighted with 90° shifted squared sine bell window functions in both the dimensions, prior to fourier transformation.
2.4 Statistical analysis (data reduction and pattern recognition)
Broad resonances were observed in the NOESY spectra due to presence of macromolecules in the native saliva and a flat base line could not be obtained for small molecules signals in the 1H NMR spectra even after phase and baseline correction (Supplementary Fig. 1). Therefore, CPMG NMR spectra were corrected for phase and baseline prior to subjecting them for further multivariate analysis. The spectra were reduced to discrete chemical shift regions (between 8.6 and 0.6 ppm) with the removal of water region from 5.15 to 4.49, area after anomeric proton of sugars 5.78–5.45 and 6.7–5.9 ppm by digitization to produce a series of sequentially integrated regions of 0.02 ppm width bins using Bruker AMIX software (Version 3.8.7, Bruker Biospin, Germany). Simple rectangular binning of these regions was performed to integrate the peak area. The histograms were normalized by dividing each integral area of the segment by the total area of the spectrum, to compensate for the differences in the overall metabolite concentrations within the samples. Subsequently, similar process were carried out by removing doublet of the methyl signal of lactate between 1.35 and 1.30 and acetate singlet between 1.95 and 1.85 ppm appearing as an intense signal. The resulting data matrices in both the cases having normalized integral values were exported into Microsoft Office Excel 2007 (Microsoft Corporation, USA). These were further imported to ‘The Unscrambler X’ Software package (Version 10.0.1, Camo USA, Norway) for multivariate principal component analysis (PCA) and partial least square discriminant analysis (PLS-DA) analysis. PCA was performed on matrices of spectra from healthy (Control) (n = 52), chronic periodontitis (Case) (n = 62) subjects for comparing their metabolic profiles. In PCA and PLS-DA, a full cross validation using leave-one-out were applied for avoiding the over fitting of the mathematical model. The cross-validation parameter Q2, indicating the predictability of the model related to its statistical validity was calculated. An additional cross validation tool, a permutation test, was performed for each model by randomizing the order of Y variables for a specified number of times (Jiang et al. 2013). The initial multivariate analysis where methyl of the lactate and acetate signals were taken into account did not provided meaningful result (Supplementary Fig. 2). After completing the multivariate analysis on the given dataset, about 75% of the data were randomly selected from Fisher and Yates table (Fisher 1957) as training set. The training set comprised Healthy (Control) (n = 37), periodontitis (Case) (n = 47) subjects NMR spectra out of the total saliva samples. Another PLS-DA model was generated with the data of the training set. The test set was not included during the construction of the PLS-DA model, but was predicted based on this model. To validate the process of prediction of any sample, a Receiver Operator Characteristic curve analysis was accomplished on the PLS-DA Factor 1 using SPSS (SPSS Inc., Version 20) software. The ROC curve analysis is considered to be a statistically valid method to evaluate the performance of biomarkers (Søreide 2009; Zweig and Campbell 1993). The results were further confirmed by performing VIP score analysis by Metaboanalyst (version 3.0).
Discriminant metabolites identified from the VIP score analysis along with acetate metabolites were quantified using NMR suite 8.1 (Chenomx) software. Univariate statistical student’s t-Test using SPSS (SPSS Inc., Version 20) software for two independent groups were performed and is presented in Table 1.
3 Result
3.1 Metabolic profile of saliva samples
Proton (1H) NMR spectra were processed (n = 114) and went under analysis obtained from healthy and periodontal subjects. Characterization of these metabolites was carried out on the basis of chemical shift, coupling constant and splitting pattern of metabolites as reported in literature (Aimetti et al. 2012; Dame et al. 2015), two-dimensional NMR spectra and by comparison with standard NMR spectra of metabolites reported in the Biological Magnetic Resonance Bank (BMRB, http://www.bmrb.wisc.edu) and Human Metabolome Data Base (HMDB, http://www.hmdb.ca) (Markley et al. 2007; Wishart et al. 2009). One and two-dimensional NMR spectra allowed identification and assignments of hundred endogenous metabolites. A representative class of CPMG 1H NMR-spectra obtained from healthy and chronic periodontal subjects are presented in Fig 1. Along with earlier metabolites, 20 new metabolites were observed in saliva samples as 2-hydroxy 3-methylvalerate, caprylate, inosine, imidazol, nicotinate, phenol and tryptophan in healthy controls. While, 3-hydroxybutyrate, acetoin, asparagine, glutamine, sucrose and xanthine in periodontitis disease sample and remaining 2-methylhistidine, desaminotyrosine, fumarate, isoleucine, maltose, trimethylamine N-oxide and uridine were present in both controls and disease state respectively (Supplementary Table 2).
3.2 PCA of healthy and periodontal case samples
As observed in the 2D scattered PC1 v PC2 score plot, there is a very significant difference between the control and diseased group Fig 2a. The PCA score plot showed 57% of explained variance. The PC1 loading plot, Fig 2b demonstrated that principally observed major metabolites in the NMR spectra are present in very high concentration diseased state and masking the effect of minor metabolites. This does not explain the exact picture of metabolomic differences between healthy and periodontal condition. This observation was further validated by VIP score Fig 4. However, PC-2 loading plot, Fig 2c demonstrated positive correlation with propionate (1.07 ppm), propylene Glycol (1.15 ppm), butyrate (1.55 ppm), 5′ APO (1.67 ppm), acetoin (2.23 ppm), valine (2.33 ppm), lactate (4.11 ppm), desaminotyrosine (7.17 ppm), tyrosine (6.87 ppm) and formate (8.45 ppm) (Aimetti et al. 2012). Negative loading with ethanol (1.19 ppm), ethane sulfonate (1.25 ppm), glutamate (2.09 ppm), NAG (2.13 ppm), glycine (3.57 ppm), alanine (3.77 ppm), phenol (6.91 ppm), histidine (7.07 ppm), phenylalanine (7.37 ppm), phenylacetate (7.33 ppm) and 3-methylhistidine (7.79 ppm) (Barnes et al. 2009).
a 2-D PCA Scatter plot showing difference between control (healthy) and diseased (periodontal) group. b PC-1 loading plot showing the principally observed major metabolites, which depicts the severe inflammation in diverse fashion. c PC-2 loading plot showing small molecular weight metabolites. Positive correlation showing upregulation and negative correlation indicating the downregulation
3.3 PLS-DA of healthy and periodontal case samples
PLS-DA facilitated better characterization of the metabolites, distinguishing between healthy and periodontal cases. Scattered plot of PLS-DA Fig 3a with R2 and Q2 values were 84 and 79% was observed. The robustness of the PLS-DA model was further grounded for training set Fig 3b and prediction of the test set was performed with nearly 100% of accuracy Fig 3c.
a 2-D PLS-DA Scatter plot showing difference between control (healthy) and diseased (periodontal) group, attributing R2 and Q2 values of 84 and 79% respectively. b 2-D PLS-DA Scatter plot of training set showing difference between control (healthy) and diseased (periodontal) group, attributing R2 and Q2 values of 70% each. c Prediction of unknown saliva samples (test set, n = 30) using PLS model which was prepared using training data set (n = 84) samples. The predictions are made on the basis of a priori cut-off value of 0.5 for class membership, using y-predicted box-plot between control (healthy) and case (periodontal) group
The VIP score plot is shown in Fig 4 and the scores of seven metabolites of more than value of 2 were considered to be discriminant. Their significance were further confirmed by statistical analysis on quantified data of these metabolites (Table 1).
3.4 ROC curve analysis
The prediction accuracy of the model was again validated with the area under the curve (AUC) value obtained from the ROC analysis Fig 5. The value of AUC was found to be identical (0.982) for the total 114 samples, which was more than 95% of confidence interval. The ROC calculates the sensitivity and specificity (1-specificity actually) for each cut-off value assuming values that are equal or above the threshold as positive and those below as negative (Hart and Kornman 1997; Xia et al. 2013). The given ROC curve analysis was found to be near accurate in its sensitivity and specificity.
4 Discussion
Various studies have demonstrated that susceptibility to chronic periodontitis has a genetic component too (Hart and Kornman 1997). Disease severity may be categorized as early, moderate and advance stage of periodontitis with 1–2, 3–4 mm and more than 5 mm of sub gingival plaque respectively. Smoking and tobacco chewing may also increase the susceptibility towards periodontitis, as these components disturbs the biofilm layer. Other than that, many layers of biofilm is protective enough for avoiding periodontitis (Chen et al. 2005). Cellular Immune system also plays a major role in this disease as various cytokines, Interleukins and TNF- alpha gives better insights and offer the area of genetic basis of the disease. Moving from dysbiosis to symbiosis or restoring the homeostasis with reference to polymicrobial community in chronic periodontitis (i.e. biofilm restoration) is the main motif towards the effective therapy. Especially the P. gingivalis proteins play crucial role for essential function for entire population as it is often marked as a key member for polymicrobial community. PC-1 demonstrates overall increase in metabolites that may be a part of overall increase as major confounders, which seems to be highly unusual. This may also be indicative towards outburst of various chemokines in order to cope up with inflammation thus giving rise to highly populated chemical environment leading to increase in concentration of salivary metabolites which can’t be treated with medication. As compared to healthy sites, diseased site have compromised defense mechanism in the periodontium. Neutrophils localization is a sign of bacterial infection but unfortunately they do not reside in periodontal tissue and rather than they use them as a passage for migration towards the site of infection. This completely resemble with the fact that innate defense mechanism might play a very crucial role in case of periodontitis (Darveau 2010). Using the local anti-inflammatory agent therapies shows marginal improvements; this may be because of less effectiveness of particular agent in multifactorial infection leading to multiple disturbances in host tissue homeostasis followed by initiation of destruction of periodontal tissue with heavy load of inflammation. This is the reason in polymicrobial community disease ultimately; flap surgery would be required in chronic periodontitis patient. PC-1 results also draws attention towards oxidative stress where major antioxidants had failed to control the inflammatory condition, which is also a part of strong cellular defense defeating at the diseased site.
PC-2 loading projects marked differences clearly for defining the periodontal condition from the healthy controls. Here we can see the increase of particular metabolites like propionate, propylene glycol, acetoin, valine, butyrate and 5- aminopentanoate indicates the population shift from homeostatic to anaerobic condition. Decreasing levels of NAG along with pyruvate, glutamate, ethane sulfonate and ethanol supports the above dysbiotic condition defining the severity of chronic periodontitis (Aimetti et al. 2012). These could be used as a possible biomarker in defining the chronic periodontal condition as they become very important factor in distinguishing the periodontal condition from the healthy one. Ethane sulfonate and some more fermented product like H2S production could exist majorly because of two prominent increase in the population of bacteria; Fusobacterium spp. and P. gingivalis producing offensive foul breath, which could be considered as characteristic symptom of periodontal disease which share the characteristic of other disease known as Halitosis (Matsuyama et al. 2005; Ratcliff and Johnson 1999). H2S production also up regulates LPS induced Interleukin 6 and 8 via activating P. gingivalis which activate more aggressive form of periodontitis (Greabu et al. 2016). As mentioned above oxidative stress could play major role, this could also be connected well with H2S production that indirectly activates neutrophils. PC-1 clearly states a burst of inflammatory responses produced in response to chronic periodontitis and above mentioned biochemical response can be connected well with the findings. Majority of anaerobic bacteria predominantly starts replacing the homeostatic one and lots of Lipopolysaccharides (LPS) exposed to the local environment, which triggers extracellular matrix (ECM) degradation and followed by osteoclastogenesis process. This favors the bone resorption stage, the final outcome in case of chronic periodontitis (Anderson 2000; Inohara and Nunez 2003; Sims and Gooi 2008). Aromatic region of the spectra shows very promising results with decreasing level of phenylalanine, phenylacetate, phenol and histidine, which establishes good correlation with purine degradation pathway getting upregulated. The decreasing level of these may taken into the consideration along with protein degradation could be the possible reason in this multifactorial inflammatory disease (Barnes et al. 2009). Formate increase also presents a strong connection with increase in dysbiotic population leading to loss of homeostatic environment. Some newly identified metabolite such as 3-hydroxybutyrate, acetoin (Johnson et al. 2008), glutamine, asparagine and xanthine (Supplementary Information) (Barnes et al. 2009) found in periodontal disease condition. It can be used for identifying and characterizing the disease specifically and may also serve as biomarkers (Zhang et al. 2012) for defining chronic periodontitis. These consequences mentioned above are most commonly irreversible in nature and medication is not helpful, henceforth flap surgery is ultimately required. This polymicrobial disease results in modulating various inflammatory cytokines, which alters the local population balance and promotes destructive inflammation that really affects the tooth biofilm a lot, a very important protective role towards the beginning of periodontal disease. Understanding the role of biofilm will surely help in near future with the fact that how these communities interact with each other and how population shift disturbs and proceed to dysbiosis along with restoration of healthy biofilm.
5 Conclusion
Present study reveals many insights in the periodontitis using high field 1H NMR spectroscopy metabolomics and the information gathered may be used in differentiating the periodontal condition from the healthy controls. It may benefit for getting the metabolic indicators being used as biomarkers for distinguishing the disease in chronic stage. Newly identified metabolites give geographical diversity and project the habitat and hygiene in first place where it could depict the healthy oral condition. Saliva could be used as a potential body fluid over the traditional (Blood, urine and CSF) in other important diseases as saliva can be easily restored in body where other body fluids may not be easily accessible. Thus this study has importance in oral diseases as well as indicative of many other diseases for use of saliva as a better diagnostic fluid in future (Figueira et al. 2016).
Abbreviations
- CPMG:
-
Carr-Purcell-Meiboom-Gill
- CSF:
-
Cerebrospinal fluid
- ECM:
-
Extracellular matrix
- LPS:
-
Lipopolysaccharides
- NMR:
-
Nuclear magnetic resonance
- NOESY:
-
Nuclear overhauser effect spectroscopy
- NAG:
-
N-Acetyl glucosamine
- PCA:
-
Principal component analysis
- PLS-DA:
-
Partial least square discriminant analysis
- ROC:
-
Receiver operating characteristic curve
- SD:
-
Standard deviation
- SPSS:
-
Statistical package for the social science
- TSP:
-
Trimethylsilyl propionate
- VIP:
-
Variables Importance in Projection
References
Aimetti, M., Cacciatore, S., Graziano, A., & Tenori, L. (2012). Metabonomic analysis of saliva reveals generalized chronic periodontitis signature. Metabolomics, 8, 465–474.
Anderson, K. V. (2000). Toll signaling pathways in the innate immune response. Current Opinion in Immunology, 12, 13–19.
Arakaki, A. K., Skolnick, J., & McDonald, J. F. (2008). Marker metabolites can be therapeutic targets as well. Nature, 456, 443–443.
Barnes, V., et al. (2009). Acceleration of purine degradation by periodontal diseases. Journal of Dental Research, 88, 851–855.
Brogden, K. A., & Guthmiller, J. M. (2002). Polymicrobial diseases, Washington, DC: ASM Press.
Chen, F.-M., et al. (2005). Position paper: epidemiology of periodontal diseases. Journal of Periodontology, 76, 1406–1419.
Dame, Z. T., et al. (2015). The human saliva metabolome. Metabolomics, 11, 1864–1883.
Darveau, R. P. (2010). Periodontitis: a polymicrobial disruption of host homeostasis. Nature Reviews Microbiology 8, 481–490.
Dumitrescu, A. L. (2010). Etiology and pathogenesis of periodontal disease, Berlin: Springer.
Eastoe, A. S. C. J. E. (1988). Biochemistry and oral biology, (2nd edn), Amsterdam: Elsevier.
Estrela, C. R. D. A., Pimenta, F. C., Alencar, A. H., Ruiz, L. F. N., & Estrela, C. (2010). Detection of selected bacterial species in intraoral sites of patients with chronic periodontitis using multiplex polymerase chain reaction. Journal of Applied Oral Science, 18, 426–431.
Figueira, J., Jonsson, P., Adolfsson, A. N., Adolfsson, R., Nyberg, L., & Öhman A. (2016) NMR analysis of the human saliva metabolome distinguishes dementia patients from matched controls. Molecular BioSystems, 12, 2562–2571.
Fisher, R. A., & Yates, F. (1957). Statistical tables for biological, agricultural, and medical research, (5th edn). London, Edinburgh Oliver and Boyd.
Greabu, M., A. Totan, D. Miricescu, R. Radulescu, J. Virlan, B. Calenic (2016). Hydrogen sulfide, oxidative stress and periodontal diseases: A concise review. Antioxidants, 5, 3.
Haffajee, A. D., & Socransky, S. S. (2006). Introduction to microbial aspects of periodontal biofilm communities, development and treatment. Periodontology, 42, 7–12.
Hart, T. C., & Kornman, K. S. (1997). Genetic factors in the pathogenesis of periodontitis. Periodontology, 14, 202–215.
Inohara, N., & Nunez, G. (2003). NODs: Intracellular proteins involved in inflammation and apoptosis. Nature Reviews Immunology, 3, 371–382.
Jiang, C.-Y., Yang, K.-M., Yang, L., Miao, Z.-X., Wang, Y.-H., Zhu, H.-B. (2013) A 1 H NMR-based metabonomic investigation of time-related metabolic trajectories of the plasma, urine and liver extracts of hyperlipidemic hamsters. PloS ONE, 8, e66786.
Johnson, B. T., Shaw, L. N., Nelson, D. C., & Mayo, J. A. (2008). Extracellular proteolytic activities expressed by Bacillus pumilus isolated from endodontic and periodontal lesions. Journal of Medical Microbiology, 57, 643–651.
Markley, J. L., et al. (2007) New bioinformatics resources for metabolomics. In: Pacific symposium on biocomputing, vol 12. Madison, WI, pp. 157–168.
Matsuyama, T., Kawai, T., Izumi, Y., & Taubman, M. A. (2005). Expression of major histocompatibility complex class II and CD80 by gingival epithelial cells induces activation of CD4 + T cells in response to bacterial challenge. Infection and Immunity, 73, 1044–1051.
Merritt, J., Niu, G., Okinaga, T., & Qi, F. (2009). Autoaggregation response of fusobacterium nucleatum. Applied and Environmental Microbiology, 75, 7725–7733.
Meyle, J. (1993). Neutrophil chemotaxis and serum concentration of tumornecrosis-factor-α (TNFA). Journal of Periodontal Research, 28, 491–493.
Offenbacher, S. (1996). Periodontal diseases: Pathogenesis. Annals of Periodontology, 1, 821–878.
Page, R. C., & Kornman, K. S. (1997). The pathogenesis of human periodontitis: An introduction. Periodontology, 14, 9–11.
Park, C., Yun, S., Lee, S. Y., Park, K., & Lee, J. (2012). Metabolic profiling of Klebsiella oxytoca: evaluation of methods for extraction of intracellular metabolites using UPLC/Q-TOF-MS. Applied Biochemistry and Biotechnology, 167, 425–438.
Perez-Chaparro, P., et al. (2014). Newly identified pathogens associated with periodontitis: A systematic review. Journal of Dental Research, 93, 846–858.
Ratcliff, P. A., & Johnson, P. W. (1999). The relationship between oral malodor, gingivitis, and periodontitis. A review. Journal of Periodontology, 70, 485–489.
Sims, N. A., & Gooi, J. H. (2008) Gooi bone remodeling: Multiple cellular interactions required for coupling of bone formation and resorption. In: Seminars in cell & developmental biology, vol 19. New York, Elsevier, pp. 444–451.
Søreide, K. (2009). Receiver-operating characteristic curve analysis in diagnostic, prognostic and predictive biomarker research. Journal of Clinical Pathology, 62, 1–5.
Wang, X., Yang, B., Sun, H., & Zhang, A. (2011). Pattern recognition approaches and computational systems tools for ultra performance liquid chromatography–mass spectrometry-based comprehensive metabolomic profiling and pathways analysis of biological data sets. Analytical Chemistry, 84, 428–439.
Wishart, D. S., et al. (2009). HMDB: a knowledgebase for the human metabolome. Nucleic Acids Research, 37, D603–D610.
Wolf, H. F. (2006). Etiology and pathogenesis. periodontitis-a multifactorial disease. colour atlas of dental hygiene-periodontology. (p. 22). New York: Thieme publications.
Xia, J., Broadhurst, D. I., Wilson, M., & Wishart, D. S. (2013). Translational biomarker discovery in clinical metabolomics: an introductory tutorial. Metabolomics, 9, 280–299.
Yu, F.-M., Qiao, B., Zhu, F., Wu, J.-C., & Yuan, Y.-J. (2006). Functional analysis of type II thioesterase of Streptomyces lydicus AS 4.2501. Applied Biochemistry and Biotechnology, 135, 145–158.
Zhang, A., Sun, H., & Wang, X. (2012). Saliva metabolomics opens door to biomarker discovery, disease diagnosis, and treatment. Applied Biochemistry and Biotechnology, 168, 1718–1727.
Zweig, M. H., & Campbell, G. (1993). Receiver-operating characteristic (ROC) plots: A fundamental evaluation tool in clinical medicine. Clinical Chemistry, 39, 561–577.
Acknowledgements
The authors are thankful for the grant funded by ICMR, New Delhi (Budget Allotment letter no.-5/4/2-7-12-NCD-II). Manvendra Pratap Singh also expresses his gratitude for the award of fellowship. Manuscript Communication Number: IU/R&D/2017/-MCN000134.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no potential conflict of interest. The disclosure of potential conflict of interest in the prescribed format has been obtained from all the authors.
Ethical approval
The study was ethically approved and the work was performed in strict accordance with the guidelines of Institutional Ethical Committee. The selected patients were fully informed and their consents were taken well beforethe investigations (Ethical approval No.- B-07: CBMR/EMP/IEC/3/26-02-2011). The authors: Manvendra Pratap Singh, Mona Saxena, C.S. Saimbi, Jamal M. Arif and Raja Roy are aware of ethical policy.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Singh, M.P., Saxena, M., Saimbi, C.S. et al. Metabolic profiling by 1H NMR spectroscopy of saliva shows clear distinction between control and diseased case of periodontitis. Metabolomics 13, 137 (2017). https://doi.org/10.1007/s11306-017-1245-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-017-1245-4