Abstract
Introduction
While green tea is known to protect skin from ultraviolet (UV) light, underlying damage-repair mechanisms remain unclear.
Objectives
The major objective of this study was to investigate, using multi-omics analysis, the metabolic mechanisms associated with the effects of a diet supplemented with green tea (SGT) in UVB-damaged mice.
Methods
Six to eight weeks old female Skh:HR-1 mice were randomly divided into three experimental groups i.e., non-irradiated with control diet (NOR), UVB-irradiated with control diet (UND), and UVB-irradiated with SGT (UGD), and subjected to experimental conditions over 10 weeks. The skin samples were analyzed by metabolomics, transcriptomics, and in silico modeling.
Results
Our results revealed that SGT significantly alleviated UVB-induced metabolite alterations (aspartic acid, ornithine, ascorbic acid, ethanolamine, and C20:0-lysophosphatidylethanolamine) and gene expressions (keratin sulfate biosynthesis/degradation, fatty acid oxidation, and steroid metabolism) in the skin. Among these changes, key metabolic pathways, including ascorbate metabolism and the urea cycle, were the major pathways mitigated through SGT diet among UGD group mice. Additionally, SGT treatment also affected serum and hepatic lysophospholipid levels through attenuating and intensifying UVB-induced metabolic changes, respectively.
Conclusion
Our results suggested that the SGT diets primarily influenced ascorbate metabolism and the urea cycle in UVB-irradiated mouse skin, alleviating deleterious UVB-induced skin wrinkles, epidermal thickening, and collagen-fiber destruction.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Skin aging is accelerated by endogenous and exogenous factors, such as inflammation, hormonal imbalance, exposure to ultraviolet (UV) radiation, and smoking. These stressors cause oxidative stress that can potentially damage the membranes, lipids, proteins, and DNA of skin cells. Dietary supplements, such as collagen, green tea, probiotics, unsaturated fatty acids, and vitamins, improve the quality of skin via skin-barrier homeostasis (Draelos 2010). In our previous study, we observed that supplemented green tea (GTS) significantly attenuated UVB-induced harmful changes in mouse skin viz., wrinkles, erythema, skin hydration, and elasticity through mending skin metabolites vital for its structure and function (Jung et al. 2015). However, the underlying genomic-metabolic mechanisms correlating the GTS administration and its components leading to skin repair following the UVB irradiation are largely remain unexplored. Further underpinning the skin health effects of GTS, we conceive a hypothesis that GTS administration potentially attenuates UVB induced mammalian skin damage through mending the biochemical pathways critical for skin repair and maintenance, in either direct or indirect way.
An increasing number of studies integrated multiple omics data, including genomics, transcriptomics, proteomics, and metabolomics, to comprehensively model cellular functions (Li et al. 2016; Hyotylainen et al. 2016). Recently, studies used multi-omics approaches to reveal biomarkers for various disease states involving cancer, neurodegenerative disorders, stress, and the effects of bioactive ingredients (Liu and Zhang 2015; Pineda et al. 2015; Boccio et al. 2016; Mastrokolias et al. 2016). The metabolome consists of products derived from metabolic responses and is a sensitive measure of phenotype and associated perturbations. Moreover, combination with other omics data, such as that of the transcriptome and proteome, allows additional insight into pathway modulation due to synergistically described pathophysiological conditions (Boccio et al. 2016; Mastrokolias et al. 2016). In order to process high-throughput omics data, large-scale computational models, such as genome-scale models (GEMs), can also be used to describe the metabolic states of an organism (Lee et al. 2005).
In this study, we performed complementary investigation of the metabolome and transcriptome to reveal the effects of a diet supplemented with green tea (SGT) on the metabolic mechanisms involved with UVB-induced alterations in skin conditions. Furthermore, we also conducted endogenous metabolomics analysis of the serum and liver to determine additional influences of a SGT on mice and relation with the skin condition.
2 Materials and methods
2.1 Chemicals and materials
Acetonitrile, dichloromethane, methanol, and water were obtained from Fisher Scientific (Pittsburgh, PA, USA). Pyridine, formic acid, n-methyl-n-(trimethylsilyl) trifluoroacetamide (MSTFA), methoxyamine hydrochloride, and standard compounds were purchased from Sigma-Aldrich (St. Louis, MO, USA). The green tea extract (GTE) was provided by AmorePacific Corp. (Seoul, Korea) and mainly contains four kinds of catechins, including epicatechin, epigallocatechin, epicatechin gallate, and epigallocatechin gallate. The catechin content of GTE was analyzed and quantified by triple-quadrupole mass spectrometry (MS) (Supplementary Table 1). GTE contained 50% total catechins and 4.5% caffeine.
2.2 Animal experiments
Six to eight weeks old female Skh:HR-1 mice were purchased from Orient Bio (Seongnam, Korea). Mice were housed under controlled conditions as followed: temperatures (24 ± 2 °C), light (12-h light/dark cycle), and humidity (55 ± 10%). After acclimation (1 week), the mice were randomly divided into three experimental groups. These groups were non-irradiated with control diet (Ain-93G) (NOR; n = 12), UVB-irradiated with control diet (UND; n = 12), and UVB-irradiated with SGT (Ain-93G + 1% GTE) (UGD; n = 12). For UVB irradiation, four fluorescent lamps (TL 20 W/12 RS SLV; peak emission: 315 nm; wavelength: 290–390 nm; Philips, Amsterdam, Netherlands) were used, and intensity was monitored with a UV meter (VARIOCONTROL v.2.03; Waldmann, Villingen-Schwenningen, Germany). Mice were exposed to UVB radiation three times every week by moving freely in the cage at a distance of 30 cm from the lamps. The UVB irradiation began with doses of 75 mJ/cm2 [one minimal erythema dose (MED)] and was elevated weekly to four MED and maintained subsequently. Food intake and the body weight of mice were measured weekly, with no significant differences observed among the groups. After 10 weeks, mice were sacrificed and their dorsal skin were collected and stored at −80 °C. All animal experimental procedures were approved by the Institutional Animal Care and Use Committee of Gyeonggi Institute of Science & Technology (permit number: 2013-12-0006) and were performed accordance with their guidelines.
2.3 Clinical and histological assessment
Skin wrinkles of hairless mice were determined using the Visioline VL 650 (Courage + Khazaka Electronic GmbH, Köln, Germany). Total area, number, mean length, and depth of wrinkles were analyzed by skin-replica assessment, which was created by applying silicon rubber (SILFLO; J&S Davis, Stevenage, England) to the dorsal skin of hairless mice. For histological assessment, excised skin and liver tissues of mice were fixed in 10% neutral-buffered formalin, followed by embedding in paraffin, frozen, and sectioned (5 μm). The sections were stained with hematoxylin and eosin and masson’s trichrome solutions. The epidermal thickness was measured at five different sites, and a mean value was calculated. The stained sections were observed using an ECLIPSE Ti-E inverted microscope (Nikon, Tokyo, Japan) and processed with NIS-Elements BR 3.00 software (Nikon).
2.4 Sample preparation and metabolite profiling
For metabolite profiling, the skin and serum extracts were prepared. Before skin extraction, 2 × 2 cm sections of mouse central-dorsal skin tissue were finely chopped. Methanol (1 mL) was added to the finely chopped skin tissues, which were then homogenized using an MM400 mixer mill (Retch, Haan, Germany) at a frequency of 30 s−1 for 15 min at room temperature. After homogenization, the suspension was kept at 4 °C for 30 min and then centrifuged at 12,578×g for 10 min at 4 °C. The supernatant was filtered through a 0.2-μm polytetrafluoroethylene (PTFE) filter and dried using a speed vacuum concentrator (Modulspin 31; Biotron, Gyeonggi-Do, Korea). For serum extraction, 600 μL methanol was added to 200 μL serum, and the mixture was sonicated with shaking for 15 min. After centrifugation at 12,578×g for 10 min at 4 °C, the supernatant was filtered using a 0.2-μm PTFE filter and dried using a speed vacuum concentrator (Biotron). The liver-tissue extracts were prepared by adding 1 mL of methanol:water (1:1, v/v) to liver tissue (150 mg ± 1 mg) using a mixer mill with a frequency of 30 s−1 for 15 min. After homogenization, the suspension was centrifuged at 12,578×g for 10 min at 4 °C, filtered through a 0.2-μm PTFE filter, and dried using a speed vacuum concentrator (Biotron). Metabolite profiling was performed using two different high throughput analytical approaches i.e., ultra-performance liquid chromatography quadrupole time-of-flight MS (UPLC-Q-TOF-MS) and gas chromatography (GC)-TOF-MS, owing to the chemical nature of metabolites. The dried-skin and serum extracts were re-dissolved with methanol, and liver extracts were re-dissolved with 50% methanol for UPLC-Q-TOF-MS analysis. The UPLC chromatographic separation was carried out on a Waters ACQUITY BEH C18 column (i.d., 100 mm × 2.1 mm, 1.7 μm particle size; Waters Corp.) with 5 μL of injection volume. The column temperature and flow rates were set at 37 °C and 0.3 mL/min, respectively. Binary solvent system was consisting of 0.1% v/v formic acid in water (A) and 0.1% v/v formic acid in acetonitrile (B). The chromatographic run was programmed with 5% of B for initial 1 min followed by gradient increase to 100% of B over 9 min. Total run time was 14 min including re-equilibration to the initial condition of the column. The MS analysis conditions were similar to those described previously by Jung et al. 2015. For GC-TOF-MS analysis, the dried samples were prior derivatized in two consecutive steps involving; (a) oximation with 50 μL methoxyamine hydrochloride (20 mg/mL in pyridine) at 30 °C for 90 min, and (b) silylation using 50 μL MSTFA at 37 °C for 30 min. The operational conditions for GC-TOF-MS analysis were identical to those described in our previous study (Jung et al. 2015).
2.5 Targeted skin-ceramide analysis
The mouse epidermis was separated as described previously (Shin et al. 2014), and epidermis samples (5 mg) were homogenized in a tissue lyser (30 Hz for 30 s; Qiagen, Hilden, Germany) with 500 μL methanol. The homogenized samples were centrifuged at 12,578×g for 10 min at 4 °C. The precipitates were again homogenized with 500 μL dichloromethane:methanol (3:1, v/v), and the homogenized samples were centrifuged. The resulting supernatants were transferred into clean tubes and dried under vacuum. Dried samples were resuspended in 100 μL chloroform:methanol (1:9, v/v) and diluted 10-fold with chloroform:methanol (1:9, v/v) containing 7.5 mM ammonium acetate. Skin-ceramide profiling was performed on an LTQ XL mass spectrometer (Thermo Fischer Scientific, West Palm Beach, FL, USA) equipped with a TriVersa NanoMate (Advion Biosciences, Ithaca, NY, USA) using previously described analytical conditions (Shin et al. 2014). Skin ceramides were tentatively identified by comparison with the MS/MS-fragmentation patterns of commercially available ceramide standards and our in-house ceramide library (Shin et al. 2014).
2.6 Data processing and multivariate statistical analysis
The datasets from GC-TOF-MS and UPLC-Q-TOF-MS were acquired and preprocessed using LECO Chroma TOF software (v.4.44; LECO Corp.) and MassLynx software (v.4.1; Waters Corp., Milford, MA, USA), respectively. These raw data were converted into the NetCDF format (*.cdf). Using the Metalign software package (http://www.metalign.nl), retention-time correction, peak detection, and alignment were processed and exported resulting data to an Excel file (Microsoft Corp., Redmond, WA, USA). Unit variance scaling (autoscale) and mean cantering in a column-wise fashion were applied to datasets. The multivariate statistical analyses were performed using SIMCA-P+ (v.12.0; Umetrics, Umea, Sweden). The significantly discriminant variables among the experimental groups were selected depending on the variable importance in projection (VIP) values. The discriminant metabolites were putatively identified by comparing their retention time, mass spectra, and mass fragment patterns with those for commercial standard compounds analyzed under identical conditions and various available databases, including the National Institutes of Standards and Technology (NIST) library, the Human Metabolome Database (HMDB; http://www.hmdb.ca/), and Wiley 8. The data significance was determined through Duncan’s multiple-range tests and analysis of variance using PASW Statistics 18 software (SPSS Inc., Chicago, IL, USA). Correlation coefficients were also calculated by PASW statistics 18 software, and a correlation network was constructed using Cytoscape software (version 3.4.0, http://www.cytoscape.org/).
2.7 Microarray analysis
Quantitative analysis of RNA expression was conducted using an Affymetrix GeneChip mouse genome 430 A 2.0 array (Affymetrix, Santa Clara, CA, USA). Total RNA was isolated from the dorsal skin of three mice from each group using RNAiso Plus (Takara, Shiga, Japan). RNA quality and quantity was measured using a NanoDrop 2000 (Thermo Fisher Scientific) to ensure the integrity of the RNA. All experiments following the RNA isolation were conducted by DNPbiotec (Daegu, Korea).
2.8 Identification of differentially expressed genes
Differentially expressed genes between NOR versus UND and UND versus UGD groups were identified based on a modified t statistic (Wettenhall and Smyth 2004) implemented using the limma package within the R environment. Subsequently, p-values were adjusted for multiple-hypothesis testing using Benjamini and Hochberg’s method (Benjamini and Hochberg 1995), and the false discovery rate was controlled at 5%.
2.9 Reporter-metabolite identification
Reporter metabolites in NOR versus UND and UND versus UGD groups were identified based on a previously published method (Zelezniak et al. 2010). Briefly, each metabolite in the mouse GEM was scored based on the p-values of neighboring differentially expressed enzymes using the p-value associated with differential expression of corresponding genes. In the case of isozymes or enzyme complexes, the lowest p-value for the isozyme or enzyme subunit was used. Subsequently, the p-values were converted into Z-scores using an inverse normal cumulative distribution function (CDF) for each enzyme, i, as follows:
Once each enzyme was Z-scored, the Z-score for each metabolite (Z metabolite ) was calculated. To do so, the aggregated Z-scores of k neighboring enzymes were computed as follows:
Finally, the metabolite Z-scores were corrected for background distribution by subtracting the mean (µ k ) and dividing by the standard deviation (σ k ) of the original Z-score for the Z metabolite .
The corrected Z-scores were once again transformed into p-values using normal CDF. Metabolites with p < 0.05 were classified as statistically enriched reporter metabolites. The COBRA toolbox (Schellenberger et al. 2011) was used to identify the reporter metabolites from a mouse GEM.
3 Results and discussion
3.1 Study design and histopathology of skin tissues
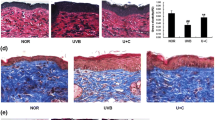
The female albino hairless mice (Skh:HR-1) were subjected to comprehensive unbiased skin metabolomics and transcriptomics studies towards evaluating the effects of SGT against UVB induced photoaging. The animal experiments were spanned for 10 weeks for each mice group i.e., NOR, UND, UGD. After 10 weeks, detailed histopathology examination of the skin tissues of the treated mice groups exhibited the deleterious effects of UVB irradiation viz., severe deep wrinkles, increased total wrinkle area, mean length, and wrinkle depth on the dorsal skin of mice (Fig. 1a, b). Additionally, an enhanced epidermal thickness and collagen-fiber destruction were observed among UND mice groups as compared to NOR-groups (Fig. 1c, d). Intriguingly, the UVB induced deleterious effects were significantly alleviated in mice groups subjected to SGT diet (UGD).
3.2 SGT-induced alterations in the skin metabolome of UVB-irradiated mice groups
The significantly discriminated endogenous metabolome altered through UVB irradiation on mice groups with or without SGT diets were determined through comprehensive metabolite profiling coupled with targeted ceramide analysis. In PCA-score plots derived from GC-TOF-MS and UPLC-Q-TOF-MS datasets, all three experimental groups were well clustered across the principal components (Supplementary Fig. 1a and b). Further, the clear grouping patterns for each experimental mice group were observed in corresponding PLS-DA score plots (Fig. 2a, b). The fitness and prediction accuracy of the PLS-DA model signified by R2X(cum), R2Y(cum),and Q2 (cum), as well as the p-values determined through cross-validation analyses are summarized in Supplementary Table 3. Thirty-four skin associated metabolites were selected and identified as significantly discriminant among the NOR, UND, and UGD groups based on VIP values > 0.7 and p < 0.05. Additionally, 49 ceramides were targeted for analysis, showing significantly different levels among the NOR, UND, and UGD groups. The relative levels of 10 amino acids, 5 organic compounds, 1 nucleobase, 2 carbohydrates, 5 fatty acids, 1 bile acid, 10 lipids, and 49 ceramides were converted to respective fold changes and are presented in Table 1. We observed that SGT diets significantly attenuated various UVB-induced skin-metabolite alterations. In particular, skin metabolites, including aspartic acid (1), ornithine (6), ascorbic acid (11), ethanolamine (12), and lysoPE (20:0) (30) showed the most significant changes. The levels of skin metabolites, including ornithine (6), ascorbic acid (11), ethanolamine (12), and lysoPE (20:0) (30), increased in the UND group as compared with those observed in the NOR group, whereas changes in these metabolite levels were attenuated in the UGD group. By contrast, the levels of aspartic acid (1) in skin showed a reciprocal pattern. In comparison with our previous study (Jung et al. 2015), the levels of most skin lysophospholipids and ceramides, which are related to skin structure, showed a similar attenuating patterns effected by SGT diet, while the levels of amino acids and fatty acids were conversely varied compared to our previous study (Table 1). In contrast to some skin ailments viz., psoriasis and atopic dermatitis, the UVB irradiation induce ceramide generation in keratinocytes as well as murine skin through the activation of sphingomyelinases and non-bound free ceremides, respectively (Meckfessel and Brandt 2014; Magnoni et al. 2002; Takagi et al. 2004). Functionally, the ceramides are the important lipids components of the epidermis, which plays an important role in skin barrier functions (Meckfessel and Brandt 2014). In the present study, we observed a similar increasing pattern of skin ceramides through UVB irradiation in UND group. However, no significant changes in ceramides levels were observed in the skin tissue of UVB irradiated mice fed on GTS diet i.e., UGD (Table 1). Among those, ceramides containing non-hydroxy fatty acid dihydrosphingosine and phytosphingosine were observed remarkably higher in UND groups as compared to UGD group. Kobayashi et al. (2016) have reported that catechins effect strong inhibition of secretory sphingomyelinase activity which in turn decreases plasma ceramide concentrations, however, the direct correlations establishing the link between ceramide levels and catechins in GTS are lacking. We construe that the varying patterns of skin-metabolite alterations in the present study and those reported previously might be attributed to the different experimental conditions. In general, we observed a common attenuating pattern induced by SGT diet on the levels of metabolites reportedly altered through UVB exposures (Jung et al. 2015). In particular, ascorbic acid (11) levels in skin, a biomarker determined in a previous study, showed a similar patterns (twofold increases in the UND group as compared with the NOR group, with relatively decreased levels in UGD mice group). The skin metabolites that showed attenuating patterns as result of a SGT diet on UVB-exposed mice were used for further analyses and integration with transcriptome data.
3.3 The differentially expressed skin transcriptome mediated by SGT in UVB-irradiated mice group
We performed microarray analysis to identify differentially expressed genes in UVB-irradiated mouse skin and/or the skin of mice fed a SGT. Skin-transcriptome data was mapped onto a mouse GEM (Sigurdsson et al. 2010) using the gene-protein-reaction association. Previously, integrating transcriptome data with GEM has successfully been applied to microbes and plants to improve predictions of metabolic behaviors under certain stimuli (Akesson et al. 2004; Mohanty et al. 2016). The expression levels of 1366 metabolic genes pertaining to 2011 reactions were associated with the model and used for further analyses. We explored the expression patterns associated with individual reactions and metabolite levels by calculating the differential expression of each reaction under various treatments in a pairwise manner using a modified t statistic adjusted for multiple-hypothesis testing. This analysis identified significantly altered pathways related to UVB irradiation and a SGT, thereby highlighting the possible attenuating effect of the SGT on UVB-irradiated skin in key metabolic pathways of gene expressions, such as keratin sulfate biosynthesis/degradation, fatty acid oxidation, and steroid metabolism (Supplementary Fig. 2). Skin keratin sulfate, sterols, lipids, and amino acids were closely related to skin-barrier functions, such as moisture retention (Meguro et al. 1999; Katagiri et al. 2003; Oh et al. 2011; Yahagi et al. 2011). Glycosaminoglycans, such as keratin sulfates, comprise the core proteins in proteoglycans, which may contribute to formation of the extracellular matrix, including collagen fibers and elastic fibers (Reinboth et al. 2002; Oh et al. 2011). Here, chronic UVB irradiation disrupted collagen fiber formation, whereas mice in the UGD group were less affected by UVB irradiation, showing a consistency between phenotype and genotype (Fig. 1d). From the list of reactions with the most significant transcriptional changes, we then identified reporter metabolites (Patil and Nielsen 2005) to determine metabolic hotspots in the mouse-skin network and their corresponding transcriptional-regulatory mechanisms. The list of statistically enriched reporter metabolites for each treatment is provided in Table 2. The top-ranked reporter metabolites were mostly involved in steroid metabolism, fatty acid metabolism, glycan biosynthesis and degradation, nucleotide metabolism, and inositol metabolism. These reporter metabolites and the related gene-expression patterns were used for constructing metabolic pathways for integration into skin-metabolome data.
3.4 Complementary identification using integrative metabolomic approach for regulatory pathways in mouse skin affected by a SGT
Multi-omics analysis combining the metabolomic and transcriptomic data using the in silico modeling was performed to comprehensively elucidate the molecular mechanisms affected by SGT in UVB-irradiated mouse skin. A multi-omics analysis can provide comprehensive insight and better understanding of model systems. The transcriptomic response represents an activated metabolic network and directly corresponds to metabolic changes perturbed through altered cellular states (Ippolito et al. 2005; Joyce and Palsson 2006). Based on the list of identified skin metabolites (those that showed attenuation patterns linked with the effects of SGT on UVB-induced skin metabolome alterations; Table 1), selected reporter metabolites (Table 2), and the related gene-expression patterns, a skin-related metabolic pathway was proposed (Fig. 3). Integrating skin metabolites, reporter metabolites, and related gene-expression patterns allowed the construction of a more precise skin-related metabolic pathway. We could observe that metabolome and transcriptome data filling the gaps in corresponding skin metabolism. Further, we showed the gene expression covering metabolic space in skin metabolic pathways. Based on this pathway, we conjecture that UVB-induced alterations in gene expression and metabolites levels associated with ascorbate metabolism, amino acid metabolism, steroid metabolism, nucleotide metabolism, and the urea cycle were highly attenuated by a SGT diet. Further, we observed the overlaps between several reporter metabolites derived from transcriptomics and metabolomics approaches viz., myo-inositol (ascorbate metabolism), citrulline (urea cycle), ornithine (urea cycle), and urea (urea cycle). The gene-expression patterns associated with UVB-induced changes in the metabolism of myo-inositol to ascorbic acid were highly attenuated by a SGT. Ascorbic acid can be synthesized in mice from glucose and regulates collagen biosynthesis (Murad et al. 1981). It is a cofactor for the enzyme prolylhydroxylase, which generate hydoxyproline, a metabolite vital for stable collagen structure (Murad et al. 1981). Furthermore, ascorbic acid is also one of the low molecular weight endogenous antioxidants along with Vitamin E, uric acid, ubiquinol, and glutathione. In addition to these, skin also has endogenous enzymatic antioxidants such as superoxide dismutase, catalase, and glutathione peroxidase. In skin, these antioxidants together play an important role in protection of the cells against oxidative stresses induced by UV radiation (Shindo et al. 1993). However, when skin photoaging occurs, the expression of endogenous antioxidant enzymes in stratum corneum and epidermis were significantly reduced (Pandel et al. 2013). In the present study, we observed a significant increase in the relative levels of skin ascorbic acid by chronic UVB irradiation, while SGT alleviated this marked alteration. Overall, the changes in ascorbate metabolism may significantly influence the collage construction as well as the cell’s defence mechanisms against oxidative stress induced by UVB irradiation.
A schematic diagram of the proposed skin-related metabolic pathway based on information regarding skin metabolites (metabolomics), reporter metabolites (transcriptomics), and related gene-expression patterns. All selected metabolites and genes exhibited attenuating patterns associated with a SGT on UVB-induced changes. The dashed line indicates multiple steps. Black square boxes with green characters indicate skin metabolites derived from metabolomics analysis; blue characters indicate reporter metabolites from transcriptome analysis using in silico modeling; yellow square boxes indicate an overlap of metabolites between skin metabolites and reporter metabolites. The pathway was modified from the reported KEGG (http://www.genome.jp/kegg/) pathway. EC numbers for enzymes are as follows: 1.1.1.170 [NAD(P) dependent steroid dehydrogenase-like], 1.1.3.8 [gulonolactone (L-) oxidase], 1.13.99.1 [myo-inositol oxygenase], 1.14.11.2 [procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha II polypeptide], 1.14.13.15 [cytochrome P450, family 27, subfamily a, polypeptide 1], 1.14.13.39 [nitric oxide synthase 1, neuronal], 1.14.13.70 [cytochrome P450, family 51], 1.3.1.21 [7-dehydrocholesterol reductase], 1.3.1.70 [transmembrane 7 superfamily member 2], 1.3.5.1 [succinate dehydrogenase complex, subunit A, flavoprotein (Fp)], 1.4.3.4 [monoamine oxidase B], 2.1.3.3 [ornithine transcarbamylase], 2.3.1.65 [bile acid-Coenzyme A: amino acid N-acyltransferase], 2.5.1.21 [farnesyl diphosphate farnesyl transferase 1], 2.6.1.1 [glutamate oxaloacetate transaminase 2, mitochondrial], 2.6.1.5 [tyrosine aminotransferase], 2.7.1.36 [mevalonate kinase], 3.1.1.17 [regucalcin], 3.1.3.3 [phosphoserine phosphatase], 3.1.3.5 [5′ nucleotidase, ecto], 3.5.3.11 [agmatine ureohydrolase (agmatinase)], 3.6.1.6 [ectonucleoside triphosphate diphosphohydrolase 4], 4.1.1.- [pyridoxal-dependent decarboxylase domain containing 1], 4.1.1.15 [glutamate-ammonia ligase (glutamine synthetase)], 4.1.1.17 [ornithine decarboxylase, structural 1], 4.1.1.28 [dopa decarboxylase], 4.1.1.33 [mevalonate (diphospho) decarboxylase], 4.2.1.11 [enolase 3, beta muscle], 5.1.99.4 [C1q and tumor necrosis factor related protein 3], and 5.4.99.7 [lanosterol synthase]
Further, SGT also attenuated UVB-induced metabolic changes and gene expression related to the urea cycle (Fig. 3). In urea cycle, citrulline is a precursor of arginine, and ornithine is the immediate metabolite derived from arginine. Ornithine and arginine are known to have various pharmacological effects (Shi et al. 2002). According to other studies, supplementation of ornithine and arginine positively effects wound healing by increasing wound breaking strength and scar collagen deposition. These effects are dependent and independent on nitric oxide synthesis, respectively. The expression of nitric oxide synthase (NOS) vital for enzymatic conversion of arginine to nitric oxide and citrulline (Shi et al. 2000, 2002; Debats et al. 2009), was inversely effected by UVB irradiation and SGT. In addition, urea is naturally occurring endogenous metabolite in the skin and acts a natural moisturizing factor that enhances stratum corneum hydration and permeability barrier function (Grether-Beck et al. 2012; Bjorklund et al. 2013). According to Grether-Beck et al. (2012), topical application of urea regulates epidermal-permeability barrier functions and antimicrobial-peptide gene expression in the epidermis as a small-molecule regulator. Additionally, ornithine decarboxylase, which converts ornithine to putrescine through an enzymatic reaction, in the urea cycle contributes to UVB-induced skin cancer development (Elmets and Athar 2010). In Fig. 3, not only metabolites levels of citrulline, ornithine, and urea, but also gene expression levels of nitric oxide synthase, ornithine transcarbamylase, and ornithine decarboxylase were highly attenuated by SGT, altered significantly by UVB irradiation. However, the studies linking the alteration of skin urea cycle and improvement of photo-damaged skin are not widely reported. However, pertaining to the observed changes in overall metabolite levels and gene expressions for urea cycle, we can arguably correlate the alleviating effects of SGT with UVB induced skin photoaging. The two metabolic pathways related to ascorbate metabolism and the urea cycle are highly related to skin structure and barrier function. Therefore, our results suggested that these metabolic pathways might be involved in mechanisms associated with a SGT alleviating UVB-related skin damage.
3.5 The effects of a SGT on endogenous serum and liver metabolites in UVB-irradiated mice and correlation with skin metabolites
Chronic UVB irradiation induces metabolite changes not only in the skin, but also throughout the body, including the serum and liver. Most of previous studies on UV radiation have focused on skin tissue, but these works also focussed on the effects of UV irradiation on non-skin tissues including, blood and internal organs. Svobodova et al. (2011) reported that acute exposure of UVA and UVB causing alterations of oxidative stress related parameters in skin, erythrocytes, and liver such as catalase, glutathione, superoxide dismutase. Goerz et al. (1996) reported that UVA and UVB exposure on rat skin modified P450 isoenzymes activity, especially of P450 1A1 in skin and in liver. In addition, skin releases anti-inflammatory mediator including vitamin D and nitric oxide by UV exposure, these are crucial in prevention of liver inflammation by circulation. Recently, Park et al. (2014) reported that chronic UVB radiation altered not only skin metabolites but also hepatic metabolites in mice, which indicated that UV radiation also affected internal organ. Only a limited number of studies reported the effects of UV radiation on internal organ. However, UV radiation induced damaged biomolecules and signal molecules may reach the blood circulation and in this way affect blood cells and internal organs. To determine the effects of a SGT on serum and the liver of UVB-irradiated mice, we measured biochemical parameters and performed metabolite profiling. According to Fig. 1e, no macroscopic differences in liver tissues were observed among groups; however, the relative levels of triglycerides were significantly decreased in the UGD group (Supplementary Table 1) relative to the levels observed in other groups. According to PCA and PLS-DA, all three experimental groups were clearly separated from one another (Supplementary Fig. 1c–f and Fig. 2c–f). We selected distinguishing metabolites among the NOR, UND, and UGD groups and summarized these in Supplementary Table 2. Among these metabolites, those exhibiting significant alterations are shown in Fig. 4, which shows that a SGT resulted in the emergence of two different patterns of effects on endogenous metabolites altered by UVB radiation. A SGT significantly attenuated changes in most serum metabolites that were induced by UVB irradiation, similar alleviating pattern with those of skin metabolites. However, most hepatic metabolite changes induced by UVB irradiation were significantly intensified by a SGT. Most serum lysoPCs, such as C16:1 (40, 41), C20:4 (55, 56), and C22:6 (59), except (18:2) (46, 47), were increased in the UND group as compared to levels observed in the NOR group, whereas these changes were attenuated in the UGD group. By contrast, levels of liver metabolites were significantly altered by UVB irradiation, with a SGT significantly intensifying these changes. The majority of the enhanced endogenous hepatic metabolites were amino acids, cholesterol, and lipids, which are closely related to metabolic liver functions. These metabolites included aspartic acid (3), phenylalanine (6), 2-ketoglutaric acid (11), cholesterol (33), lysoPC (16:0) (36), lysoPE (16:0) (38), lysoPC (18:1) (44), lysoPC (20:4) (55), lysoPC (22:5) (57, 58), and lysoPC (22:6) (59, 60). Lysophospholipids control initiation of the adaptive immune response and induce humoral and cellular immune responses. Additionally, saturated acyl lysoPCs stimulate pro-inflammatory cytokines, which induces inflammation, while polyunsaturated acyl lysoPCs exhibit anti-inflammatory effects (Perrin-Cocon et al. 2006; Hung et al. 2012; Olofsson et al. 2008). Our results showed that levels of saturated acyl lysophospholipids, such as lysoPC (16:0), lysoPE (16:0), and lysoPC (18:0), increased in both the UND and UGD groups, whereas levels of polyunsaturated acyl lysophospholipids decreased in both the UND and UGD groups. These results indicated that both UVB irradiation and a SGT might cause inflammation in the liver; however, the altered levels of serum lysophospholipids by UVB irradiation were attenuated by SGT, which was inconsistent with those of hepatic metabolites (Fig. 4 and Supplementary Table 2). Notably, the attenuating effects associated with serum lysoPCs may be closely related to the anti-inflammatory effects of green tea. In addition, correlation analysis among skin, serum, and liver metabolites showed that a number of serum metabolites were highly correlated with skin metabolites including aspartic acid, ascorbic acid, ethanolamine, citrulline, urea, which showed most remarkable alleviation in UVB-induced changes through SGT diet (Supplementary Fig. 3).
4 Concluding remarks
We performed a comprehensive and integrated metabolomic-transcriptomic analysis to investigate the effects of SGT diet on skin metabolic mechanisms altered through UVB-irradiation in mice. Our results revealed that SGT highly alleviated UVB-induced alterations of metabolites and gene expressions associated with ascorbate metabolism, amino acid metabolism, steroid metabolism, nucleotide metabolism, and urea cycle. Especially, ascorbate metabolism and urea cycle were selected as key metabolic pathways by integrated analysis based on metabolomics, transcriptomics, and in silico modeling results. These metabolic pathways could constitute a repair mechanism related to green tea on chronic UVB-induced skin damage. Additionally, chronic UVB irradiation also influenced the levels of serum and hepatic lysophospholipids, SGT attenuated and intensified those of changes, respectively. Among them serum metabolites were highly correlated with skin biomarkers. As a result, our study suggested that integrated metabolomic analysis was effective at determining key metabolic pathways from large amounts of data, as well as a variety of changes caused by specific stimuli.
References
Åkesson, M., Förster, J., & Nielsen, J. (2004). Integration of gene expression data into genome-scale metabolic models. Metabolic Engineering, 6(4), 285–293.
Benjamini, Y., & Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological), 289–300.
Björklund, S., Engblom, J., Thuresson, K., & Sparr, E. (2013). Glycerol and urea can be used to increase skin permeability in reduced hydration conditions. European Journal of Pharmaceutical Sciences, 50(5), 638–645.
Boccio, P., Rossi, C., Ioia, M., Cicalini, I., Sacchetta, P., & Pieragostino, D. (2016). Integration of metabolomics and proteomics in multiple sclerosis: From biomarkers discovery to personalized medicine. Proteomics Clinical Applications, 10(4), 470–484.
Debats, I. B. J. G., Wolfs, T. G. A. M., Gotoh, T., Cleutjens, J. P. M., Peutz-Kootstra, C. J., & Van der Hulst, R. R. W. J. (2009). Role of arginine in superficial wound healing in man. Nitric oxide: biology and chemistry/official journal of the Nitric Oxide Society, 21(3), 175–183.
Draelos, Z. D. (2010). Nutrition and enhancing youthful-appearing skin. Clinics in Dermatology, 28(4), 400–408.
Elmets, C. A., & Athar, M. (2010). Targeting ornithine decarboxylase for the prevention of nonmelanoma skin cancer in humans. Cancer Prevention Research, 3(1), 8–11.
Goerz, G., Barnstorf, W., Winnekendonk, G., Bolsen, K., Fritsch, C., Kalka, K., et al. (1996). Influence of UVA and UVB irradiation on hepatic and cutaneous P450 isoenzymes. Archives of Dermatological Research, 289(1), 46–51.
Grether-Beck, S., Felsner, I., Brenden, H., Kohne, Z., Majora, M., Marini, A., et al. (2012). Urea uptake enhances barrier function and antimicrobial defence in humans by regulating epidermal gene expression. Journal of Investigative Dermatology, 132(6), 1561–1572.
Hung, N. D., Sok, D. E., & Kim, M. R. (2012). Prevention of 1-palmitoyl lysophosphatidylcholine-induced inflammation by polyunsaturated acyl lysophosphatidylcholine. Inflammation Research, 61(5), 473–483.
Hyötyläinen, T., Jerby, L., Petäjä, E. M., Mattila, I., Jäntti, S., Auvinen, P., et al. (2016). Genome-scale study reveals reduced metabolic adaptability in patients with non-alcoholic fatty liver disease. Nature Communications, 7.
Ippolito, J. E., Xu, J., Jain, S., Moulder, K., Mennerick, S., Crowley, J. R., et al. (2005). An integrated functional genomics and metabolomics approach for defining poor prognosis in human neuroendocrine cancers. Proceedings of the National Academy of Sciences of the United States of America, 102(28), 9901–9906.
Joyce, A. R., & Palsson, B. Ø (2006). The model organism as a system: Integrating’omics’ data sets. Nature Reviews Molecular Cell Biology, 7(3), 198–210.
Jung, E. S., Park, H. M., Lee, K. E., Shin, J. H., Mun, S., Kim, J. K., et al. (2015). A metabolomics approach shows that catechin-enriched green tea attenuates ultraviolet B-induced skin metabolite alterations in mice. Metabolomics, 11(4), 861–871.
Katagiri, C., Sato, J., Nomura, J., & Denda, M. (2003). Changes in environmental humidity affect the water-holding property of the stratum corneum and its free amino acid content, and the expression of filaggrin in the epidermis of hairless mice. Journal of Dermatological Science, 31(1), 29–35.
Kobayashi, K., Ishizaki, Y., Shosuke, K. O. J. O., & Kikuzaki, H. (2016). Strong inhibition of secretory sphingomyelinase by catechins, Particularly by (–)-epicatechin 3-O-gallate and (–)-3′-O-methylepigallocatechin 3-O-gallate. Journal of nutritional science and vitaminology, 62(2), 123–129.
Lee, S. Y., Lee, D. Y., & Kim, T. Y. (2005). Systems biotechnology for strain improvement. Trends in Biotechnology, 23(7), 349–358.
Li, J., Ren, S., Piao, H. L., Wang, F., Yin, P., Xu, C., et al. (2016). Integration of lipidomics and transcriptomics unravels aberrant lipid metabolism and defines cholesteryl oleate as potential biomarker of prostate cancer. Scientific Reports, 6.
Liu, Q., & Zhang, B. (2015). Integrative omics analysis reveals post-transcriptionally enhanced protective host response in colorectal cancers with microsatellite instability. Journal of Proteome Research, 15(3), 766–776.
Magnoni, C., Euclidi, E., Benassi, L., Bertazzoni, G., Cossarizza, A., Seidenari, S., & Giannetti, A. (2002). Ultraviolet B radiation induces activation of neutral and acidic sphingomyelinases and ceramide generation in cultured normal human keratinocytes. Toxicology in vitro, 16(4), 349–355.
Mastrokolias, A., Pool, R., Mina, E., Hettne, K. M., van Duijn, E., van der Mast, R. C., et al. (2016). Integration of targeted metabolomics and transcriptomics identifies deregulation of phosphatidylcholine metabolism in Huntington’s disease peripheral blood samples. Metabolomics, 12(8), 137.
Meckfessel, M. H., & Brandt, S. (2014). The structure, function, and importance of ceramides in skin and their use as therapeutic agents in skin-care products. Journal of the American Academy of Dermatology, 71(1), 177–184.
Meguro, S., Aral, Y., Masukawa, K., Uie, K., & Tokimitsu, I. (1999). Stratum corneum lipid abnormalities in UVB-lrradiated skin. Photochemistry and Photobiology, 69(3), 317–321.
Mohanty, B., Kitazumi, A., Cheung, C. M., Lakshmanan, M., Benildo, G., Jang, I. C., et al. (2016). Identification of candidate network hubs involved in metabolic adjustments of rice under drought stress by integrating transcriptome data and genome-scale metabolic network. Plant Science, 242, 224–239.
Murad, S., Grove, D., Lindberg, K. A., Reynolds, G., Sivarajah, A., & Pinnell, S. R. (1981). Regulation of collagen synthesis by ascorbic acid. Proceedings of the National Academy of Sciences of the United States of America, 78(5), 2879–2882.
Oh, J. H., Kim, Y. K., Jung, J. Y., Shin, J. E., Kim, K. H., Cho, K. H., et al. (2011). Intrinsic aging-and photoaging-dependent level changes of glycosaminoglycans and their correlation with water content in human skin. Journal of Dermatological Science, 62(3), 192–201.
Olofsson, K. E., Andersson, L., Nilsson, J., & Björkbacka, H. (2008). Nanomolar concentrations of lysophosphatidylcholine recruit monocytes and induce pro-inflammatory cytokine production in macrophages. Biochemical and Biophysical Research Communications, 370(2), 348–352.
Pandel, R., Poljšak, B., Godic, A., & Dahmane, R. (2013). Skin photoaging and the role of antioxidants in its prevention. International Scholarly Research Notices Dermatology, 2013.
Park, H. M., Shon, J. C., Lee, M. Y., Liu, K. H., Kim, J. K., Lee, S. J., et al. (2014). Mass spectrometry-based metabolite profiling in the mouse liver following exposure to ultraviolet B radiation. PloS ONE, 9(10), e109479.
Patil, K. R., & Nielsen, J. (2005). Uncovering transcriptional regulation of metabolism by using metabolic network topology. Proceedings of the National Academy of Sciences of the United States of America, 102(8), 2685–2689.
Perrin-Cocon, L., Agaugué, S., Coutant, F., Saint-Mézard, P., Guironnet-Paquet, A., Nicolas, J. F., et al. (2006). Lysophosphatidylcholine is a natural adjuvant that initiates cellular immune responses. Vaccine, 24(9), 1254–1263.
Pineda, S., Real, F. X., Kogevinas, M., Carrato, A., Chanock, S. J., Malats, N., et al. (2015). Integration analysis of three omics data using penalized regression methods: An application to bladder cancer. PLoS Genetics, 11(12), e1005689.
Rajnochová Svobodová, A., Galandáková, A., Šianská, J., Doležal, D., Ulrichová, J., & Vostálová, J. (2011). Acute exposure to solar simulated ultraviolet radiation affects oxidative stress-related biomarkers in skin, liver and blood of hairless mice. Biological and Pharmaceutical Bulletin, 34(4), 471–479.
Reinboth, B., Hanssen, E., Cleary, E. G., & Gibson, M. A. (2002). Molecular Interactions of Biglycan and Decorin with Elastic Fiber Components BIGLYCAN FORMS A TERNARY COMPLEX WITH TROPOELASTIN AND MICROFIBRIL-ASSOCIATED GLYCOPROTEIN 1. Journal of Biological Chemistry, 277(6), 3950–3957.
Schellenberger, J., Que, R., Fleming, R. M., Thiele, I., Orth, J. D., Feist, A. M., et al. (2011). Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2. 0. Nature Protocols, 6(9), 1290–1307.
Shi, H. P., Efron, D. T., Most, D., Tantry, U. S., & Barbul, A. (2000). Supplemental dietary arginine enhances wound healing in normal but not inducible nitric oxide synthase knockout mice. Surgery, 128(2), 374–378.
Shi, H. P., Fishel, R. S., Efron, D. T., Williams, J. Z., Fishel, M. H., & Barbul, A. (2002). Effect of supplemental ornithine on wound healing. Journal of Surgical Research, 106(2), 299–302.
Shin, J. H., Shon, J. C., Lee, K., Kim, S., Park, C. S., Choi, E. H., et al. (2014). A lipidomic platform establishment for structural identification of skin ceramides with non-hydroxyacyl chains. Analytical and Bioanalytical Chemistry, 406(7), 1917–1932.
Shindo, Y., Witt, E., & Packer, L. (1993). Antioxidant defense mechanisms in murine epidermis and dermis and their responses to ultraviolet light. Journal of Investigative Dermatology, 100(3), 260–265.
Sigurdsson, M. I., Jamshidi, N., Steingrimsson, E., Thiele, I., & Palsson, B. Ø (2010). A detailed genome-wide reconstruction of mouse metabolism based on human Recon 1. BMC Systems Biology, 4(1), 1.
Takagi, Y., Nakagawa, H., Kondo, H., Takema, Y., & Imokawa, G. (2004). Decreased levels of covalently bound ceramide are associated with ultraviolet B-induced perturbation of the skin barrier. Journal of investigative dermatology, 123(6), 1102–1109.
Wettenhall, J. M., & Smyth, G. K. (2004). limmaGUI: A graphical user interface for linear modeling of microarray data. Bioinformatics (Oxford, England), 20(18), 3705–3706.
Yahagi, S., Koike, M., Okano, Y., & Masaki, H. (2011). Lysophospholipids improve skin moisturization by modulating of calcium-dependent cell differentiation pathway. International Journal of Cosmetic Science, 33(3), 251–256.
Zelezniak, A., Pers, T. H., Soares, S., Patti, M. E., & Patil, K. R. (2010). Metabolic network topology reveals transcriptional regulatory signatures of type 2 diabetes. PLoS Computational Biology, 6(4), e1000729.
Acknowledgements
This study was supported by the Bio & Medical Technology Development Program of the National Research Foundation funded by the Korean government, MSIP (2016M3A9A5923160); and the Korea Food Research Institute (E0164503-01).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The all authors declare that they have no conflict of interest.
Ethical Approval
All procedures performed in studies involving animals were in accordance with the ethical standards of the institution or practice at which the studies were conducted.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jung, E.S., Park, H.M., Hyun, S.M. et al. Integrative metabolomic analysis reveals diet supplementation with green tea alleviates UVB-damaged mouse skin correlated with ascorbate metabolism and urea cycle. Metabolomics 13, 82 (2017). https://doi.org/10.1007/s11306-017-1218-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-017-1218-7