Abstract
Introduction
Leishmaniasis is a parasitic neglected disease affecting millions of people worldwide. Clinical practice resorts to long and costly treatments with a therapeutic arsenal limited to highly toxic drugs, often associated to adverse side effects. Additionally, resistant strains are reported to be increasing.
Aim
In this work, the mechanistic action of a drug candidate (methydehydrodieugenol B), isolated from twigs of Nectandra leucantha, towards Leishmania infantum was studied by a global metabolomics approach using GC-MS and RPLC-MS platforms.
Method
L. infantum promastigotes were grown in culture medium for 72 h and treated with methydehydrodieugenol B at 58.18 μg.mL-1 concentration; after 48 h treatment, enzyme activity was quenched, cells washed and frozen until analysis. For GC-MS analysis (Fiehn’s method), 1:1 methanol:water extracts were prepared and derivatized with O-methoxyamine in pyridine at room temperature for 90 min, followed by silylation with BSTFA/1% TMCS at 40 °C for 30 min. Pure methanolic extracts were also prepared and analyzed directly by RPLC-MS with a acetonitrile/water mobile phase acidulated with formic acid and gradient elution.
Result
Several amino acids, fatty acids, carbohydrates, and glycerolipids were found as discriminant metabolites, mostly decreased in treated samples. Due to the complexity of the parasite metabolism and the great diversity of altered metabolites, a multi-target mechanism was assigned to the drug candidate, where changes in the cell energy sources and in the lipid composition of the parasite plasma membrane were prominent.
Conclusion
These results contributed to elucidate the broad action of methyldehydrodieugenol B against Leishmania, paving the way in the search of novel alternative therapies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Leishmaniasis is one of the seventeen prioritized neglected diseases listed by the World Health Organization (WHO) (WHO 2016; Canuto et al. 2015). It is a poverty-associated infection, commonly found in tropical and subtropical countries, caused by protozoan parasites from different Leishmania species that multiply inside macrophages and dendritic cells of mammals. Transmitted by sand flies of the genus Phlebotomus and Lutzomyia during blood feeding, the parasites cause three main clinical manifestations: the visceral form (VL), which affects the host internal organs such as liver, spleen, and bone marrow, being fatal if untreated, the cutaneous form (CL), the most common form of the disease, associated with severe skin lesions, and the mucocutaneous form (MCL), the least common form, which affects mucous membranes, such as nose, mouth, etc., causing disfiguring ulcers (WHO 2016; Von Stebut 2015). Updated numbers of Leishmaniasis infection, revised in February 2015 by WHO, indicates 12 million cases worldwide with 0.7 to 1.3 million new cases of CL and 0.2 to 0.4 million new cases of VL each year, resulting in 20,000 to 30,000 deaths worldwide annually (WHO 2016).
The treatment consists of the oral administration and/or topical application of a limited therapeutic arsenal comprised of pentavalent antimonials, amphotericin B (amphoB), pentamidine, miltefosine (MT), and drug combinations. Besides the high cost and toxicity of such treatments, associated with severe adverse effects (Alvar et al. 2006), problems with parasite resistance caused by long periods of administration have resulted in losses of drug effectiveness (Singh 2006). Therefore, there is an urgent demand for alternative lines of therapy and the discovery of novel, less toxic, and more effective drugs continues to progress as a multifaceted area of investigation (Pécoul 2004; Singh et al. 2012).
Two important research strategies for the development of new drugs are based on computational modeling and the screening of active compounds in natural products. Computational modeling enables the development of new drugs by searching alternative (target) compounds according to prior information of mechanism of action and physicochemical properties, based on structure-activity relationships (Persico et al. 2016), as it can be seen in the work of Singh and Mandlik (2015). Screening of active compounds in natural products is another attractive and promising way for drug design, once a large number of molecules isolated from insects, marine invertebrates, amphibians and especially plants as well as synthetic analogs have demonstrated antimicrobial, antifungal, antiviral, anti-inflammatory and antiparasitic activities (Daly 2004; de Oliveira et al. 2015; El-Shitany et al. 2015; Rodrigues et al. 2016). Literature reviews have demonstrated the elevated potential of plants to provide potent metabolites with antileishmanial activity (Rocha et al. 2005; Schmidt et al. 2012a, b). Such compilations open a myriad of possibilities regarding parasite treatments. However, in the search of new biochemical death routes in parasites, additional systematic studies to understand the mechanism of action of bioactive metabolites are mandatory, and can contribute to the selection of new lead compounds.
Metabolomics, the study of metabolites produced or modified by a perturbation in an organism (either genetic, environmental, diet-related or drug-induced, among others) (Fiehn 2002), has become a promising tool to address such needs with great speed and efficacy (Ellis et al. 2007). Two approaches can be envisioned in such studies: untargeted metabolomics (scan of all detectable metabolites in intra- and/or extra-cellular matrices) and targeted metabolomics (quantification of preselected metabolites) (Villas-Bôas et al. 2005). Mass Spectrometry (MS) and Nuclear Magnetic Resonance (NMR) are the most commonly used analytical platforms in metabolomics. In general, MS is coupled with a separation technique, such as Gas Chromatography (GC), Liquid Chromatography (LC), or Capillary Electrophoresis (CE) (Kuehnbaum and Britz-McKibbin 2013). The complex nature of biological samples, comprising metabolites from a variety of chemical classes with distinct physicochemical properties (polarity, solubility, volatility, etc.) and varying abundances, imposes a high demand on the analytical technique performance. It is well accepted that no single platform is able to scan the entire metabolome, therefore multiplatform assemblies are usually preferred, and their use has become a common practice, especially in untargeted metabolomics studies. Since a distinct selectivity is provided by each analytical technique, the complementarity of information acquired from the use of multiplatform approaches is readily recognized, resulting in improved metabolic coverage (Putri et al. 2013).
Metabolomics applied to neglected diseases, including Leishmaniasis, have been revised periodically (Canuto et al. 2015; Paget et al. 2013; Barret et al. 2010; Creek et al. 2012; Scheltema et al. 2010; Watson 2010). In these reviews, the majority of studies focused on biomarker discovery for diagnostic purposes, and on the search of alternative therapies, although temporal progression of infection and the contrasting of behavior and infectivity of different parasites strains at metabolic level have also been addressed. The efficacy of current treatments and novel therapies as well as drug resistance under the perspective of metabolomics continue to be a niche of intense investigation as reported by selected reviews (Vincent and Barrett 2015; Berg et al. 2013; Creek and Barrett 2014). Such studies can provide important insights into the mechanism by which a given drug works against parasites, since they reveal neat changes at the cellular level, and no prior knowledge of drug action is cogitated at this stage.
The mechanism of drug action and resistance of Leishmania parasites towards several drugs is an emergent issue and have been studied using untargeted metabolomics approaches (t’Kind et al. 2010; Canuto et al. 2012, 2014; Berg et al. 2013, 2015; Vincent et al. 2014; Rojo et al. 2015; Shaw et al. 2015). In this context, information acquired from analytical multiplatforms (GC-MS, LC-MS and CE-MS) has been used to unveil drug action and resistance of MT/L. donovani (Canuto et al. 2014) and trivalent antimonial/L. infantum systems (Rojo et al. 2015); the latter has also been investigated by CE-MS single platform using sheathless interfacing (Canuto et al. 2012). In these reports the main observations were a multi-target metabolic alteration affecting mostly the redox system and changes in the cytoplasmic membrane composition. t´Kind et al. and Berg et al. further addressed L. donovani resistance towards antimonials and found changes in the transport of metabolites due to modifications of the cytoplasmic membrane (t’Kind et al. 2010; Berg et al. 2013). Alterations in lipid metabolism and induction of reactive oxygen species (ROS) were observed in another work, by interrogation of a group of L. infantum resistant parasites to antimonials and a group of resistant parasites to MT, compared to controls samples (Vincent et al. 2014). Drug combinations (antimonials, MT, amphoB and paramomycin) (Berg et al. 2015) and multi-resistance studies (antimonials and MT) (Shaw et al. 2015) were also evaluated in L. donovani, leading to similar metabolic perturbations.
The metabolism of Leishmania parasites is very complex and yet not completely elucidated. The overall results on drug action and resistance of Leishmania parasites towards specific drugs and combinations discussed so far converged to a few common conclusions. These findings include alterations in the bioenergetics metabolism, and redox system, resulting in protection against oxidative stress to sustain cell survival, as well as changes in the lipid metabolism imparting alterations in the cytoplasmic membrane fluidity, which affects drug permeability and uptake.
In order to explore new avenues for Leishmaniasis treatment issues such as drug action and resistance, efficacy, toxicity, and cost, the development of new therapies are clearly on demand. In this context, the selection of substances with established anti-parasitic activity, isolated from natural products, seems to be an excellent starting point to drug discovery. In this work, new insights into the mechanistic action of a bioactive neolignan, methyldehydrodieugenol B, isolated from the plant Nectandra leucantha, against L. infantum promastigotes were derived from an untargeted metabolomics approach using GC-MS and RPLC-MS (reversed-phase liquid chromatography-mass spectrometry). Leishmania infantum has been selected here as model organism because it is the only etiologic agent of visceral leishmaniasis in Brazil and also in the Mediterranean region. It causes a fatal disease if untreated and there is a great interest to find novel drugs against this species in Brazil and Europe. To our knowledge, no such studies have been compiled in the literature so far.
2 Materials and methods
2.1 Chemicals
M-199 medium, N,O-bis(trimethylsilyl)trifluoroacetamide (BSTFA) plus 1% (v/v) trimethylchlorosilane (TMCS), O-methoxyamine hydrochloride, pyridine (silylation grade), dimethyl sulfoxide (DMSO), acetonitrile (MS grade), heptane, formic acid (FA, LC-MS grade), and C13 methyl ester were purchased from Sigma-Aldrich (Germany). Other chemicals include calf serum (Cultilab, Brazil), hemin (Himedia, Brazil), methanol (LC-MS grade, JT Baker, Mexico), n-hexane (Labsynth Ltda, Brazil), ethyl acetate (Labsynth Ltda, Brazil), sodium chloride (Synth, Brazil), sodium dihydrogen phosphate (Merck, Germany), and potassium monohydrogen phosphate (QM, Brazil). Water was purified by a deionization system (Milli-Q, Millipore, USA).
2.2 Isolation of methyldehydrodieugenol B
The neolignan methyldehydrodieugenol B was isolated from twigs of Nectandra leucantha (da Costa-Silva et al. 2015). Briefly, 320 g of air-dried twigs were exhaustively extracted in n-hexane. After concentration under reduced pressure, part of the crude extract (9 g) was percolated through a SiO2 chromatography column and eluted with increasing amounts of EtOAc in n-hexane and MeOH in EtOAc to afford 12 fractions. Fraction III (40 mg) exhibited stronger antiparasitic activity and was purified by preparative TLC in SiO2 column using 7:3 (v/v) n-hexane:EtOAc as eluent to afford 25 mg of methyldehydrodieugenol B. The structure of this compound was elucidated based on NMR and MS data analysis (spectral information is presented in the Supplementary Material S1).
2.3 Leishmania parasites: growth and collection
Leishmania (L.) infantum (MHOM/BR/1972/LD) promastigotes were cultured in 1000 mL sterile flasks containing M-199 medium supplemented with 10% (v/v) calf serum, 0.25% (v/v) hemin, and 5% human male urine (Singh et al. 2000) and kept at 26 °C under slow rotation in a shaker (New Brunswick Scientific, USA). Cell maintenance was carried out twice a week by transferring a small aliquot (5%) of the current cell culture into a fresh medium. NB2 biosafety level is needed to work with promastigotes of Leishmania.
For the metabolomics studies, six independent flasks (three for control and three for treated samples) were prepared in consecutive weeks to generate three biological replicates for each studied group at log phase (48 h after cell transfer). The detailed procedure is described as follows: 4 × 107 promastigotes/mL were grown in M-199 supplemented medium (described above) for approximately 72 h, reaching 8 × 107 promastigotes/mL. The promastigotes were transferred to new flasks with fresh culture medium and incubated either with 4.70 μL methyldehydrodieugenol B in DMSO (stock solution is 12.4 mg mL−1) reaching the final concentration of 58.18 μg mL−1, which corresponds to the IC50 in promastigotes (Grecco et al. 2016, submitted)—named treated samples, TRT—or with the same volume (4.70 µL) of DMSO in the absence of drug—named control samples, CTR. The percentage DMSO in the culture media was kept at 0.47%; above 0.5% cell integrity may be compromised. Based on the IC50 determination protocol (da Costa-Silva et al. 2015) 48 h incubation was used for treatment. After the incubation period, control and treated samples were immediately quenched in dry ice and the culture medium was removed by centrifugation (1000 g for 10 min at 4 °C). Cells were washed twice with cold PBS buffer to eliminate extracellular content, and after cell counting in a Neubauer chamber, cells were divided into several Eppendorf tubes containing 1 × 108 promastigotes each. The supernatant was removed by suction. The cell pellets were frozen at −80 °C for about 3 months until analysis. The biomass was determined by the pellet weight.
At the end, for the metabolomics studies, sixty sample pellets generated from three independent biological replicates, resulting in 10 samples per culture flask (control and treated), were used in each analytical multiplatform: 15 CTR and 15 TRT for GC-MS and 15 CTR and 15 TRT for RPLC-MS.
2.4 Intracellular metabolite extraction
For GC-MS analysis, pellets were extracted by the addition of 500 µL cold 1:1 methanol:water solution and the cells were lysed by an ultrasound probe for 1 min at 30% (OMNI Ultrasonic Ruptor 400, USA), followed by centrifugation at 10,000 g for 10 min (Excelsa 3, Fanen Ltda, Brazil). The supernatants were removed and 150 µL were used as individual samples; 200 µL were also collected and mixed with all other samples to obtain the pooled quality control sample (QC). The QC was homogenized and divided into 150 µL portions. A blank solution was prepared by using the solvent extractor at the same extraction conditions. Samples, blank solution and QC portions were evaporated to dryness in a SpeedVac (Thermo Scientific SPD1010, USA), operated at 35 °C, maintained at −20 °C overnight, and submitted to the derivatization protocol described below prior to chromatographic analysis.
For RPLC-MS analysis, pellets were extracted with 500 µL cold pure methanol and the cells lysed by the ultrasound probe (1 min at 30%). After centrifugation (10,000 g for 10 min), 200 µL of the supernatant were collected and pooled to prepare the QC sample. A blank solution was prepared using the solvent extractor at the same extraction conditions. The remaining supernatants (150 µL from each sample), QC and blank solution were directly analyzed by RPLC-MS. All prepared samples were maintained at -20 °C overnight prior to chromatographic analysis.
2.5 GC-MS derivatization
In the methoximation step, 10 µL O-methoxyamine (15 mg mL−1) in pyridine were added to the dried samples, sonicated for 10 s (Quimis, Brazil) and vortexed for 20 s (Quimis, Brazil). Samples were maintained in the dark for 90 min at room temperature. In the silylation step, 10 µL BSTFA plus 1% (v/v) TMCS were added to the samples; the mixtures were then homogenized and placed into a thermostatic bath (ECO SILVER RE 1225, Lauda, Brazil) for 30 min at 40 °C. The derivatized samples were ressuspended in 100 µL heptane containing 10 mg mL−1 C13 methyl ester (internal standard), homogenized and analyzed.
2.6 GC-MS equipment
GC-MS analyses were performed in a gas chromatography system (model 7890 A, Agilent Technologies, CA, USA) coupled to a single quadrupole mass spectrometer (model 5975 C inert XL, Agilent Technologies). The separation method was adapted from Kind et al. 2009 with minor modifications. A HP-5ms column (30 m length, 0.25 mm i.d., with a 0.25 µm film composed of 95% dimethyl and 5% diphenyl polysiloxane, from Agilent Technologies) was used. Helium at 1.0 mL/min flow rate was used as carrier gas. The GC injector was maintained at 250 °C and samples were injected with 1:10 split, using He at 10 mL/min. The temperature gradient was as follows: initial oven temperature of 60 °C maintained for 1 min, increased to 300 °C at 10 ºC/min rate, followed by cooling to 60 °C and maintained for 5 min. The run time was 25 min. The temperatures of the detector transfer line, filament source, and the quadrupole were maintained at 290, 230 and 150 °C, respectively. The electron ionization source used −70 eV energy. The MS was operated in scan mode in the range of 50–600 m/z. The software Mass Hunter B.07.01 (Agilent Technologies) was used for instrument operation and data acquisition.
2.7 RPLC-MS equipment
RPLC-MS analyses were performed using a binary ultra-fast liquid chromatography system (Prominence, Shimadzu Co., Japan) coupled to an ion trap-time-of-flight mass spectrometer (Shimadzu Co., Japan) via electrospray ionization (ESI). The separation method was adapted from Ciborowski et al. 2010 and Canuto et al. 2014, with minor modifications. A C18 column (2.1 mm × 5 cm, 0.5 µm, Discovery, Supelco) maintained at 40 °C was used. The mobile phase was: (A) deionized water acidulated with 0.1% (v/v) FA and (B) acetonitrile acidulated with 0.1% (v/v) FA. The gradient elution was: 25 to 100% B in 20 min, 100% B for 2 min, 100 to 25% B in 2 min and 25% B for 5 min for column conditioning. Flow rate was constant at 0.2 mL/min. The MS was operated in the positive mode ionization with 3.5 kV at the ESI interface, 1.66 kV as the detector voltage, and 200 °C dry gas. Spectra were acquired from m/z 80 to 1000. The software LabSolutions (LCMS solution 3.60.361 version, Shimadzu Co., Japan) was used for instrument auto tuning (error < 3 ppm; resolution better than 10,000 at m/z 1000), operation, and data acquisition.
2.8 Data treatment and multivariate analysis
GC-MS raw data files were acquired in centroid mode and converted into *.mzData (using Qualitative Analysis Mass Hunter software B05.00, Agilent Technologies) and RPLC-MS raw data files were acquired in profile mode and converted into *mzXML (using LabSolutions software, Shimadzu Co.) for further processing by publically available XCMS software package (version 1.24.1) (Smith et al. 2006) running at R platform (3.1.0, R Foundation for Statistical Computing) (R Core Team).
For GC-MS data processing, signals were extracted from 6 to 25 min, initial data were not examined due to solvent peak interference. XCMS parameters were: matched filter method, fwhm (full width at half maximum of model peak) = 4, snthresh (signal-to-noise cutoff) = 1.5, max (maximum number of groups to identify in a single m/z slice) = 30, bw (bandwidth) = 2, and mzwid (width of overlapping m/z slices) = 0.25. Other parameters were kept at default values. For RPLC-MS data processing, signals were extracted from 5 to 22 min, excluding features in dead volume and post-column conditioning. XCMS parameters were: centWave method, snthresh = 2.0, bw = 2, mzwid = 0.01, minfrac (minimum fraction of samples necessary in one group to be a valid group) = 0.25, and default parameters for obiwarp method in retcor, used in the alignment of profile MS data, in which the algorithm calculates the retention time deviation for each sample centralizing in one of them. For GC and LC data processing, after second grouping, fillPeaks was used to integrate areas of missing values, and median fold change normalization was performed (in-house developed algorithm) (Veselkov et al. 2011), followed by biomass normalization (pellet weight of each sample, and for QCs, the average weight was used).
Multivariate analyses, PCA (Principal Component Analysis) and PLS-DA (Partial Least Squares Discriminant Analysis), performed in SIMCA P+ software (12.0.1 version, Umetrics, CA, USA), were used to indicate the differences between the studied groups. VIP scores from PLS-DA were used to select the significant discriminant entities. Univariate analysis of the selected VIPs were performed as follows: test of normality (Lilliefors test) and Mann–Whitney U test, using Statistica software, and false discovery rate (FDR) using BH procedure (Benjamini and Hochberg 1995), calculated in R platform.
For GC-MS data, metabolites identification was performed using Fiehn RTL Library (FiehnLib) (Kind et al. 2009) and NIST MS, 2.0 g library (National Institute of Standards and Technology Mass Spectra Library) after peak deconvolution in AMDIS 2.69 (Automated Mass Spectral Deconvolution and Identification System) software. Peaks were assigned with basis on retention time and mass spectra fragmentation pattern, after retention index and retention time analyses. To confirm peak assignment, match factor (NIST) and NET factor (AMDIS) were greater than 700 and 85, respectively. In addition, identification of a few metabolites was confirmed by the analysis of authentic analytical standards used previously in the optimization of the GC-MS derivatization procedure.
For RPLC-MS, statistical significant mass-to-charge ratios (m/z) were searched in public databases: Human Metabolome Database, HMDB (http://www.hmdb.ca/), Kyoto Encyclopedia of Genes and Genome, KEGG (http://www.genome.jp/kegg/) and Metlin (metlin.scripps.edu) using [M + H]+, [M + 2H]2+ and [M + Na]+ as possible adducts and 5 ppm as maximum error. Putative metabolites described in Leishmania’s metabolic pathways were selected.
3 Results and discussion
Drug research in the area of neglected diseases has been intensified in recent years employing different strategies, with metabolomics based research occupying an important niche (Barrett et al. 2010). In order to gain a deeper understanding of the mechanism involved in the action of plant metabolites against Leishmania, an untargeted multiplatform metabolomics approach has been proposed in this work. A bioactive neolignan identified as methyldehydrodieugenol B (Fig. 1), isolated from Nectandra leucantha, has been shown to display anti-promastigotes and anti-amastigotes activities towards L. donovani (da Costa-Silva et al. 2015) and also L. infantum (Grecco et al. 2016, submitted). For the latter, the compound under investigation exhibited a low IC50 (58.18 μg mL−1 in promastigotes) and a selectivity index above 10, showing it has a considerable potential as candidate active principle in future developments of pharmaceutical formulations against Leishmaniasis. In addition, the compound can be isolated from the plant in large amounts and it was therefore selected here in conjunction with L. infantum promastigotes for the metabolomics study. Although it is essential that the potential activity of anti-Leishmania drugs be verified in amastigotes, it has been described in the literature that the interpretation of the mechanistic action of methyldehydrodieugenol B using macrophage-infected cells can be misleading, since the drug can interfere with the immune response of the macrophage to the parasite or the amount of NO produced (da Costa-Silva et al. 2015). Furthermore, as pointed out by Vincent and Barret (2015), it is acceptable to work with promastigotes in exploratory screening studies.
Aiming the detection of the largest possible amount of metabolites, a systematic sample preparation strategy was studied, including: (i) growth stages for Leishmania parasites before drug incubation (48, 72 or 96 h), (ii) number of parasite washes with PBS (1 to 6 times), and (iii) number of parasites per sample (1, 2, 4, 6, 8, and 10 × 107 promastigotes/mL). Optimized conditions at this stage include: cells were grown in fresh culture medium until 72 h before drug incubation (occurred in the next 48 h). After removal of culture medium, cells were washed twice with cold PBS, to eliminate extracellular content. Sample pellets were aliquoted in separate eppendorfs at 1 × 108 promastigotes/mL and frozen until analyses.
To establish the best conditions for intracellular metabolites extraction, eleven mixtures (ternary and binary mixtures, and pure solvents) were tested as follows: (A) 100% MeOH, (B) 100% H2O, (C) H2O:CHCl3 (1:9, v/v), (D) MeOH:CHCl3 (1:9, v/v), (E) MeOH:H2O (1:1, v/v), (F) H2O:CHCl3 (1:1, v/v), (G) MeOH:CHCl3 (1:1, v/v), (H) MeOH:H2O:CHCl3 (6:3:1, v/v/v), (I) MeOH:H2O:CHCl3 (2:4:4, v/v/v), (J) MeOH:H2O:CHCl3 (6:1:3, v/v/v) and (K) MeOH:H2O:CHCl3 (4:3:3, v/v/v), and evaluated according to the number of molecular features and/or metabolites extracted from the cells (GC-MS generated 29 identified metabolites and ca. 2000 molecular features; RPLC-MS generated 77 identified metabolites and ca. 1300 molecular features). Optimal conditions for metabolite extraction were: 500 µL 1:1 MeOH:H2O for GC-MS and pure MeOH for RPLC-MS were added to the sample pellet containing 1 × 108 promastigotes; cells were lysed using a ultrasound probe for 1 min at 30% power.
Optimization of GC-MS derivatization steps included varying the volume of O-methoxyamine in pyridine (10 or 30 µL), time of reaction (90 or 120 min), and reaction temperature (37 or 60 °C) for the methoxymation step, and the volume of BSTFA + TMCS (10 or 30 µL), time of reaction (30 or 60 min), and reaction temperature (40 or 70 °C) for the silylation step. These studies were conducted with authentic standards (24 metabolites involved in Leishmania metabolism). Many authors have used analogous conditions for derivatization (García and Barbas 2011): either room temperature for 16 h or shorter periods with higher temperature for the methoxymation step and high temperatures (70 °C) in short periods (1 h) for the silylation step. Our results showed that methoxymation can be completed in a much shorter time at room temperature (90 min) and silylation is performed at shorter periods (30 min) with lower temperatures (40 °C) as compared to stated literature conditions. Detailed conditions of the derivatization procedure employed here are listed in the experimental section. At optimal conditions no losses of the selected metabolites due to evaporation or degradation were observed.
In the metabolomics studies, sixty sample pellets generated from three independent biological replicates were used in each group (15 CTR and 15 TRT) for GC-MS and (15 CTR and 15 TRT) for RPLC-MS. Both GC-MS and RPLC-MS methods employed in this work were adapted from literature. GC-MS analyses followed Fiehn’s method (Kind et al. 2009) and RPLC-MS followed Barba’s method (Ciborowski et al. 2010 and Canuto et al. 2014), with minor modifications. After analysis, the resulting mass chromatograms were inspected and two control samples were discarded as outliers due to inconsistent chromatographic profile.
Data generated from both GC-MS and RPLC-MS experiments were treated using the XCMS package (Smith et al. 2006). For GC-MS data, XCMS parameters were optimized according to the minimum missing values and number of identified metabolites by varying fwhm from 2 to 6, snthresh at 0.5, 1.0, 1.5, 2.0 and 3.0, max from 10 to 50, bw at 1, 2, 3, 5 and 10, and mzwid at 0.1, 0.25 and 0.50. For RPLC-MS data, XCMS parameters were optimized by variation of snthresh at 2.0, 2.5 and 5.0, bw at 2, 5 and 10, mzwid at 0.01 and 0.05, and minfracat 0.25, 0.50 and 0.75. Optimal conditions for GC-MS and RPLC-MS are shown in the experimental section.
Multivariate analyses were performed using the SIMCA P+ software (Umetrics). PCA models were used to verify instrumental performance during the analytical sequence by inspection of Quality Control (QC) samples. QCs, pool of equal volumes of all samples under consideration in this study, were clustered together in the PCA plot (Fig. 2), attesting stable instrumental conditions during data acquisition. In both datasets, GC-MS and RPLC-MS, coefficients of variation (CV) of the intensities of selected m/z’s were calculated for the QC samples, showing that more than 75% of the molecular features and identified metabolites presented CV < 30%. PLS-DA models were further built with QC prediction, to confirm QC low dispersion: 96% prediction capability for GC-MS and 99% for RPLC-MS were obtained.
PCA models (Pareto scaling) built for L. infantum samples treatment with methyldehydrodieugenol B. a GC-MS analysis quality parameters: explained variance R2 = 0.913 and predicted variance Q2 = 0.804, and b RPLC-MS analysis quality parameters: explained variance R2 = 0.540 and predicted variance Q2 = 0.459. Datasets were normalized by median and biomass. Red dots are QC, quality control samples, black squares are CRT, control samples, and blue diamonds are TRT, treated samples
After QC evaluation, CTR and TRT samples were re-processed in XCMS, using optimal parameters. In order to select discriminant metabolites (GC-MS) and/or molecular features (RPLC-MS), new PLS-DA models (samples only) were built (Fig. 3) and validated by leave-group-out and 100 permutations procedures. Analysis of Variance (ANOVA) shows high significance for both models, with F = 160.01 and p value = 1.97 × 10−16, for GC-MS and F = 456.88 and p value = 1.51 × 10−21, for RPLC-MS. The criterion used in the selection of significant molecular features was based on the variable importance projection (VIP) scores larger than 1.
PLS-DA models (Pareto scaling) built for L. infantum samples treatment with methyldehydrodieugenol B. a GC-MS analysis quality parameters: explained variance R2 = 0.971, predicted variance Q2 = 0.963, ANOVA F = 160.01 and p value = 1.97 × 10−16, and b RPLC-MS analysis quality parameters: explained variance R2 = 0.991, predicted variance Q2 = 0.998, ANOVA F = 456.88 and p value = 1.51 × 10−21. Datasets were normalized by median and biomass. Black squares are CRT, control samples, and blue diamonds are TRT, treated samples
The statistical significance was also evaluated by univariate analysis. Lillifoers procedure attested the non-normality of the data, therefore Mann–Whitney method was used to calculate p-values. The FDR analysis (BH procedure), on both datasets considering all extracted molecular features, estimates a false positive rate of 8% within the GC-MS dataset and 3% within the LC-MS dataset (both at 95% confidence level).
For the GC-MS dataset, metabolites were readily identified, since it relies on build-in libraries (Fiehn and NIST); for the RPLC-MS dataset, the putative metabolites were compiled as m/z values for further identification in public accessible databases (HMDB, KEGG and Metlin).
Table 1 shows the significant metabolites found by GC-MS and RPLC-MS, their respective chemical classes and fold changes. Complementary information, such as monoisotopic mass, molecular formula, retention time, identification parameters, univariate parameters results, and %CV of intensities obtained from the QC samples, is available in Table S2.1 (Supplementary Material S2). According to the Metabolomics Standards Initiative (MSI) metabolites identifications in this work correspond to level 1 for GC-MS and level 2 (putatively annotated compounds) for RPLC-MS (Salek et al. 2013).
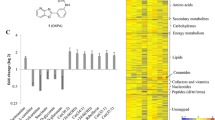
Twenty metabolites identified by GC-MS and twenty-eight putative metabolites detected by RPLC-MS were altered when TRT and CTR samples were contrasted. The altered metabolites show high significance, with some compounds detected only in treated samples, which suggests that either they appear only after drug treatment or that their intensities in control samples were below the signal-to-noise cutoff of 2.0 (RPLC-MS), as set by the centWave method used during XCMS data processing). To highlight the alterations, presented as fold changes in Table 1, the Supplementary Material S3 provides bar charts with respective errors of the identified and putatively identified significant metabolites. Large errors can be associated to intra-flask variation of the biological replicates. The metabolites of Table 1 represent twelve classes of metabolites, wherein the four largest groups are amino acids, peptides and analogues (17%), fatty acids and conjugates (17%), carbohydrates and conjugates (13%), and glycerolipids, glycerophospholipids and derivatives (13%). No common metabolite was found by both techniques, showing the complementarity of the analytical platforms used in this work. In addition, the observed discriminant metabolites covered a wide range of polarities, from polar amino acids and sugars to strongly non-polar glycerolipids and fatty acids.
Further inspection of Table 1 allows inferences on the behavior of the tested methyldehydrodieugenol B against Leishmania at the metabolic level. From this compilation, it may be suggested that this neolignan acts as a multi-target compound, affecting different central metabolic pathways localized in distinct compartments of promastigotes. Carbohydrate, amino acids, polyamine and lipid metabolism are affected, although at different degrees.
Reduction in the available energy and carbon sources in neolignan-treated promastigotes (Table 1; Fig. 4) is suggested by the strong decrease in the amount of metabolites from the degradation of carbohydrates (such as glucose-6-phosphate, fumarate, malate, pentoses) and in the amount of glucogenic amino acids (such as alanine, valine or phenylalanine) in treated promastigotes. Additionally, a slight increase in the amount of disaccharides, most likely sucrose, and pentose alcohols was also detected. The increment of sucrose, internalized by an H+ symport system (Singh and Mandal 2011), and 3-ketosucrose can be attributed to a lower activity of the internal sucrase of Leishmania (Singh and Mandal 2016), contributing to the reduction of fructose and glucose available. Increase in pentose alcohols (ribitol/arabitol/xylitol; indistinguishable by GC-MS) may be a consequence of the inhibition of ribose-5-phosphate isomerase or transaldolase (Fig. 4b) and it is associated to disorders of sugar metabolism in mammals (van der Knaap et al. 1999; Verhoeven et al. 2001; Huch et al. 2004; Stone et al. 2014). Alternatively, inhibition of NADPH-linked pentitol dehydrogenases, which convert polyols to keto sugars for further incorporation in the pentose phosphate or glycolytic pathways, could explain the increment, as described for other organisms (Fig. 4b). However, no specific polyoldehydrogenases were described in Leishmania, although the uptake of sugar alcohols and a wide variety of other substances was previously reported (Callahan and Beverley 1992; Uzcategui et al. 2008). In the context of low carbon source in neolignan-treated promastigotes, it should be noted that glycerol-3-phosphate is also accumulated. Glycerol can be taken from the medium by the parasite and enter in the gluconeogenesis pathway after its conversion into glycerol-3-phosphate by glycerol kinase (Murray et al. 2005; Rodríguez-Contreras 2014). Impairment of the gluconeogenesis can explain the accumulation of glycerol-3-phosphate, as well as the increment of fructose 1,6 bisphosphate (Fig. 4b). Likewise, lipid metabolism can explain this increment, including the synthesis of triacylglycerol, since glycerol-3-phosphate is the substrate of acyltransferase, the first key enzyme of the synthesis in Leishmania (Zufferey and Mamoun 2005). Of note, the amount of triacylglycerol increases in drug-treated promastigotes (Table 1), pointing out to the decrease of the lipase activity, and consequently, in fatty acid ß-oxidation. Although fatty acid ß-oxidation is almost irrelevant in dividing parasites, it is more important in non-dividing promastigotes (Creek et al. 2012). However, the fold change of triacylglycerol in drug-treated parasites could not be estimated, since it was not detected in the untreated promastigotes.
Representation of a Arginine and Glutathione metabolism and b Glycolysis and Gluconeogenesis pathways for the metabolomics evaluation of the methyldehydrodieugenol B treatment in L. infantum. Red metabolites are increased and blue metabolites are decreased in treated samples. Glc—glucose, PI—phosphatidylinositol, Fru—fructose, P—phosphate, S-adoMet—S-adenosyl methionine, AcCoA—acetyl coenzime A, OH-Pro—hydroxyproline, Ala—alanine, DHAP—dihydroxyacetone phosphate, GAP—glyceraldehyde 3-phosphate, and TCA cycle—tricarboxylic acid cycle. Highlighted enzymes: (ALDO) fructose-bisphosphatealdolase, (araB) ribulokinase, (ARD1) arabinitol dehydrogenase, (ARD) arabinitol 2-dehydrogenase, (da1D) arabinitol 4-dehydrogenase, (FBPase) fructose 1,6-biphosphatase, (GK) glycerol 3-kinase, (GPI) glucose 6-phosphate isomerase, (PKFK) phosphofructokinase, (rbtD) ribitol 2-dehydrogenase, (RPI) ribose 5-phosphate isomerase, (RPE) ribulose 5-phosphate epimerase, (SRM) spermidine synthase, (XYLB) xylulokinase, and (6PGDH) 6-phosphogluconate dehydrogenase. Keto and aldopentoses (xylulose/ribulose/ribose), and pentoses alcohols (arabitol/ribitol) are indistinguishable by the analytical methodology
The lipid composition of the membranes is likewise altered in 48 h-time neolignan-treated promastigotes. Leishmania is able to scavenger fatty acids and lipids from the medium, as well as synthesized them, except for cholesterol and sphingomyelin, which have to be taken from the medium (Zhang and Beverley 2010; Ramakishnan et al. 2013). Nevertheless, phospholipid and glycolipid synthesis are altered in treated promastigotes, as suggested by the general reduction of carbon and energy sources, as described above, and by the low amount of building block elements, such as myo-inositol or unsaturated long chain fatty acids. Myo-inositol is synthesized by Leishmania from glucose-6-phosphate (Fig. 4b) or, mediated by a myo-inositol/ H+transporter, taken up from the medium and employed in the synthesis of phosphatidylinositol, glycosylphosphatidyl inositol (GPI)-anchored molecules or lipophosphoglycan (LPG), abundant molecule localized at the cell surface of Leishmania (Zhang and Beverley 2010). A prominent 35 fold increment of acyl-lysophosphatidylcholine (LysoPC 14:1) and decrease of other LysoPC (18:3, 20:1, 20:3) as well as the decrease of free unsaturated (mono and polyunsaturated) long chain fatty acids observed in drug-treated promastigotes are most likely due to the inhibition of the endoplasmatic reticulum-based elongase and desaturase pathways described in Leishmania (Lee et al. 2007; Creek et al. 2012; Ramakrishnan et al. 2013). This can affect the acyl composition of the membrane lipids, as well as other components of the cell membranes, such as glycosylinositolphospholipids (GIPLS) or GPI-anchors. The increment of lysophosphatidylcholine may also have an unknown function in treated parasites. In the case of the intracellular amastigotes, lysophosphatidylcholine was described to facilitate the proliferation of the parasites by neutralizing oxidative and nitrosative stresses (Tounsi et al. 2015). A significant increment of sphingomyelin (d18:1/22:1) and its metabolic derivative, D-erythro-sphingosylphosphorylcholine, (12 and 77%, respectively) was observed in drug-treated promastigotes. Despite the absence of sphingomielin synthesis, a potent sphigomyelinase activity, which is dependent on inositol phosphosphingolipid phospholipase C-like protein, is present in promastigotes. Products of the degradation of sphingomyelin mediated many signaling pathways, including Ca2+ mobilization, cell growth, apoptosis, stimulation of prostaglandin 12 production. In Leishmania, it was associated with proliferation of amastigotes in mammalian host. The role of D-erythro-sphingosylphosphorylcholine in methyldehydrodieugenol B treated-promastigotes is unknown.
Overall, changes in the composition of the biological membranes, including the decrease in the amount of cholesterol measured in drug-treated promastigotes, will affect the flexibility and fluidity of the membrane, which may modify membrane function, besides disturbing signaling pathways dependent on phospholipids and sphingolipids in the plasma membrane.
Finally, polyamine synthesis in promastigotes was also affected by methyldehydrodieugenol B. Polyamines play vital roles in a variety of cellular processes, such as growth and differentiation, making the four enzymes involved in the polyamine synthesis good targets for drug development. Accumulation of putrescine (1.4 fold change) suggests the reduction on the activity of spermine synthase, which catalyzes the synthesis of spermidine from putrescine and S-adenosyl methionine (Fig. 4a). Putrescine and spermidine can be taken from the medium by the putrescine-spermidine transporter, as described for L. major (Hasne and Ullman 2005), and although spermidine is considered essential for growth and survival in all trypanosomatids, putrescine is also critical for cellular proliferation, as shown recently (Boitz et al. 2017). Of note, spermine synthase is in the main route of trypanothione synthesis and its inhibition affects the parasite protection against oxidative/nitrosative stress (Barret 1997; Balaña-Force et al. 2012). Also, a slight decrease in the amount of cadaverine (0.8 fold change), which is metabolized to homotrypanothione, an analogue of trypanothione, was found in drug-treated parasites. The data suggests that synthesis of polyamine and trypanothione does not seen strongly affected by methyldehydrodieugenol B treatment.
For an overall analysis of metabolite pathways, the Integrated Molecular Pathway Level Analysis (IMPaLA) was employed (Kamburov et al. 2011). By using the pathway over-representation analysis feature of IMPaLA, it was found that biosynthesis of unsaturated fatty acids, arginine and proline metabolism, as well as alpha linolenic acid and linoleic acid metabolism were the most significant altered pathways, with p-values of 3.22 × 10−6, 1.02 × 10−5, and 6.23 × 10−5, respectively. Those pathways integrate the data obtained by GC-MS and RPLC-MS analysis conjunctly, in which for the fatty acids metabolism (oleic acid, linolenic acid, linoleic acid, 13-docosenoic acid, and eicosadienoic acid) are decreased in treated parasites; in arginine and proline metabolism it was observed an accumulation of putrescine and S-adenosylhomocysteine, and depletion of trans-4-hydroxy-l-proline, aminobutyric acid, and fumaric acid; and for linolenic and linoleic acid pathways, the three identified metabolites (linolenic acid, linoleic acid, tetracosahexaenoic acid) were decreased in treated cells. All results generated by IMPaLA are compiled in better detail in the Supplementary Material S4 and corroborate with the findings already discussed.
4 Conclusions
The altered metabolites indicate a multi-target effect of the neolignan methyldehydrodieugenol B in promastigotes from L. infantum. Since large pathways of the metabolism were affected, the determination of the primary targets of the studied compound is a difficult task. Neolignan-treatment altered drastically main metabolic routes in Leishmania, as pointed out by the drastic reduction of sugars, amino acids and intermediates of lipid metabolism. Polyamine synthesis is less affected. As a result, carbon and energy sources, response to oxidative/nitrosative stressor synthesis of important lipid components of membranes may be compromised. Although accumulation of metabolites suggests the inhibition of specific enzymes in main routes, the modifications may have affected almost all intermediate metabolism in Leishmania. In addition, inhibition of metabolites or lipids uptake cannot be ruled out by the presented data. Nevertheless, all these findings contributed to improve the understanding of the broad action of methyldehydrodieugenol B against Leishmania.
References
Alvar, J., Croft, S., & Olliaro, P. (2006). Chemotherapy in the treatment and control of Leishmaniasis. Advances in Parasitology, 61, 223–274.
Balaña-Fouce, R., Calvo-Álvarez, E., Álvarez-Velilla, R., Prada, C. F., Pérez-Pertejo, Y., & Reguera, R. M. (2012). Role of trypanosomatid’s arginase in polyamine biosynthesis and pathogenesis. Molecular and Biochemical Parasitology, 181(2), 85–93.
Barret, M. P. (1997). The pentose phosphate pathway and parasitic protozoa. Parasitology Today, 13(1), 11–16.
Barrett, M. P., Bakker, B. M., & Breitling, R. (2010). Metabolomic systems biology of trypanosomes. Parasitology, 137(9), 1285–1290.
Benjamini, Y., & Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of Royal Statistical Society. Series B, 57(1), 289–300.
Berg, M., Mannaert, A., Vanaerschot, M., Vander Auwera, G., & Dujardin, J. C. (2013). (Post-) Genomic approaches to tackle drug resistance in Leishmania. Parasitology, 140(12), 1492–1505.
Berg, M., García-Hernández, R., Cuypers, B., Vanaerschot, M., Manzano, J. I., Poveda, J. A., Ferragut, J. A., Castanys, S., Dujardin, J. C., & Gamarro, F. (2015). Experimental resistance to drug combinations in Leishmania donovani: metabolic and phenotypic adaptations. Antimicrobial Agents and Chemotherapy, 59(4), 2242–2255.
Berg, M., Vanaerschot, M., Jankevics, A., Cuypers, B., Maes, I., Mukherje, S., Khanal, B., Rijal, S., Roy, S., Opperdoes, F., Breitling, R., & Dujardin, J. C. (2013). Metabolic adaptations of Leishmania donovani in relation to differentiation, drug resistance, and drug pressure. Molecular Microbiology, 90(2), 428–442.
Boitz, J. M., Gilroy, C. A., Olenyik, T. D., Paradis, D., Perdeh, J., Dearman, K., Davis, M. J., Yates, P. A., Li, Y., Riscoe, M. K., Ullman, B., & Roberts, S. C. (2017). Arginase is essential for survival of Leishmania donovani promastigotes but not intracellular amastigotes. Infection and Immunity. doi:10.1128/IAI.00554-16.
Callahan, H. L., & Beverley, S. M. (1992). A member of the aldoketo reductase family confers methotrexate resistance in Leishmania. The Journal of Biological Chemistry, 267(34), 24165–24168.
Canuto, G. A. B., Castilho-Martins, E. A., Tavares, M. F. M., López-Gonzálvez, A., Rivas, L., & Barbas, C. (2012). CE-ESI-MS metabolic fingerprinting of Leishmania resistance to antimony treatment. Electrophoresis, 33(12), 1901–1910.
Canuto, G. A. B., Castilho-Martins, E. A., Tavares, M. F. M., Rivas, L., Barbas, C., & López-Gonzálvez, A. (2014). Multi-analytical platform metabolomic approach to study miltefosine mechanism of action and resistance in Leishmania. Analytical and Bioanalytical Chemistry, 406(14), 3459–3476.
Canuto, G. A. B., da Cruz, P. L. R., Faccio, A. T., Klassen, A., & Tavares, M. F. M. (2015). Neglected diseases prioritized in Brazil under the perspective of metabolomics. A review. Electrophoresis, 36(18), 2336–2347.
Ciborowski, M., Rupérez, F. J., Martínez-Alcázar, M. P., Angulo, S., Radziwon, P., Olszanski, R., Kloczko, J., & Barbas, C. (2010). Metabolomic approach with LC-MS reveals significant effect of pressure on diver’s plasma. Journal of Proteome Research, 9(8), 4131–4137.
Creek, D., Anderson, J., McConville, M. J., & Barret, M. P. (2012). Metabolomic analysis of trypanosomatid protozoa. Molecular and Biochemical Parasitology, 181(2), 73–84.
Creek, D. J., & Barrett, M. P. (2014). Determination of antiprotozoal drug mechanisms by metabolomics approaches. Parasitology, 141(1), 83–92.
da Costa-Silva, T. A., Grecco, S. S., de Sousa, F. S., Lago, J. H. G., Martins, E. G. A., Terrazas, C. A., Varikuti, S., Owens, K. L., Beverley, S. M., Satoskar, A. R., & Tempone, A. G. (2015). Immunomodulatory and antileishmanial activity of phenylpropanoid dimers isolated from Nectandra leucantha. Journal of Natural Products, 78(4), 653–657.
Daly, J. W. (2004). Marine toxins and nonmarine toxins: convergence or symbiotic organisms? Journal of Natural Products, 67(8), 1211–1215.
de Oliveira, D. R., Tintino, S. R., Braga, M. F., Boligon, A. A., Athayde, M. L., Coutinho, H. D., de Menezes, I. R., & Fachinetto, R. (2015). In vitro antimicrobial and modulatory activity of the natural products silymarin and silibinin. Biomed Research International. doi:10.1155/2015/292797.
Ellis, D. I., Dunn, W. B., Grifin, J. L., Allwood, J. W., & Goodacre, R. (2007). Metabolic fingerprinting as a diagnostic tool. Pharmacogenomics, 8(9), 1243–1266.
El-Shitany, N. A., Shaala, L. A., Abbas, A. T., Abdel-Dayem, U. A., Azhar, E. I., Ali, S. S., van Soest, R. W., & Youssef, D. T. (2015). Evaluation of the anti-inflammatory, antioxidant and immunomodulatory effects of the organic extract of the red sea marine sponge Xestospongia testudinaria against carrageenan induced rat paw inflammation. PLoS One, 10(9), e0138917.
Fiehn, O. (2002). Metabolomics—the link between genotypes and phenotypes. Plant Molecular Biology, 48(1–2), 155–171.
García, A., & Barbas, C. (2011). Gas chromatography-mass spectrometry (GC-MS)-based metabolomics. In T. O. Metz (Ed.), Metabolic Profiling, Methods in Molecular Biology (pp. 191–204). New York: Springer.
Grecco, S. S., Sousa, F. S., Jerz, G., Jones, P. G., da Costa-Silva, T. A., Tempone, A. G., Martins, E. G. A., & Lago, J. H. G. (2016). Antileishmanial and antitrypanosomal neolignans from leaves of Nectandra leucantha Ness and Mart (Lauraceae). Planta Medica, submitted.
Hasne, M. P., & Ullman, B. (2005). Identification and Characterization of a Polyamine Permease from the Protozoan Parasite Leishmania major. The Journal of Biological Chemistry, 280(15), 15188–15194.
Huch, J. H., Roos, B., Jakobs, C., van der Knaap, M. S., & Verhoeven, N. M. (2004). Evaluation of pentitol metabolism in mammalian tissues provides new insight into disorders of human sugar metabolism. Molecular Genetics and Metabolism, 82(3), 231–237.
Kamburov, A., Cavill, R., Ebbels, T. M., Herwig, R., & Keun, H. C. (2011). Integrated pathway-level analysis of transcriptomics and metabolomics data with IMPaLA. Bioinformatics, 27(20), 2917–2918.
Kind, T., Wohlgemuth, G., Lee, D. Y., Lu, Y., Palazoglu, M., Shahbaz, S., & Fiehn, O. (2009). FiehnLib: Mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Analytical Chemistry, 81(24), 10038–10048.
Kuehnbaum, N. L., & Britz-McKibbin, P. (2013). New advances in separation science for metabolomics: resolving chemical diversity in a post-genomic era. Chemical Reviews, 133(4), 2437–2468.
Lee, S. H., Stephens, J. L., & Englund, P. T. (2007). A fatty-acid synthesis mechanism specialized for parasitism. Nature Reviews Microbiology, 5(4), 287–297.
Murray, H. W., Berman, J. D., Davies, C. R., & Saravia, N. G. (2005). Advances in leishmaniasis. Lancet, 366(9496), 1561–1577.
Paget, T., Haroune, N., Bagchi, S., & Jarroll, E. (2013). Metabolomics and protozoan parasites. Acta Parasitology, 58(2), 127–131.
Pécoul, B. (2004). New drugs for neglected diseases: from pipeline to patients. PLoS Medicine, 1(1), e6.
Persico, M., Di Dato, A., Orteca, N., Cimino, P., Novellino, E., Fattorusso, C (2016). Use of integrated computational approaches in the search for new therapeutic agents. Molecular Informatics, 35(8–9), 309–325.
Putri, S. P., Yamamoto, S., Tsugawa, H., & Fukusaki, E. (2013). Current metabolomics: Technological advances. Journal of Bioscience and Bioengineering, 116(1), 9–16.
R Core Team. (2016). R: a language and environmental for statistical computing. Vienna: R Foundation for Statistical Computing. http://www.R-project.org/.
Ramakrishnan, S., Serricchio, M., Striepen, B., & Bütikofer, P. (2013). Lipid synthesis in protozoan parasites: a comparison between kinetoplastids and apicomplexans. Progress Lipid Research, 52(4), 488–512.
Rocha, L. G., Almeida, J. R. G. S., Macêdo, R. O., & Barbosa-Filho, J. M. (2005). A review of natural products with antileishmanial activity. Phytomedicine, 12(6–7), 514–535.
Rodrigues, L. C., Barbosa-Filho, J. M., de Oliveira, M. R., Néris, P. L. N., Borges, F.V.P., & Mioso, R. (2016). Synthesis and antileishmanial activity of natural dehydrodieugenol and its mono and dimethyl ethers. Chemistry & Biodiversity, 13(7), 870–874.
Rodriguez-Contreras, D., & Hamilton, N. (2014). Gluconeogenesis in Leishmania mexicana: Contribution of glycerol kinase, phosphoenolpyruvate carboxykinase, and pyruvate phosphate dikinase. The Journal of Biological Chemistry, 289(47), 32989–33000.
Rojo, D., Canuto, G. A. B., Castilho-Martins, E. A., Tavares, M. F. M., Barbas, C., López-Gonzálvez, Á., & Rivas, Luis (2015). A multiplatform metabolomics approach to the basis of antimonial action and resistance in Leishmania infantum. PLoS ONE, 10(7), e0130675.
Salek, R. M., Steinbeck, C., Viant, M. R., Goodracre, R., & Dunn, W. B. (2013). The role of reporting standards for metabolite annotation and identification in metabolomic studies. Gigascience, 2(1), doi:10.1186/2047-217X-2-13.
Scheltema, R. A., Decuypere, S., t’Kindt, R., Dujardin, J. C., Coombs, G. H., & Breitling, R. (2010). The potential of metabolomics for Leishmania research in the post-genomics era. Parasitology, 137(9), 1291–1302.
Schmidt, T. J., Khalid, S. A., Romanha, A. J., Alves, T. M., Biavatti, M. W., Brun, R., da Costa, F. B., de Castro, S. L., Ferreira, V. F., de Lacerda, M. V., Lago, J. H., Leon, L. L., Lopes, N. P., das Neves Amorim, R. C., Niehues M., Ogungbe, I. V., Pohlit, A. M., Scotti, M. T., Setzer, W. N., de N C Soeiro, M., Steindel, M., & Tempone, A. G. (2012a). The potential of secondary metabolites from plants as drugs or leads against protozoan neglected diseases—part I. Current Medicinal Chemistry, 19(14), 2128–2175.
Schmidt, T. J., Khalid, S. A., Romanha, A. J., Alves, T. M., Biavatti, M. W., Brun, R., da Costa, F. B., de Castro, S. L., Ferreira, V. F., de Lacerda, M. V., Lago, J. H., Leon, L. L., Lopes, N. P., das Neves Amorim, R. C., Niehues M., Ogungbe, I. V., Pohlit, A. M., Scotti, M. T., Setzer, W. N., de N C Soeiro, M., Steindel, M., & Tempone, A. G. (2012b). The potential of secondary metabolites from plants as drugs or leads against protozoan neglected diseases—part II. Current Medicinal Chemistry, 19(14), 2176–2228.
Shaw, C. D., Lonchamp, J., Downing, T., Imamura, H., Freeman, T. M., Cotton, J. A., Sanders, M., Blackburn, G., Dujardin, J. C., Rijal, S., Khanal, B., Illingworth, C. J. R., Coombs, G. H., & Carter, K. C. (2015). In vitro selection of miltefosine in promastigotes of Leishmania donovani from Nepal: genomic and metabolomics characterization. Molecular Microbiology, 99(6), 1134–1148.
Singh, S., & Mandlik, V. (2015). Structure based investigation on the binding interaction of transport proteins in leishmaniasis: insights from molecular simulation. Molecular Biosystems, 11(5), 1251–1259.
Singh, A., & Mandal, D. (2011). A novel sucrose/H+ symport system and an intracellular sucrase in Leishmania donovani. International Journal of Parasitology, 41(8), 817–826.
Singh, A., & Mandal, D. (2016). Purification and characterization of a novel intracellular sucrase enzyme of Leishmania donovani promastigotes. Biochemistry Research International. doi:10.1155/2016/7108261.
Singh, N. (2006). Drug resistance mechanisms in clinical isolates of Leishmania donovani. The Indian Journal of Medicine Research, 123(3), 411–422.
Singh, N., Kumar, M., & Singh, R. K. (2012). Leishmaniasis: Current status of available drugs and new potential drug targets. Asian Pacific Journal of Tropical Medicine, 5(6), 485–497.
Singh, S., Mohapatra, D. P., & Sivakumar, R. (2000). Successful replacement of fetal calf serum with human urine for in vitro culture of Leishmania donovani. The Journal of Communicable Diseases, 32(4), 289–294.
Smith, C. A., Want, E. J., Tong, G. C., Abagyan, R., & Siuzdak, G. (2006). XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Analytical Chemistry, 78(3), 779–787.
Stone, V., Kudo, K. Y., August, P. M., Marcelino, T. B., & Matté, C. (2014). Polyols accumulated in ribose-5-phosphate isomerase deficiency increase mitochondrial superoxide production and improve antioxidant defenses in rats’ prefrontal cortex. International Journal of Developmental Neuroscience, 37(34), 21–25.
t’Kind, R., Scheltema, R. A., Jankevics, A., Brunker, K., Rijal, S., Dujardin, J. C., Breitling, R., Watson, D. G., Coombs, G. H., & Decuypere, S. (2010). Metabolomics to unveil and understand phenotypic diversity between pathogen populations. PLoS Neglected Tropical Diseases, 4(11), e904.
Tounsi, N., Meghari, S., Moser, M., & Djerdjouri, B. (2015). Lysophosphatidylcholine exacerbates Leishmania major-dendritic cell infection through interleukin-10 and a burst in arginase1 and indoleamine 2,3-dioxygenase activities. International Immunopharmacology, 25(1), 1–9.
Uzcategui, N. L., Zhou, Y., Figarella, K., Mukhopadhyay, R., & Bhattacharjee, H. (2008). Alteration in glycerol and metalloid permeability by a single mutation in the extracellular C-loop of Leishmania major aquaglyceroporin LmAQP1. Molecular Microbiology, 70(6), 1477–1486.
van der Knaap, M. S., Wevers, R. A., Struys, E. A., Verhoeven, N. M., Pouwels, P. J., Engelke, U. F., Feikema, W., Valk, J., & Jakobs, C. (1999). Leukoencephalopathy associated with a disturbance in the metabolism of polyols. Annals of Neurology, 46(6), 925–928.
Verhoeven, N. M., Huck, J. H., Roos, B., Struys, E. A., Salomons, G. S., Douwes, A. C., van der Knaap, M. S., & Jakobs, C. (2001). Transaldolase deficiency: liver cirrhosis associated with a new inborn error in the pentose phosphate pathway. American Journal of Human Genetics, 68(5), 1086–1092.
Veselkov, K. A., Vingara, L. K., Masson, P., Robinette, S. L., Want, E., Li, J. V., Barton, R. H., Boursier-Neyret, C., Walther, B., Ebbels, T. M., Pelczer, I., Holmes, E., Lindon, J. C., & Nicholson, J. K. (2011). Optimizing preprocessing of ultra-performance liquid chromatography/mass spectrometry urinary metabolic profiles for improved information discovery. Analytical Chemistry, 83(15), 5864–5872.
Villas-Bôas, S. G., Mas, S., Akesson, M., Smedsgaard, J., & Nielsen, J. (2005). Mass spectrometry in metabolome analysis. Mass Spectrometry Reviews, 24(5), 613–646.
Vincent, I. M., & Barrett, M. P. (2015). Metabolomic-based strategies for anti-parasite drug discovery. Journal of Biomolecular Screening, 20(1), 44–55.
Vincent, I. M., Weidt, S., Rivas, L., Burgess, K., Smith, T. K., & Ouellette, M. (2014). Untargeted metabolomic analysis of miltefosine action in Leishmania infantum reveals changes to the internal lipid metabolism. International Journal for Parasitology, 4(1), 20–27.
Von Stebut, E. (2015). Leishmaniasis. Journal of the German Society of Dermatology, 13(3), 191–201.
Watson, D. G. (2010). The potential of mass spectrometry for the global profiling of parasite metabolomes. Parasitology, 137(9), 1409–1423.
WHO. (2016). World Health Organization, Leishmaniasis. Accessed February, from http://www.who.int/mediacentre/factsheets/fs375/en/.
Zhang, K., & Beverly, S. M. (2010). Phospholipid and sphingolipid metabolism in Leishmania. Molecular and Biochemical Parasitology, 170(2), 55–64.
Zufferey, R., & Mamoun, C. B. (2005). The initial step of glycerolipid metabolism in Leishmania major promastigotes involves a single glycerol-3-phosphate acyltransferase enzyme important for the synthesis of triacylglycerol but not essential for virulence. Molecular Microbiology, 56(3), 800–810.
Acknowledgements
G.A.B.C. is thankful to Sao Paulo Research Foundation (FAPESP 2012/04601-6) for a doctoral fellowship. A.G.T., M.J.M.A., and M.F.M.T. acknowledge FAPESP for financial support (Grants 2012/18756-1, 2012/07361-6, 2014/25494-9, and 2015/50075-2).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest regarding this work.
Research involving human participants and/or animals
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Canuto, G.A.B., Dörr, F., Lago, J.H.G. et al. New insights into the mechanistic action of methyldehydrodieugenol B towards Leishmania (L.) infantum via a multiplatform based untargeted metabolomics approach. Metabolomics 13, 56 (2017). https://doi.org/10.1007/s11306-017-1193-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-017-1193-z