Abstract
In sweet cherry, as in most non-climacteric species, abscisic acid (ABA) plays a major role in the control of fruit ripening and color development. Although the ABA treatment of sweet cherry fruits has been reported to upregulate anthocyanin pathway-related genes or ABA pathway-related genes, the temporality of molecular and physiological events occurring during color development and the ABA control of these events during the color initiation are lacking in this species. In this work, we analyzed variations in the Index of Absorbance Difference (IAD), a maturity index, and total anthocyanins along with changes in transcript abundance of ABA and anthocyanin pathway-related genes, from light green to red fruit stages. PavNCED1 and ABA signaling pathway-related genes upregulated when fruits transitioned from light green to pink stage, whereas anthocyanin pathway-related transcripts increased from pink to the red stage, together with increases in the anthocyanin content and IAD, suggesting sequentiality in molecular and physiological events during color development. Additionally, ABA applied at color initiation in planta advanced IAD, increased anthocyanin content, and yielded darker fruits at harvest. These changes were accompanied by changes in the transcript accumulation of ABA and anthocyanin pathway-related genes. This in planta treatment of sweet cherry fruits with ABA confirms that ABA is a central player in the control of color initiation in sweet cherries, associated with the transcript accumulation of genes involved in ABA homeostasis and signaling, which is followed by the up-regulation of anthocyanin pathway-related genes and color development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Maturity timing is an important parameter for fruit production and genetic breeding, tightly associated with ripening processes (Hardner et al. 2019), including pink color development, which can be affected by environmental factors (Guo et al. 2018). Ripening is a complex process that combines physiological and molecular events, including cellular modifications occurring in the fruits, adjustments in the source-sink balance, and changes in the fruit hormone content (Coombe 1976). For physiological changes to occur coordinately with the embryo development, the ripening process must be highly regulated to start at a precise moment. Ripening is associated with fruit growth resumption, chlorophyll metabolization, fruit softening, accumulation of secondary metabolites, color change, sugar content increase, and acidity decline (Giovannoni 2001; McAtee et al. 2013). Several hormones are involved in triggering ripening in non-climacteric fruits (McAtee et al. 2013; Cherian et al. 2014). Among these hormones, abscisic acid (ABA) seems to have a key role in promoting ripening since this phytohormone increases when softening begins and pink color initiates; moreover, ABA treatment influences the ripening-related parameters such as soluble solids and anthocyanin content (Kondo and Inoue 1997; Wheeler et al. 2009; Jia et al. 2011; Sutthiwal 2012; Kuhn et al. 2014). Other hormones possibly participate through complex networks, with many of them interacting with the ABA pathway (Zhang et al. 2009; Böttcher et al. 2010; Guo et al. 2018; Kuhn et al. 2020). Therefore, studying ABA’s role during fruit ripening may provide a better understanding of the regulation of this process.
ABA is involved in controlling the timing and intensity of several plant responses (e.g., stomatal closure, transduction of abiotic signals, and seed dormancy), where it can delay or advance the occurrence of these processes (Umezawa et al. 2010). For instance, cyp707a2-1 and cyp707a2-2 mutants presenting increased ABA content exhibited delayed seed germination (Kushiro et al. 2004). In the context of fruit ripening, ABA accelerates the fruit color development in grape (Vitis vinifera L.; Wheeler et al. 2009). In sweet cherry fruits, RNA interference (RNAi) lines with downregulated expression of PavCYP707A2 (coding for a putative ABA 8′-hydroxylase, possibly involved in ABA catabolism) had an increase in the transcript abundance of some anthocyanin pathway-related genes and this was correlated with increased ABA content at harvest (Li et al. 2015). However, in this study the direct effect of ABA on the color initiation timing was not assessed, nor the physiological changes occurring concomitantly with the advanced transcript accumulation of anthocyanin pathway-related genes (only harvest evaluation; see Table 1).
During non-climacteric fruit ripening, ABA exerts its action through a dynamic balance in its production, degradation, transport, and the ability of the fruit cells to perceive and transduce ABA signals (Li et al. 2011; Leng et al. 2014). In sweet cherry, ABA levels increase at color initiation, coinciding with fruit growth activation, sugar and anthocyanin content increase, and the reduction of firmness (Kondo and Gemma 1993; Ren et al. 2011; Luo et al. 2014; Teribia et al. 2016). ABA treatments affect several ripening-related parameters, such as sugar content, firmness, fruit size, and acidity (Luo et al. 2014). Regarding color development, exogenously applied ABA increases fruit color and anthocyanin concentration (Ren et al. 2011; Luo et al. 2014; Shen et al. 2014). This is significantly associated with the upregulation of chalcone synthase (PavCHS), chalcone isomerase (PavCHI), dihydroflavonol 4-reductase (PavDFR), flavanone 3β-hydroxylase (PavF3H), anthocyanidin synthase (PavANS), and UDP-glucose:flavonoid 3-O-glucosyltransferase (PavUFGT) genes, possibly involved in anthocyanin biosynthesis in sweet cherry (Liu et al. 2013; Shen et al. 2014).
The relevance of ABA in color development was further confirmed in sweet cherry RNAi lines with downregulated expression of PavNCED1, coding for a putative 9-cis-epoxy-carotenoid dioxygenase, which had lower ABA content, resulting in less colored fruits, with reduced anthocyanin concentration (Shen et al. 2014). In contrast, PavCYP707A2 interference lines, with increased ABA levels, had more anthocyanins in the fruit than control lines (Li et al. 2015). Regarding ABA perception and signaling pathways, putative pyrabactin resistance-like (PavPYL) ABA receptors, subfamily 2 of SNF1-related kinases (PavSnRK2s), and type 2C protein phosphatases (PavPP2Cs) genes change their transcript abundance during sweet cherry fruit ripening, and some of them are ABA-responsive (Wang et al. 2015; Shen et al. 2017), but their expression during this process has not been associated with color development yet, and ABA pathway gene profiling has been correlated with fruit size variations (Wang et al. 2015). Together, this evidence shows that some aspects of ripening including color initiation regulation by ABA have been elucidated in sweet cherry fruits; however, they have not been integrated within a single study. Furthermore, most of these studies analyzed the effect of ABA treatment performed ex planta, in which the shoots or fruits were removed from the tree and treated with ABA in controlled conditions. Table 1 summarizes the research published on sweet cherry fruit development and ripening, including the quantification of physiological parameters, the ABA treatment, and the gene expression profiling of ABA and anthocyanin pathway genes reported to date in this species.
The timing of fruit maturity is an important trait for sweet cherry producers (Hardner et al. 2019). As the ability of ABA to control not only color intensity but also its timing has not been well established in in planta experiments in this species, our study aims to shed light on the temporality of physiological and molecular events associated with color initiation and the role of ABA in its timing regulation in sweet cherry fruits in planta (with the fruits on the trees under natural conditions). To describe this sequence of events, we first focused on maturity index IAD variations and anthocyanin accumulation in relationship to transcript abundance changes associated with color development. Then, we monitored the effect the in planta of ABA treatment at the transcript and physiological levels. To our knowledge, this is the first attempt to unravel the temporality of color development, ABA and anthocyanin gene expression and to address the role that ABA plays in its regulation in sweet cherry in a single study.
Material and methods
Plant material
Fruits of Prunus avium L. cv. Lapins from adult trees grown in a commercial orchard located at Rengo, Chile (Long: O 70° 43′ 6.78″, Lat: S 34° 27′ 16.92″), were used. Experiments and quantifications were performed during the 2017–2018 season. Full flowering (50% open flowers in the trees) was set at zero time 0 DAFB (days after full bloom), as in previous phenological studies performed in sweet cherry (Chavoshi et al. 2014; Fadón et al. 2015). Phenology was conducted on these trees from the green stage until harvest (Table S4). Four trees were used for IAD curve and transcript profiling during development (Table S2), where LG is 35 DAFB, P1 is 56 DAFB, P2 is 60 DAFB, and R is 67 DAFB, and four other trees were used for anthocyanin concentration estimation during development (Table S3), where LG is 40 DAFB, P2 is 61 DAFB, and R is 67 DAFB.
Physiological evaluations
For the growth curve, 80 fruits from four trees (20 fruits per tree) were each measured every 2–5 days. Equatorial diameter from the suture was measured with a caliper. For the IAD curve, 80 fruits from four control trees (20 fruits per tree) and 80 fruits from four ABA-treated trees (20 fruits per tree) were measured every 2–5 days. For this, both sides of the fruits were measured using VIS/NIF device Cherry Meter (T.R. ® Turoni, Italy), and a value from 0 to 2.5 was obtained as an average of both cheeks. IAD and width were non-destructive assessments performed during fruit development.
For physiological assessment at harvest, fruits were collected at two harvest dates, each of them one day before the commercial harvests. The first and second harvests were at 69 and 74 DAFB, respectively. At both harvest dates, color distribution using CTIFL scale, fruit IAD, soluble solid content, acidity, firmness, width, and weight were quantified (Chavoshi et al. 2014; Fig S1). For the first harvest, 25 fruits were randomly collected from n = 4 trees; whereas for the second harvest, 15 fruits were randomly collected from the same trees. For color distribution, CTIFL sweet cherry color chart (CTIFL, France) was used to allocate the 25 fruits in one to four color categories (1 the lightest; 4 the darkest). For width and IAD curves, fruits were measured, as described above. For weight, a portable mini-scale was used. For firmness, a durometer (Durofel T.R. ® Turoni, Italy) was pressed on both cheeks of the fruits, and a value from 1 to 100% was obtained for each fruit as an average of both cheeks. For soluble solid content and acidity, the five most homogenous fruits (in color and size) were selected a measured using a PAL-BX|ACID Pocket Sugar and Acidity Meter (ATAGO USA, Inc.), which measures degrees Brix and acidity as total malic acid content. The effect of ABA is presented as a percentage change (%) and was calculated using:
where “par” is the value for the parameters measured.
ABA treatment
For the ABA treatment, eight trees were randomly selected, four control trees, and four ABA trees, and their fruits were used for IAD and growth curves and for qPCR and ABA analysis (sampling in Table S2). ABA (S-abscisic acid, ProTone® SL, Valent) was applied in a final concentration of 400 ppm to four trees with a 10-L hand sprayer when fruits were starting to color (56 DAFB; November 16, 2017; 10% pink-blush straw yellow fruits and 90% straw yellow fruits in the tree). Control trees were untreated. Fruits of ABA-treated and control trees were measured in the tree or collected and frozen in liquid nitrogen, then stored at − 80°C until further analyses. Fruits were also collected at harvest for ripening parameter assessments (Fig. S1a).
Anthocyanin extraction and quantification
Anthocyanins were measured during development and also in control and ABA-treated fruits at the 1st harvest. For total anthocyanin estimation, three replicates (n = 3 trees), as a pool of eight fruits each, were used. Endocarps and seeds were removed from the fruits in the field with a sharp blade, frozen in liquid nitrogen, and stored at – 80 °C for further analyses. A mechanical blender was used to grind the frozen tissue with liquid nitrogen. Following grinding, tissue was freeze-dried. Approx. 100 mg of samples was transferred into 15-mL tubes and mixed with 4 mL 80% methanol. Then, the samples were sonicated, mixed on an orbital shaker for 20 min, and left overnight in the dark at 4 °C. Next, the samples were centrifuged (10 min, 4000 rpm, 4 °C) and filtered using a 0.22-μm PFTE membrane. Samples were injected into a UHPLC-MS/MS Waters Acquity system (Milford, MA, USA). The separation of anthocyanins was performed using a Waters Acquity HSS T3 column 1.8 μm, 100 mm × 2.1 mm, at 40 °C. Anthocyanins were analyzed as described by Arapitsas et al. (2012). Finally, anthocyanins were detected using mass spectrometry on a Waters Xevo TQMS instrument equipped with an ESI source. Mass Lynx Target Lynx Application Manager was used for data processing. The sum of all anthocyanins detected was used as an estimation of total (cumulative) anthocyanins. Student’s T-test at p < 0.05 was used for establishing the significance of variation between control and ABA-treated fruits.
ABA quantification
ABA was measured in the control and ABA-treated samples collected four days after the treatment. For the extraction, 10 mg of mesocarp- and exocarp-enriched frozen and ground tissue was mixed with an 80% methanol-1% acetic acid solution, containing internal deuterated standard [2H6]ABA (OlChemim Ltd., Olomouc, Czech Republic) and shaken for 1 h at 4 °C. The extracted fraction was kept at − 20°C O.N. and then centrifuged, and the supernatant was dried and dissolved in a 1% acetic acid solution.
A reverse-phase column (Oasis HLB) was used for the separation of the compounds, according to Seo et al. (2011). The dried eluate was dissolved in 5% acetonitrile-1% acetic acid solution. Next, samples were injected into an autosampler and reverse-phase UHPLC chromatography columns, 2.6 μm Accucore RP-MS, 100 mm × 2.1 mm (Thermo Fisher Scientific, San Diego, CA, USA). For ABA separation, a gradient of acetonitrile (2–55%) containing 0.05% acetic acid, at a rate of 400 μL/min over 22 min, was used. Finally, ABA was detected in a Q-Exactive mass spectrometer (Orbitrap detector; ThermoFisher Scientific, San Diego, CA, USA). Targeted selected ion monitoring and electrospray ionization (ESI) in the negative mode were used for detection (Seo et al. 2011). For the ABA concentration determination, a calibration curve was generated using the Xcalibur 4.0 and TraceFinder 4.1 SP1 software. Student’s T-test at p-value < 0.05 was used for establishing the significance of variation between control and ABA-treated fruits.
RNA extraction and RT-qPCR analysis
For RT-qPCR analyses, three biological replicates (n = 3 trees), as a pool of eight fruits each, were used. Total RNA was extracted from sweet cherry fruit samples enriched in mesocarp-exocarp using a modified version of the CTAB method described by Meisel et al. (2005). Genomic DNA traces were eliminated by using DNase TURBO™ (Thermo Fisher Scientific, CA, USA) treatment, according to the manufacturer’s specifications. The reverse transcription reaction was performed on 1 μg of total RNA, using the First Strand cDNA synthesis System Kit (Thermo Fisher Scientific, Carlsbad, CA, USA), according to the manufacturer’s specifications. RNA purity was estimated using A260/230 and A260/A280 with values around 2.0 for each one. The RNA integrity was assessed using RNA electrophoresis for ribosomal band resolution, using MOPS buffers.
Relative quantifications of sweet cherry transcripts were determined by real-time PCR (RT-qPCR) using two technical replicates for each biological replicate. Primers were selected from Alkio et al. (2014), Li et al. (2015), and Wang et al. (2015). Additionally, a BLAST was performed against the sweet cherry genome and sequences from NCBI databases, to confirm that these primers align only to the genes of interest. The efficiency of these primers for a qRT-PCR reaction was determined by the LinRegPCR program (Ruijter et al. 2009). The list of the primers used in this study can be found in Supplementary Table S1.
For The RT-qPCR reaction was performed on the QIAGEN Rotor-Gene Q, using the Rotor-Gene Q Series software version 2.1.0. The reactions were carried out using the Fast Plus system EvaGreen® qPCR Master Mix (Biotum, CA, USA) according to the manufacturer’s specifications. RT-qPCR analyses were performed using the conditions recommended in “Minimum information established for qRT-PCR experiments” (MIQE, Bustin et al. 2009) and “Golden Rules of Quantitative PCR” (Udvardi et al. 2008). Primer efficiency was taken into account for relative transcript abundance calculations, as indicated by Pfaffl (2001). PavCAC, PavTEF2, and PavACT1 were used as reference genes (Alkio et al. 2014), and for the relative abundance calculations, PavCAC was used, presenting the smallest variation between samples (< 1.5 Cq).
Quantitative transcript abundance during development was set to 1.0 at 35 DAFB for ABA signaling pathway-related genes and at 57 DAFB for ABA homeostasis and anthocyanin pathway-related genes, while in the ABA treatment experiment, it was set to 1.0 in control.
For testing the significance of variations between different developmental stages (LG, P1, P2, R) of ABA signaling and anthocyanin pathway-related genes, multiple LSD-Fisher, using Bonferroni correction for nonparametric data, with p < 0.05, was used, and for the ABA homeostasis genes the effect of the ABA treatment, the Mann-Whitney test for nonparametric data with p-value < 0.05 was used (Cheung and Klotz 1997). All the graphs were made with the GraphPad software Prism version 6.0e. ± SEM of three replicates is graphed for each value.
Results
Color, anthocyanin content, and transcript abundance variations during sweet cherry fruit development
To understand the temporality associated with the onset of sweet cherry fruit ripening and color development, we focused on establishing the order of molecular and physiological events. Hence, in the first place, we characterized the fruit color variations. For this, a color scale from light green to dark red was obtained based on the fruit color phenology of Lapins variety, where P1 is the stage where fruits are mainly yellow with some of them having pink blushes (10% of the fruits in the tree), while P2 is the stage when 25% of the fruits in the tree have a pink blush (Fig. 1a). The maturity index IAD and the total anthocyanin content in the fruits were assessed at different stages of color development (Figs. 1a and 2a, b). We observed an increase in IAD and total anthocyanin content from the pink 2 stage (P2) to the red stage (R) (Figs. 1a and 2a, b). These results were complemented by assessing the relative transcript abundance of three sets of genes at different stages of color development: anthocyanin pathway-related genes PavCHS, PavF3H, and PavUFGT (Fig. 1b); ABA signaling pathway-related genes PavPP2C3, PavPP2C4, PavSnRK2.1, PavSnRK2.2, and PavSnRK2.3 (Fig. 1c); and ABA homeostasis-related genes PavNCED1 and PavCYP707A2 (Fig. 1d). We found a significantly higher relative transcript abundance in the anthocyanin pathway-related genes at the R stage than at the LG and P1 stages (Fig. 1b). We also observed a statistically higher relative transcript abundance of ABA signaling pathway-related genes at pink stages P1 and P2 in relation to the previous LG stage, and at the R stage compared with the P2 stage (Fig. 1c). Finally, sustained increase and decrease in the transcript levels of putative ABA homeostasis-related genes, PavNCED1 and PavCYP707A2, were observed, respectively, from the LG to the R stages (Fig. 1d). In the case of PavNCED1, there is a significantly higher relative transcript abundance at the P2 stage compared to the P1 and LG stages (Fig. 1d). Additionally, the transcript abundance of the putative orthologs of the ABA signaling pathway gene PavPYL2, the homeostasis gene PavCYP707A1, and the R2R3 transcription factor PavMYBA was analyzed during fruit development (Fig. S3). These analyses revealed that the PavMYBA and PavPYL2 transcript abundance increased linearly from the light green to pink stages of development, whereas PavCYP707A1 did not show a clear linear increase during development.
Anthocyanin and ABA-related gene expression variations during sweet cherry fruit development. a Changes in IAD and total anthocyanins on a fresh weight (FW) basis. b Transcript abundance relative to PavCAC of putative orthologs of anthocyanin pathway-related genes. c Transcript abundance relative to PavCAC of putative orthologs of ABA signaling pathway-related genes. d Transcript abundance relative to PavCAC of putative orthologs of ABA homeostasis-related genes. LG, P1, P2, and R are light green, pink 1, pink 2, and red stages, respectively. Sampling for qPCR and IAD in Table S2; sampling for total anthocyanin determination in Table S3. For total anthocyanin estimation and qPCR analyses, three replicates (n=3 trees), as a pool of eight fruits each, were used; for IAD, 20 fruits from four trees each (n = 4 trees) were measured. In a, data as mean ± SD. In b, c, and d, data as mean ± SEM. In c, data was set to 1.0 in LG. In b and d, data was set to 1.0 in P1. In b and c, multiple LSD-Fisher, with Bonferroni correction, p < 0.05 was used. In d, the Mann-Whitney test for nonparametric data with p-value < 0.05 was used. Different letters denote significant differences between groups in b and c, or between stages for a given gene (d). n.d., not detected. Color scale obtained from phenological data of Table S4 including representative pictures of each developmental stage
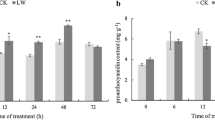
Effect of exogenous ABA on IAD, anthocyanins, and fruit color. a IAD changes during fruit ripening in control and ABA-treated fruits. b Total anthocyanins on a fresh weight (FW) basis at 1st harvest (69 DAFB; December 1, 2017). c Color distribution using CTIFL color chart at 1st harvest and 2nd harvest (74 DAFB; December 4, 2017). ABA was applied at the color initiation (arrow at 56 DAFB; November 16, 2017; P1). For a, 80 fruits from four trees (20 fruits per tree) were randomly selected. For b, pools of eight fruit per tree (n = 3 trees) were used. For c, 25 and 15 fruits per tree (n = 4 trees) were selected in the 1st and 2nd harvests, respectively; CTIFL scale from 1 to 4 was used (see Fig. S1a). In a and b, data as mean ± SD. Student’s T-test at p-value < 0.05 was used for comparing means at the indicated dates (a) or ABA versus control (b), where an asterisk denotes statistical differences between ABA-treated and control fruits
Effect of ABA on IAD, total anthocyanin, and color distribution
As changes in the transcript abundance of ABA pathway-related genes were observed (Fig. 1), and advanced transcript accumulation of some of these genes was found in PavCYP707A2 sweet cherry RNAi lines (Li et al. 2015), we evaluated whether ABA controls the timing of molecular events leading to color development. First, the effect of ABA on ripening parameters and in the fruit growth curve was analyzed. It was found that ABA modifies fruit width, fruit firmness, and fruit color, among others (Fig. S2). As shown in Fig. 2a, an advance in the IAD occurs a few days after ABA application in the field, implying a slope change between two time points in ABA-treated fruits while no difference was observed in control fruits. In this case, a significant IAD increase was observed between 67 and 69 DAFB in ABA-treated fruits (Fig. 2a).
Additionally, there are statistically significant differences in cumulative anthocyanin content in ABA-treated fruits at harvest (Fig. 2b). This is accompanied by a more intense red color distribution in ABA-treated fruits at both the 1st and 2nd harvests (Fig. 2c). These results show that ABA treatment advances maturity index IAD, which is associated with increased total anthocyanin content and darker fruits at harvest.
Effect of ABA on the transcript abundance of anthocyanin pathway-related genes and ABA pathway-related genes
To determine whether the advancement of IAD caused by ABA treatment was correlated with transcriptional changes, the same sets of genes (anthocyanin pathway-related and ABA pathway-related genes) showing sequentiality during sweet cherry fruit development (Fig. 1) were analyzed in response to exogenous application of ABA, at the P1 stage (see Table S2). The ABA treatment caused a statistically higher relative transcript abundance of anthocyanin pathway-related genes, PavF3H and PavCHS (Fig. 3a). PavUFGT did not change between ABA-treated and control fruits in our experimental conditions (Fig. 3a). In addition, significantly higher relative transcript abundance of ABA signaling pathway-related genes PavPP2C3, PavPP2C4, and PavSnRK2.1 was observed in response to ABA treatment (Fig. 3b), whereas the PavSnRK2.2 and PavSnRK2.1 transcripts did not change significantly. Finally, ABA homeostasis-related gene PavNCED1 significantly increased its transcript abundance in response to ABA, whereas PavCYP707A2 showed significantly lower transcript abundance in ABA-treated fruits compared to control fruits (Fig. 3c). Furthermore, endogenous ABA levels were higher in the ABA-treated fruits than in the control fruits (Fig. 3d). No statistically significant variation in the relative transcript abundance of PavPYL2, PavCYP707A1, and PavMYBA was detected in response to ABA treatment (Fig. S4).
Effect of ABA treatment on the ABA content and the transcript abundance relative to PavCAC of putative orthologs of ABA signaling, ABA homeostasis, and anthocyanin pathway at the onset of ripening. a Transcript abundance relative to PavCAC of putative orthologs of anthocyanin pathway-related genes. b Transcript abundance relative to PavCAC of putative orthologs of ABA signaling pathway-related genes. c Transcript abundance relative to PavCAC of putative orthologs of ABA homeostasis-related genes. d ABA content as dry weight (DW) in control and ABA-treated fruits. In a, b, and c, data are represented as mean ± SEM and set to 1.0 at the onset of ripening. In d, data is represented as mean ± DS. ABA was applied at the color initiation (56 DAFB; November 16, 2017; P1), and transcript abundance and the ABA content were evaluated 4 days later, at 60 DAFB (November 16, 2017; P2) in control and ABA-treated fruits. For qPCR analyses and ABA quantification, pools of eight fruits per tree (n = 3 trees) for control and ABA-treated trees were used. The Mann-Whitney test for nonparametric data with p-value < 0.05 was used for qPCR; ANOVA with post hoc Tukey test, p-value < 0.05 was used for ABA levels. An asterisk denotes statistical differences between ABA-treated and control fruits
Discussion
Different transcript profiling patterns underlie sweet cherry color initiation
Color initiation is related to anthocyanin accumulation in sweet cherry fruits. An increase in transcript abundance of structural genes from flavonoid and anthocyanin pathway (e.g., PavCHS, PavCHI, PavDFR, PavF3H, PavANS, and PavUFGT) has been correlated with anthocyanin accumulation (Liu et al. 2013; Shen et al. 2014). The expression of these genes is highly regulated. For instance, the transcripts of the PacCOP1 gene, possibly encoding and Ubiquitin E3 ligase, negatively correlated with mRNA levels of PacDFR and PacANS among others (Liang et al. 2020). On the other hand, hormones such as ABA upregulate the expression of these genes (Shen et al. 2014).
Regarding the ABA pathway-related gene expression, PavNCED1 (coding for putative ABA biosynthetic enzyme) increases its transcript abundance in association with ABA and anthocyanin accumulation, whereas the PavCYP707A2 transcripts, possibly encoding ABA-degrading enzymes, decrease at the same time (Luo et al. 2014). Changes in the relative expression of PavPYLs, PavSnRKs, and PavPP2Cs, coding for ABA perception and signaling pathway, occur throughout the fruit ripening process (Wang et al. 2015; Shen et al. 2017). However, given that the ABA signaling pathway and anthocyanin pathways have been analyzed separately, the sequentiality of the molecular events underlying color development has not been clearly established. For example, changes in PavSnRK2s and PP2Cs transcript abundance and ABA content increase have been described during ripening of sweet cherry; however, these changes were not associated with anthocyanin accumulation nor anthocyanin pathway-related transcription (Wang et al. 2015; Shen et al. 2017).
Here, we found that color change is accompanied by anthocyanin increase, closely reflected by the maturity index, IAD (Fig. 1a). Non-destructive IAD device excites at 560 nm (chlorophyll, anthocyanin, and other flavonoid absorption) and 640 nm (mainly chlorophyll absorption). Thus, the IAD value is obtained from the absorbance differences between 560 nm, 640 nm, and the reference value at 750 nm (Costa et al. 2016) and estimates the content of anthocyanin and other flavonoids that act as a filter for chlorophyll absorption (Betemps et al. 2012). Therefore, IAD can be used as a maturity index, where IAD values positively correlate with the anthocyanin content of cherries (Nagpala et al. 2017).
In our results, an increase in IAD and anthocyanin content is more visible from P2 to the R stage (Fig. 1a), which is accompanied by a significant increase in the transcript abundance of anthocyanin pathway-related genes, specifically from P1 to R (Fig. 1b). Luo et al. (2014) reported the same increase but earlier, from the straw-yellow to the pink stage, with minor differences in relative expression between pink and red stages. In contrast, Liu et al. (2013) and Shen et al. (2014) reported an increase in the relative expression of anthocyanin pathway-related genes between pink and red stages, in line with our results. Possibly, there are differences depending on the varieties utilized in each study. A summary of these studies in contrast with our work is summarized in Table 1.
We also observed that changes in the transcript abundance of anthocyanin structural genes are preceded by PavMYBA transcript abundance increase, occurring from LG to P1 stages (Fig S3). In contrast, Shen et al. (2014) reported that PavMYBA relative expression increases from green to yellow stage, and then from pink to red stage. It is important to mention that the results presented here refer to dark red-colored varieties since the transcript profile is different in yellow or blush varieties (Starkevič et al. 2015; Jin et al. 2016).
In this work, the anthocyanin pathway-related genes are upregulated after the increase in ABA signaling pathway-related transcript abundance occurring from LG to P1 stage (Fig. 1c). The expression of the anthocyanin pathway-related genes may depend on the ABA pathway activation. ABA signaling genes include SNF1-related kinases (PavSnRK2s) and putative type 2C protein phosphatases (PavPP2Cs). As these genes are themselves ABA-responsive (Wang et al. 2015; Shen et al. 2017), as soon as ABA induces ABA signaling pathway-related gene expression, it could transduce the ABA signal into anthocyanin pathway activation. It is worth mentioning that not only ABA influences anthocyanin biosynthesis and secondary metabolism. For instance, recent reports show that exogenous application of calcium changes the expression of several genes involved in anthocyanin biosynthesis and modifies the accumulation profile of several secondary metabolites (Michailidis et al. 2020).
Regarding ABA homeostasis-related genes, a sustained increase in PavNCED1, along with a decrease in PavCYP707A2 transcripts, was observed (Fig. 1d). These genes are possibly involved in ABA homeostasis, with PavNCED1 coding for a putative ABA biosynthetic enzyme and PAvCYP707A2 coding for a possible ABA hydroxylase. Interference lines in these genes support their functionality in sweet cherry fruits (Li et al. 2015; Shen et al. 2014). Although PavCYP707A2 changes were not significant from LG to P2 stages, there is a marked decrease up to the R stage. PavNCED1, in turn, significantly increased from LG to P2 and also from P1 to P2. Possibly, PavNCED1 and PavCYP707A2 expression changes together could generate an additive effect in which increased ABA biosynthesis combined with reduced ABA degradation could raise ABA content.
Regarding the upregulation of ABA signaling pathway-related genes (Fig. 1c), PavSnRK2s and PavPP2Cs transcripts were previously measured during sweet cherry fruit development (Wang et al. 2015; Shen et al. 2017) but not associated with the color development. We selected PavSnRK2.1, PavSnRK2.2, PavSnRK2.3, PavPP2C3, and PavPP2C4 as they were the most highly expressed ABA signaling pathway-related genes (Wang et al. 2015). Although Wang et al. (2015) did not measure the anthocyanin levels nor color changes in the fruits, the ABA content increases when most of these genes increase. It was found that both PavSnRK2s and PavPP2Cs transcripts increased at the onset of the color initiation, from LG to P1 stages (Fig. 1c), possibly in response to ABA increase, as some of them are ABA-responsive (Wang et al. 2015; Shen et al. 2017).
In Arabidopsis, SnRK2 and PP2C genes are positive and negative regulators of the ABA pathway, respectively (Umezawa et al. 2010). PP2C directly inactivates and dephosphorylates subclass III SnRK2 in vitro (Umezawa et al. 2009). Upon ABA stimulus, ABA binds to the PYR/PYL/RCAR receptors, thus preventing its interaction with PP2C inhibiting its phosphorylation activity (Park et al. 2009) and SnRK2 is then released from negative regulation by PP2C, thus allowing the ABA pathway to turn on (Umezawa et al. 2010). PavSnRK2s and PavPP2Cs are expressed at color initiation (Fig. 1c). However, PavSnRK2.3 variation is the most abrupt (Fig. 1c); hence, this could produce a rapid activation of ABA signaling, and then, a compensation mechanism, in which PavPP2Cs putative negative regulators increase their expression, turning off the ABA signaling pathway and avoiding excessive ABA signal amplification. Finally, the ABA signaling pathway would be turned off at the red stage since a decrease in PavSnRK2.3 transcript abundance is observed.
In summary, a temporality of molecular events might occur before and during color development, in which ABA biosynthesis and ABA signaling activation positively regulate anthocyanin biosynthesis.
ABA influences color initiation timing and modulates ABA pathway- and anthocyanin pathway-related gene expression
Shen et al. (2014) and Luo et al. (2014) showed increased anthocyanin content a few days after the ABA treatment. However, as only one point was assessed, it is unclear whether ABA triggers the color initiation or increases the color intensity, or both. To determine if ABA advances color initiation, we performed an IAD curve for control and ABA-treated fruits to detect the differences in situ between the fruits during development. As shown in Fig. 2a, there is a significant increase in IAD from 67 to 69 DAFB in ABA-treated fruits. Moreover, control fruits reached the 69 DAFB value of ABA-treated fruits around 72 DAFB, so ABA treatment advanced IAD by approximately 3 days. Recently, our team has reported that exogenous application of GA3 can alter the transcript abundance of ABA pathway orthologs and modulate sweet cherry fruit ripening in early- and mid-season varieties (Kuhn et al. 2020). Interestingly, GA3 treatment performs the opposite effect than ABA; i.e., in mid-season varieties, GA3 seems to delay ripening initiation whereas here we report that ABA advances IAD evolution (Fig. 2a). Interaction between both pathways might control the timing of ripening initiation by the modulation of common regulatory modules.
Besides the ABA effect on IAD, ABA treatment significantly increased the anthocyanin content in ABA-treated fruits at harvest (Fig. 2b). Additionally, a higher number of fruits were darker after ABA treatment at both harvest dates (Fig. 2c), supporting that these fruits are advanced in their color development process.
In grapevine, ABA advances color development (Wheeler et al. 2009). In peach, ABA affects ripening when applied at mid-S3, which correlated with downregulation of ripening-related genes, but it promotes ripening when applied at more advanced stages, inducing the same genes previously downregulated (Soto et al. 2013). However, we disagree with their interpretation of the results as not necessarily downregulation of gene expression would lead to advance or delay; instead, it may result only in intensity change. Therefore, to distinguish between both effects, time courses of gene expression should be performed.
Regarding the molecular features underlying this IAD advance caused by ABA, we hypothesized that transcriptional changes should occur a few days after ABA treatment. This transcriptional effect could occur in ABA homeostasis and signaling pathway-related genes and ABA response genes, such as anthocyanin pathway-related genes. In fact, in PavCYP707A2 RNAi lines, an increase in the relative expression of PavMYBA, PavUFGT, and PavANS occurred (Wang et al. 2015). We found differences in the transcript abundance of several genes in response to ABA (Fig. 3). For example, PavCHS and PavF3H significantly increase their transcript abundance in ABA-treated fruits, similar to previously reported (Luo et al. 2014; Shen et al. 2014), whereas PavUFGT transcript abundance was not affected (Fig. 3a). Li et al. (2015) found that PavMYBA, PavUFGT, and PavANS have advanced gene expression in PavCYP707A2 RNAi fruits. However, in our study, PavMYBA and PavUFGT did not present significant differences between control and treatment. As we did not perform a time course for their expression, it is difficult to say whether the apparent lack of ABA effect is due to an eventual advance rather than a variation in intensity response. For instance, PavCYP707A2 RNAi lines had slight differences in the relative expression of PavMYBA, PavUFGT, and PavANS with control lines at the full red stage, despite the differences observed at the initial red stage due to the advance in transcript accumulation (Li et al. 2015). In future work, transcript accumulation time courses may clarify which genes change in response to ABA treatment.
As putative negative regulators of the ABA pathway, PavPP2C3 and PavPP2C4 transcripts significantly increase after ABA treatment (Fig. 3b). These genes increase during color initiation (Fig. 1c), which has been previously reported for PavPP2C3 (Wang et al. 2015; Shen et al. 2017), but not for PavPP2C4 (Wang et al. 2015). Possibly, ABA increases the ABA negative regulators in order to avoid excessive ABA response amplification.
As putative positive regulators of ABA signaling, only PavSnRK2.1 increased significantly in response to ABA under our experimental conditions (Fig. 3b). At the same time, in other reports, it is downregulated (Wang et al. 2015) or not affected by ABA at the color initiation stage (Shen et al. 2017). PavSnRK2.2 expression is not significantly affected by ABA (Fig. 3b), while it decreases in previous reports (Wang et al. 2015; Shen et al. 2017). Maybe the difference between this study and those works is that they use fruit-bearing branches cut at the color change and placed in a growth chamber. In this condition, the stress associated with the cutting and eventual dehydration could increase ABA content over physiological levels, making it difficult to extrapolate results to the normal ripening processes. In grapevine berry cultures, ABA induces most of VvPP2Cs and VvSnRKs expressed during ripening at or after veraison (Gambetta et al. 2010), which is in agreement with our results. On the other hand, we observed lower transcript accumulation of PavSnRK2.3 upon ABA treatment (Fig. 3b). PavSnRK2.3 transcript abundance increases sharply during color initiation but then decreases (Fig 1c), so this could be associated with a mechanism in which this gene first activates and then ABA represses its expression in a negative feedback mechanism.
Concerning PavNCED1 and PavCYP707A2 transcripts, ABA treatment seems to mimic developmental changes, i.e., PavNCED1 transcript abundance increases during color initiation and after the ABA treatment, whereas the PavCYP707A2 transcripts decrease their transcript abundance during color initiation and are downregulated by ABA (Fig. 1d; Fig. 3c). This supports the hypothesis that ABA controls its own levels by increasing its biosynthesis and reducing its degradation (Ren et al. 2010; Luo et al. 2014). Supporting our results, the increase in PavNCED1 and the decrease in PavCYP707A2 transcripts are accompanied by increased ABA content four days after the treatment (Fig. 3d).
Taking these results together, we postulate that IAD advance in ABA-treated fruits could then be a combination of ABA accumulation advance with an ABA regulation over its own signaling, which in turn could upregulate anthocyanin pathway-related genes. Future work is needed in order to shed more light on the role of ABA on the ripening initiation timing control.
Conclusions
The evidence presented in our in planta experimental design confirms that ABA is a central player in controlling the color initiation in sweet cherries during development. This is likely associated with a sequentiality in the transcript accumulation of genes possibly involved in ABA homeostasis and signaling, as well as anthocyanin pathway-related genes. This suggests a tight control of these ABA-responsive genes to trigger the molecular and physiological responses associated with fruit ripening. Dissecting the role of ABA in the color initiation timing would be an important improvement in breeding programs looking for genetic factors involved in the maturity time trait.
References
Alkio M, Jonas U, Declercq M, Van Nocker S, Knoche M (2014) Transcriptional dynamics of the developing sweet cherry (Prunus avium L.) fruit: sequencing, annotation and expression profiling of exocarp-associated genes. Hortic Res 1(1):11. https://doi.org/10.1038/hortres.2014.11
Arapitsas P, Perenzoni D, Nicolini G, Mattivi F (2012) Study of Sangiovese wines pigment profile by UHPLC-MS/MS. J Agric Food Chem 60(42):10461–10471. https://doi.org/10.1021/jf302617e
Betemps DL, Fachinello JC, Galarca SP, Portela NM, Remorini D, Massai R, Agati G (2012) Non-destructive evaluation of ripening and quality traits in apples using a multiparametric fluorescence sensor. J Sci Food Agric 92(9):1855–1864. https://doi.org/10.1002/jsfa.5552
Böttcher C, Keyzers RA, Boss PK, Davies C (2010) Sequestration of auxin by the indole-3-acetic acid-amido synthetase GH3-1 in grape berry (Vitis vinifera L.) and the proposed role of auxin conjugation during ripening. J Exp Bot 61(13):3615–3625. https://doi.org/10.1093/jxb/erq174
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55(4):611–622. https://doi.org/10.1373/clinchem.2008.112797
Chavoshi M, Watkins C, Oraguzie B, Zhao Y, Iezzoni A, Oraguzie N (2014) Phenotyping protocol for sweet cherry (Prunus avium L.) to facilitate an understanding of trait inheritance. J Am Pom Soc (APS) 68(3):125–134. https://www.pubhort.org/aps/68/v68_n3_a2.htm
Cherian S, Figueroa CR, Nair H (2014) ‘Movers and shakers’ in the regulation of fruit ripening: a cross-dissection of climacteric versus non-climacteric fruit. J Exp Bot 65(17):4705–4722. https://doi.org/10.1093/jxb/eru280
Cheung YK, Klotz JH (1997) The Mann Whitney Wilcoxon distribution using linked lists. Statistica Sinica 7(3):805–813
Coombe BG (1976) The development of fleshy fruits. Annu Rev Plant Physiol 27(1):207–228. https://doi.org/10.1146/annurev.pp.27.060176.001231
Costa G, Vidoni S, Rocchi L (2016) Use of non-destructive devices to support pre- and postharvest fruit management. Acta Hortic 1119:329–335. https://doi.org/10.17660/ActaHortic.2016.1119.45
Fadón E, Herrero M, Rodrigo J (2015) Flower development in sweet cherry framed in the BBCH scale. Sci Hortic 192:141–147. https://doi.org/10.1016/j.scienta.2015.05.027
Gambetta GA, Matthews MA, Shaghasi TH, McElrone AJ, Castellarin SD (2010) Sugar and abscisic acid signaling orthologs are activated at the onset of ripening in grape. Planta 232(1):219–234. https://doi.org/10.1007/s00425-010-1165-2
Giovannoni J (2001) Molecular biology of fruit maturation and ripening. Annu Rev. Plant Physiol 52:725–749. https://doi.org/10.1146/annurev.arplant.52.1.725
Guo X, Wang Y, Zhai Z, Huang T, Zhao D, Peng X, Feng C, Xiao Y, Li T (2018) Transcriptomic analysis of light-dependent anthocyanin accumulation in biocolored cherry fruits. Plant Physiol Biochem 130:663–677. https://doi.org/10.1016/j.plaphy.2018.08.016
Hardner CM, Hayes BJ, Kumar S, Vanderzande S, Cai L, Piaskowski J, Quero-Garcia J, Campoy JA, Barreneche T, Giovannini D, Liverani A, Charlot G, Villamil-Castro M, Oraguzie N, Peace CP (2019) Prediction of genetic value for sweet cherry fruit maturity among environments using a 6K SNP array. Hortic Res 6:6. https://doi.org/10.1038/s41438-018-0081-7
Jia HF, Chai YM, Li CL, Lu D, Luo JJ, Qin L, Shen YY (2011) Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol 157(1):188–199. https://doi.org/10.1104/pp.111.177311
Jin W, Wang H, Li M, Wang J, Yang Y, Zhang X, Yan G, Zhang H, Liu J, Zhang K (2016) The R2R3 MYB transcription factor PavMYB10.1 involves in anthocyanin biosynthesis and determines fruit skin colour in sweet cherry (Prunus avium L.). Plant Biotechnol J 14(11):2120–2133. https://doi.org/10.1111/pbi.12568
Kondo S, Gemma H (1993) Relationship between abscisic acid (ABA) content and maturation of the sweet cherry. J Jpn Soc Hortic Sci 62(1):63–68. https://doi.org/10.2503/jjshs.62.63
Kondo S, Inoue K (1997) Abscisic acid (ABA) and 1-aminocyclopropane-l-carboxylic acid (ACC) content during growth of ‘Satohnishiki’ cherry fruit, and the effect of ABA and ethephon application on fruit quality. J Hortic Sci Biotechnol 72(2):221–227. https://doi.org/10.1080/14620316.1997.11515509
Kuhn N, Guan L, Dai ZW, Wu BH, Lauvergeat V, Gomes E, Li SH, Godoy F, Arce-Johnson P, Delrot S (2014) Berry ripening: recently heard through the grapevine. J Exp Bot 65(16):4543–4559. https://doi.org/10.1093/jxb/ert395
Kuhn N, Ponce C, Arellano M, Time A, Sagredo B, Donoso JM, Meisel LA (2020) Gibberellic acid modifies the transcript abundance of ABA pathway orthologs and modulates sweet cherry (Prunus avium) fruit ripening in early- and mid-season varieties. Plants 9:1796
Kushiro T, Okamoto M, Nakabayashi K, Yamagishi K, Kitamura S, Asami T, Hirai N, Koshiba T, Kamiya Y, Nambara E (2004) The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: key enzymes in ABA catabolism. EMBO J 23(7):1647–1656. https://doi.org/10.1038/sj.emboj.7600121
Leng P, Yuan B, Guo Y (2014) The role of abscisic acid in fruit ripening and responses to abiotic stress. J Exp Bot 65(16):4577–4588. https://doi.org/10.1093/jxb/eru204
Li C, Jia H, Chai Y, Shen Y (2011) Abscisic acid perception and signaling transduction in strawberry: a model for non-climacteric fruit ripening. Plant Signaling Behav 6(12):1950–1953. https://doi.org/10.4161/psb.6.12.18024
Li Q, Chen P, Dai S, Sun Y, Yuan B, Kai W, Pei Y, He S, Liang B, Zhang Y, Leng P (2015) PacCYP707A2 negatively regulates cherry fruit ripening while PacCYP707A1 mediates drought tolerance. J Exp Bot 66(13):3765–3774. https://doi.org/10.1093/jxb/erv169
Liang D, Zhu T, Deng Q, Lin L, Tang Y, Wang J, Wang X, Luo X, Zhang H, Lv X, Xia H (2020) PacCOP1 negatively regulates anthocyanin biosynthesis in sweet cherry (Prunus avium L.). J Photochem Photobiol B Biol 203:111779. https://doi.org/10.1016/j.jphotobiol.2020.111779
Liu Y, Shen X, Zhao K, Ben Y, Guo X, Zhang X, Li T (2013) Expression analysis of anthocyanin biosynthetic genes in different colored sweet cherries (Prunus avium L.) during fruit development. J Plant Growth Regul 32(4):901–907. https://doi.org/10.1007/s00344-013-9355-3
Luo H, Dai S, Ren J, Zhang C, Ding Y, Li Z, Sun Y, Ji K, Wang Y, Li Q, Chen P, Duan C, Wang Y, Leng P (2014) The role of ABA in the maturation and postharvest life of a nonclimacteric sweet cherry fruit. J. Plant Growth Regul 33(2):373–383. https://doi.org/10.1007/s00344-013-9388-7
McAtee P, Karim S, Schaffer R, David K (2013) A dynamic interplay between phytohormones is required for fruit development, maturation, and ripening. Front Plant Sci 4:79. https://doi.org/10.3389/fpls.2013.00079
Meisel L, Fonseca B, Gonzalez S, Baeza-Yates R, Cambiazo V, Campos R, Gonzalez M, Orellana A, Retamales J, Silva H (2005) A rapid and efficient method for purifying high quality total RNA from peaches (Prunus persica) for functional genomics analyses. Biol Res 38(1):83–88. https://doi.org/10.4067/s0716-97602005000100010
Michailidis M, Karagiannis E, Tanou G, Samiotaki M, Tsiolas G, Sarrou E, Stamatakis G, Ganopoulos I, Martens S, Argiriou A, Molassiotis A (2020) Novel insights into the calcium action in cherry fruit development revealed by high-throughput mapping. Plant Mol Biol 104:597–614. https://doi.org/10.1007/s11103-020-01063-2
Nagpala EGL, Noferini M, Farneti B, Piccinini L, Costa G (2017) Cherry-Meter: an innovative non-destructive (vis/NIR) device for cherry fruit ripening and quality assessment. Acta Hortic 1161:491–496. https://doi.org/10.17660/ActaHortic.2017.1161.78
Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TFF, Alfred SE, Bonetta D, Finkelstein R, Provart NJ, Desveaux D, Rodriguez PL, McCourt P, Zhu JK, Schroeder JI, Volkman BF, Cutler SR (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324(5930):1068–1071. https://doi.org/10.1126/science.1173041
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29(9):45–445. https://doi.org/10.1093/nar/29.9.e45
Ren J, Sun L, Wu J, Zhao S, Wang C, Wang Y, Ji K, Leng P (2010) Cloning and expression analysis of cDNAs for ABA 8′-hydroxylase during sweet cherry fruit maturation and under stress conditions. J Plant Physiol 167(17):1486–1493. https://doi.org/10.1016/j.jplph.2010.05.027
Ren J, Chen P, Dai SJ, Li P, Li Q, Ji K, Wang YP, Leng P (2011) Role of abscisic acid and ethylene in sweet cherry fruit maturation: molecular aspects. NZJ Crop Hortic Sci 39(3):161–174. https://doi.org/10.1080/01140671.2011.563424
Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, Moorman AF (2009) Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37(6):45. https://doi.org/10.1093/nar/gkp045
Seo M, Jikumaru Y, Kamiya Y. (2011) Profiling of hormones and related metabolites in seed dormancy and germination studies. Seed Dormancy, Humana Press, 99-111. https://doi.org/10.1007/978-1-61779-231-1_7
Shen X, Zhao K, Liu L, Zhang K, Yuan H, Liao X, Wang Q, Guo X, Li F, Li T (2014) A role for PacMYBA in ABA-regulated anthocyanin biosynthesis in red-colored sweet cherry cv. Hong Deng (Prunus avium L.). Plant Cell Physiol 55(5):862–880. https://doi.org/10.1093/pcp/pcu013
Shen X, Guo X, Zhao D, Zhang Q, Jiang Y, Wang Y, Peng X, Wei Y, Zhai Z, Zhao W, Li T (2017) Cloning and expression profiling of the PacSnRK2 and PacPP2C gene families during fruit development, ABA treatment, and dehydration stress in sweet cherry. Plant Physiol Biochem 119:275–285. https://doi.org/10.1016/j.plaphy.2017.08.025
Soto A, Ruiz KB, Ravaglia D, Costa G, Torrigiani P (2013) ABA may promote or delay peach fruit ripening through modulation of ripening- and hormone-related gene expression depending on the developmental stage. Plant Physiol Biochem 64:11–24. https://doi.org/10.1016/j.plaphy.2012.12.011
Starkevič P, Paukštytė J, Kazanavičiūtė V, Denkovskienė E, Stanys V, Bendokas V, Šikšnianas T, Ražanskienė A, Ražanskas R (2015) Expression and anthocyanin biosynthesis-modulating potential of sweet cherry (Prunus avium L.) MYB10 and bHLH genes. PloS one 10(5):e0126991. https://doi.org/10.1371/journal.pone.0126991
Sutthiwal S (2012) Roles of abscisic acid in fruit ripening. Walailak J Sci Tech 9(4):297–308
Teribia N, Tijero V, Munne-Bosch S (2016) Linking hormonal profiles with variations in sugar and anthocyanin contents during the natural development and ripening of sweet cherries. New Biotechnol 33(6):824–833. https://doi.org/10.1016/j.nbt.2016.07.015
Udvardi MK, Czechowski T, Scheible WR (2008) Eleven golden rules of quantitative RT-PCR. The Plant cell 20(7):1736–1737. https://doi.org/10.1105/tpc.108.061143
Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K (2009) Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci U S A 106(41):17588–17593. https://doi.org/10.1073/pnas.0907095106
Umezawa T, Nakashima K, Miyakawa T, Kuromori T, Tanokura M, Shinozaki K, Yamaguchi-Shinozaki K (2010) Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol 51(11):1821–1839. https://doi.org/10.1093/pcp/pcq156
Wang Y, Chen P, Sun L, Li Q, Dai S, Sun Y, Kai W, Zhang Y, Liang B, Leng P (2015) Transcriptional regulation of PaPYLs, PaPP2Cs and PaSnRK2s during sweet cherry fruit development and in response to abscisic acid and auxin at onset of fruit ripening. Plant Growth Regul 75(2):455–464. https://doi.org/10.1007/s10725-014-0006-x
Wheeler S, Loveys B, Ford C, Davies C (2009) The relationship between the expression of abscisic acid biosynthesis genes, accumulation of abscisic acid and the promotion of Vitis vinifera L. berry ripening by abscisic acid. Aust J Grape Wine Res 15(3):195–204. https://doi.org/10.1111/j.1755-0238.2008.00045.x
Zhang M, Yuan B, Leng P (2009) The role of ABA in triggering ethylene biosynthesis and ripening of tomato fruit. J Exp Bot 60(6):1579–1588. https://doi.org/10.1093/jxb/erp026
Acknowledgements
We would like to thank Natalia Molina for her technical assistance in the field during sweet cherry fruit development and at harvest.
Availability of data and material
All relevant data are within the manuscript and its Supporting Information files.
Code availability
Not applicable
Funding
This study was funded through the project CONICYT FONDECYT/Regular No. 1171016 in collaboration with the project CONICYT FONDECYT/Regular No. 1161377. Nathalie Kuhn was supported by CONICYT FONDECYT de Postdoctorado 2018, No. 3180138. Alson Time was supported by Conicyt Scholarship Grant No. 21190238.
Author information
Authors and Affiliations
Contributions
Nathalie Kuhn: conceptualization, methodology, investigation, visualization, writing–original draft. Claudio Ponce: conceptualization, methodology, investigation, visualization, writing–original draft. Macarena Arellano: investigation, writing–review and editing. Alson Time: Investigation, writing–review and editing. Salvatore Multari: investigation, writing–review and editing. Stefan Martens: investigation, supervision, writing–review and editing. Ester Carrera: investigation, writing–review and editing. Boris Sagredo: conceptualization, writing–review and editing. José Manuel Donoso: conceptualization, writing–review and editing. Lee A. Meisel: conceptualization, methodology, visualization, supervision, project administration, funding acquisition, writing–review and editing
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethics approval
Not applicable
Consent to participate
Not applicable
Consent for publication
Not applicable
Data Archiving Statement
All relevant data are within the manuscript and its Supporting Information files.
Additional information
Communicated by C. Dardick
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Communicated by C. Dardick
Supplementary information
Fig. S1
Schematic representation of the fruit sampling for quality parameters measurement at harvest and the experimental design of the ABA treatment. (a), Twenty-five fruits of (n=4) trees were collected on November 29th, 2017 (1st harvest; 69 DAFB), and December 1st , 2017 (2nd harvest; 75 DAFB) for the analysis at harvest. 25 and 15 fruits per tree (n=4 trees) were used for quantifying the parameters in the 1st and 2nd harvest, respectively. The five most homogeneous fruits among the 25 or 15 were used for SSC (soluble solid content) and acidity determination. A. Refractometer; B. Durometer; C. Cherry meter; D. CTIFL color chart for color distribution; E. Fruit Caliper; F. Scale. (b), Diagram showing the number of trees treated with ABA and the control trees. Additionally, a picture of representative fruits minutes prior the ABA treatment (56 DAFB; November 16th, 2017; P1) is included. (PNG 1583 kb)
Fig. S2
Effect of ABA on ripening related parameters. (a), Fruit growth curve of ABA and control trees. The arrows indicate ABA treatment at 56 DAFB, as well as the 1st and 2nd fruit harvests. The asterisks indicate statistical differences at the indicated dates determined with T-student test, p-value < 0.05. (b), Percentage change of fruit quality parameters (width, weight, firmness, soluble solids, and acidity) in response to ABA treatment at 1st and 2nd harvest. (c), Representative pictures of the control and ABA-treated fruits at 2nd harvest. (PNG 820 kb)
Fig. S3
Transcript abundance relative to PavCAC of putative orthologs of ABA signaling pathway during fruit development. (a), PavPYL2 gene, (b), PavCYP707A1 gene, (c), PavMYB gene, encoding putative R2R3 MYB transcription factor . LG, P1, P2, and R are Light green, Pink 1, Pink 2, and Red stages, respectively (see Table S2). Pools of eight fruits per tree (n=3 trees) were used. Data were represented as mean ± SEM. (PNG 587 kb)
Fig. S4
Effect of ABA treatment on the transcript abundance relative to PavCAC of putative orthologs of the ABA signaling pathway. (a), PavPYL2 gene, (b), PavCYP707A1 gene, and (c), PavMYBA gene, encoding putative R2R3 MYB transcription factor. Pools of eight fruits per tree (n=3 trees) for control and ABA-treated trees were used. Data were represented as mean ± SEM and the values were set to 1.0 in Control. (PNG 200 kb)
ESM 5
(DOCX 14 kb)
ESM 6
(DOCX 14 kb)
ESM 7
(DOCX 14 kb)
ESM 8
(DOCX 12 kb)
Rights and permissions
About this article
Cite this article
Kuhn, N., Ponce, C., Arellano, M. et al. ABA influences color initiation timing in P. avium L. fruits by sequentially modulating the transcript levels of ABA and anthocyanin-related genes. Tree Genetics & Genomes 17, 20 (2021). https://doi.org/10.1007/s11295-021-01502-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-021-01502-1