Abstract
To further understand the basis of abscisic acid (ABA) signal transduction in regulating sweet cherry fruit development and ripening, and also the interaction between ABA and indole acetic acid (IAA) at the onset of fruit ripening, full-length or partial cDNA of three PaPYLs (ABA receptor), six PaPP2Cs (type 2C protein phosphatase) and six PaSnRK2s (subfamily 2 of SNF1-related kinases) were identified from sweet cherry. Multiple alignments and phylogeny analyses showed that most of the functional residues or domains were well conserved within each gene family; all PaPP2Cs belonged to the group A PP2C; PaSnRK2.4 and PaSnRK2.5 belonged to subclass III of SnRK2. PaPYL2/3, PaPP2C3/4/6 and PaSnRK2.4 were highly expressed at the early stages of fruit development. Moreover, PaPYL2, PaPP2C3/4, PaSnRK2.4 also had a high expression level at the onset of fruit ripening. Exogenous ABA treatment at 28 days after full bloom decreased the IAA level and promoted fruit ripening by increasing anthocyanin and soluble solids content. However, although IAA treatment induced the ABA accumulation, it delayed the onset of fruit ripening. Most of the PaPYLs and PaSnRK2s didn’t strongly respond to ABA and IAA treatments at 28 days after full bloom, while expressions of PaPP2C3, PaPP2C5 and PaPP2C6 were significantly induced by exogenous ABA and PaPYL1 was significantly induced by exogenous IAA.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sweet cherry (Prunus avium L.), because of its beauty and sweetness, has become a kind of most highly prized and popular fruit all around the world. Although it has many attractive features to the customers, several problems exist in the sweet cherry production and industry, such as fruit cracking and low storability. Many efforts have been put into solving these problems, however, a lack of knowledge on the sweet cherry fruit development and ripening mechanism, especially at the molecular level, has become a main limitation.
Phytohormones have long been known to involve in the regulation of fruit development and ripening (Crane 1969; Nitsch 1988). Among the five major phytohormones: abscisic acid (ABA), auxin, gibberellin, ethylene and cytokinin, both ABA and auxin (mainly refers to IAA) have been shown to modulate the growth and development of fruit at various stages (Ozga and Rienecke 2003). Recently, by means of silencing the key genes in ABA biosynthesis and signal transduction pathway respectively, ABA has been proved to directly regulate the fruit ripening in both climacteric (tomato) and non-climacteric fruit (strawberry) via affecting the cell wall degradation and fruit coloring (Jia et al. 2011; Sun et al. 2012). In sweet cherry, a typical non-climacteric fruit, ABA has also been reported to involve in regulating fruit maturation and ripening. Firstly, the endogenous ABA level and the expression level of the key genes involved in ABA biosynthesis (PacNCED1) and oxidative catabolism (PacCYP707As) were related to the sweet cherry fruit development and ripening process (Kondo and Gemma 1993; Setha et al. 2005; Ren et al. 2010). The ABA level peak which appeared at the beginning of sweet cherry fruit softening was thought to be the signal that triggered fruit ripening (Setha et al. 2005; Ren et al. 2010). Secondly, exogenous ABA application to the sweet cherry fruit at the second rapid growth phase can not only accelerate fruit softening and increase the maturation index levels, but also enhance the fruit quality by increasing the content of total sugar and anthocyanin (Kondo and Gemma 1993; Ren et al. 2010). Furthermore, a latest study reported that ABA can directly regulate sweet cherry fruit coloration via affecting the expression level of the PacMYBA which is a transcription factor that interacts with several anthocyanin-related bHLH transcription factors to further activate the promoters of key genes in anthocyanin biosynthesis pathway (Shen et al. 2014). Besides the effects on pericarp, ABA can also affect the lignification process thereby regulating the seed development in sweet cherry (Kondo and Inoue 1997). On the other hand, besides ABA, auxin can also affect the fruit development of sweet cherry. Applying the synthetic auxin at the beginning of pit-hardening can lead to appreciable and significant increase in fruit size and total yield (Stern et al. 2007).
Phytohormones, both the endogenous and the synthetic, have been proved to have mass of effects on the development and ripening of many kinds of fruits (Given et al. 1988; Zhang et al. 2009; Jia et al. 2011), including sweet cherry (Shen et al. 2014), and at most of the time, their biological functions must depend on the perception of their specific receptors and transduction of their signaling pathways. There are two kinds of ABA signal transduction pathways that have been reported: one is the PYR/PYL/RCARs (ABA receptor, hereafter the PYLs)—PP2C (ABA signal negative regulators, type 2C protein phosphatases)—SnRK2 (ABA signal positive regulators, subfamily 2 of SNF1-related kinases) pathway, in which the signal transduction depends on the phosphorylation/dephosphorylation of these three core components (Ma et al. 2009; Melcher et al. 2009; Nishimura et al. 2009; Park et al. 2009; Umezawa et al. 2009): ABA can be sensed and bound by PYLs which belongs to the START protein superfamily through their ligand-binding pockets; with the binding of ABA, PYL closes two highly conserved β-loops around the entry of the ligand-binding pocket and forms a ‘cap-lock’ interface which in turn binds to PP2C phosphatase activity site and directly inhibits the activity of PP2C; then SnRK2s are released from the dephosphorylating of PP2C and can phosphorylate downstream proteins or transcription factors that trigger the expression of ABA-responsive genes. Another pathway is the magnesium chelatase H subunit (CHLH) pathway, in which the CHLH as an ABA receptor antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition (Shen et al. 2006; Wu et al. 2009). In tomato and strawberry, the PYL–PP2C–SnRK2 and CHLH pathway have been shown to be involved in the regulation of fruit ripening: in tomato the expression of the three core components (PYL, PP2C and SnRK2) genes were observed to coincide with the fruit development process (Sun et al. 2011); in strawberry, down-regulation of the PYR1 and CHLH genes in fruit by virus induced gene silencing (VIGS), significantly inhibited the fruit coloration process (Chai et al. 2011; Jia et al. 2011). However, in sweet cherry, although the CHLH gene has been cloned and studied, its function in fruit is more likely to involve in stress response, but not in the regulation of fruit ripening (Ren et al. 2011). Therefore, the involvement of ABA signal transduction pathway during fruit ripening is still unclear in sweet cherry.
In this study, based on sequence similarity and alignment with Arabidopsis homologues, three PaPYLs, six PaPP2Cs and six PaSnRK2s were identified from cDNA contig database which was generated from sweet cherry RNA-seq short reads pool. Expression patterns of these genes during sweet cherry fruit development and ripening and in response to exogenous ABA and IAA treatment were characterized. We hope these results could lay foundation for further studies on the molecular mechanism of ABA in regulating sweet cherry fruit development and ripening.
Materials and methods
In silico analysis
The PYL, PP2C and SnRK2 full-length cDNA sequences and the corresponding amino acid sequences of Arabidopsis were obtained from NCBI (http://www.ncbi.nlm.nih.gov/) as reported in previous studies (Huai et al. 2008; Ma et al. 2009; Melcher et al. 2009; Park et al. 2009; Shen et al. 2010; Umezawa et al. 2010). To identify sweet cherry orthologues of selected Arabidopsis genes, BLASTN searches were conducted in the sweet cherry contig database with the Arabidopsis cDNA sequences as queries. The sweet cherry contig database was obtained from Dr. Merianne Alkio (Alkio et al. 2012). All the nucleotide sequences that we used are listed in the Supplementary data. The identified sweet cherry sequences were further verified by Align Sequences Protein BLAST with the deduced sweet cherry protein sequences as query and the homologous Arabidopsis proteins as subject sequences. Homology alignments were performed within each gene family by comparing the sweet cherry deduced amino acid sequences with the orthologues in Arabidopsis, tomato, cucumber and grape, using ClustalX software with default option. Alignment results were visualized and edited by BOXSHADE 3.21 software (http://www.ch.embnet.org/software/BOX_form.html) and Jalview software (Waterhouse et al. 2009). The phylogenetic trees were constructed by using the Neighbor-Joining (N-J) method in ClustalX software with the bootstrap analysis setting at 1,000 replicates for evaluating the reliability of different phylogenetic groups. For the PYL gene family, phylogeny analysis was performed by using full-length of deduced amino acid sequences. For the PP2C and SnRK2 gene families, because some genes (PaPP2C2, PaSnRK2.4/2.5/2.6) are partial, phylogeny analyses were carried out by analyzing the conserved domains. Tree files were visualized and edited using MEGA 4.0.2 software.
Plant materials
Fruits from Prunus avium L. cv. Sato Nishiki were collected from adult trees grown in an experimental orchard at China Agricultural University (Beijing, P. R. China) in the spring of 2013. Fruits were collected at 10, 13, 17, 21, 25, 29, 33, 35, 37 and 40 days after full bloom (DAFB), respectively. Pulp was immediately separated using a knife, frozen in liquid nitrogen and stored at −80 °C for subsequent analysis.
Exogenous ABA and IAA treatments
Bearing branches with the same bearing ability were cut at 28 DAFB and the shoot was placed in 1 mM ABA (Sigma, A1049) solution, 1 mM IAA (Sigma, S12886) solution or distilled water as control in a tissue culture room at 25 ± 2 °C with a 16 h light/8 h dark cycle for 3 days. The treatment solution was freshly prepared every day and put in triangular flask covered with aluminum foil. Pulp and peel from each treatment were separated and collected after 3 days of treatment, frozen in liquid nitrogen and stored at −80 °C for subsequent analysis. Each treatment was repeated three times.
Determination of ABA and IAA content
Determination of ABA and IAA was conducted according to the method described by Yu et al. (2008). Three grams powders of each sample was extracted in 40 ml 80 % methanol (v/v) which contained 4 mg 2, 6-di-tert-butyl-4-methylphenol at −20 °C in dark for 18 h. The extract was then centrifuged at 10,000×g for 20 min, 4 °C and the supernatant saved. The pellet was re-extracted with 40 ml 80 % methanol (v/v) at −20 °C in dark for 2 h. Then the extract was centrifuged at 10,000×g for 20 min, 4 °C and the supernatant saved. The pellet was reextracted a third time with 20 ml 80 % methanol (v/v) at −20 °C in dark for 2 h. After that the extract was centrifuged at 10,000×g for 20 min, 4 °C and the supernatant saved. All the supernatants were collected and evaporated to dryness at 40 °C with rotary evaporator. Residue was washed with 10 ml petroleum ether and then dissolved in 10 ml 0.02 M phosphate buffer solution (pH 8.0). The phosphate buffer was decolorised with 10 ml petroleum ether for three times and then added 0.2 g insoluble PVPP, mixed at 0 °C for 15 min. Then PVPP was detached by vacuum filtration. The phosphate buffer was then extracted three times by ethyl acetate at pH 3.0 (ethyl acetate phase retained). The ethyl acetate phase was evaporated to dryness at 40 °C and dissolved in 1 ml 50 % methanol (v/v) for HPLC analysis. 20 µl of each sample was separated by HPLC (1200 Series; Agilent Technologies, Santa Clara, CA, USA) on a 4.8 × 150 mm C18 column (Agilent Technologies) at a flow rate of 0.8 ml min−1. The solvents were 0.8 % (v/v) glacial acetic acid (solvent A) and 100 % methanol (solvent B). Elution was by means of a changeable gradient of solvent B: 0–4 min 45 %, 4–5 min from 45 to 50 %, 5–17 min from 50 to 60 %, 17–18 min 60 % and 18–19 min from 60 to 45 %. ABA and IAA retention time was determined with standard substance [(±)-abscisic acid, Sigma A1049; IAA, Sigma SI2886] at wavelength of 260 nm. Each sample had three biological replicates.
Measurement of single fruit weight, fruit transverse, vertical diameter and soluble solids content
Single fruit weight was measured periodically during fruit development using an electronic balance. Fruit transverse and vertical diameter were measured at the same time. Soluble solids content was obtained by determining the juice of pulp with a Pocket Refractometer Pal-1 (ATAGO, TOKYO; units = %Brix).
Anthocyanin analysis
1 g peel was extracted with 0.1 % HCl-methanol for 4 h and centrifuged at 12,000 rpm at 4 °C. Absorbance of the extracted solution was determined at 530 and 657 nm. The formula A = A530–0.25 A657 was used to compensate for the contribution of chlorophyll and its degradation products to the absorption at 530 nm (Rabino and Mancinelli 1986). Anthocyanin content was a relative value and we set A = 0.01 equal to one unit (U).
RNA extraction and real-time PCR analysis
Total RNA was extracted from 1.0 g pulp using the hot borate method (Wan and Wilkins 1994). Genomic DNA was eliminated using RNase Free DNase I kit (Takara, Dalian, China) according to the manufacturer’s recommendations. The quality of every RNA sample was assessed by agarose gel electrophoresis. Band intensities were measured using the Image J software. The intact total RNA run on a denaturing gel should have sharp, clear 28S and 18S rRNA bands. The 28S rRNA band should be approximately twice as intense as the 18S rRNA band. Then RNA sample with good quality was quantified by NanoDrop 1000 Spectrophotometers (Thermo Scientific). The cDNA was synthesized from the total RNA using the PrimeScript™ RT reagent kit (Takara, Dalian, China) according to the manufacturer’s instructions. Amplification of specific regions of targeted genes and real-time detection of amplification production was conducted using a Rotor-Gene 3000 system (Corbett Research, Australia) with specific primers (Supplementary Table S1). Real-time PCR was performed using the SYBR Premix Ex TaqTM kit (TaKaRa, Dalian, China). Reactions contained 1 μl of primer mix (containing 5 μM of each forward and reverse primer), 2 μl cDNA template, 12.5 μl SYBR Premix Ex TaqTM (2×) mix and 9.5 μl water to make a total volume of 25 μl. Reactions were carried out under the following conditions: 95 °C/30 s (1 cycle); 95 °C/15 s, 60 °C/20 s; 72 °C/15 s (40cycles).
PCR amplification of a single product of the correct size for PaPYLs, PaPP2Cs and PaSnRK2s were confirmed by agarose gel electrophoresis and double-strand sequencing. Plasmids containing each specific gene were used in standard curve assays. Relative fold expression for the genes were calculated using Rotor-Gene 6.1.81 software. According to Ren et al. (2010), the PacACT1 (FJ560908) gene was selected as an internal control gene and its expression level was not only used to normalize the difference among of the template added in different PCR tubes, but also to adjust the variations that were generated during the whole process from RNA extraction to reverse transcription (relative quantification).
Results
Identification of sweet cherry orthologues of Arabidopsis genes involved in ABA signal transduction
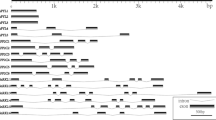
In order to isolate the PYLs, PP2Cs and SnRK2 s involved in the ABA signal transduction from the sweet cherry contig database, BLASTN searches were conducted in the sweet cherry contig database with the Arabidopsis cDNA sequences as queries. In order to further verifying the sequences obtained from sweet cherry, BLASTP searches were conducted using the deduced sweet cherry protein sequences as query and the corresponding Arabidopsis protein sequences as subject. Finally, three PaPYLs, six PaPP2Cs and six PaSnRK2s which showed high similarity in BLASTP analysis were identified from the sweet cherry contig database (Tables S2–S4). Based on multiple alignments of these deduced protein sequences (Fig. 1a–c; Fig. S1), most of the functional residues or domains were well conserved within each gene family. Phylogeny analyses revealed that, within the PYL gene family, PaPYL1 and PaPYL3 belonged to subfamily I, while PaPYL2 belonged to subfamily II (Fig. 1a; Fig. S2). Within the PP2C gene family, all the PaPP2Cs were classified into the group A in both of the phylogeny analyses using full-length sequences and conserved regions and kept a significant distance from SlPP2C, SlPP2C3 and SlPP2C4 (Fig. 1b; Fig. S3 and S4). Within the SnRK2 gene family, as we only obtained the C-terminal sequences for PaSnRK2.4 and PaSnRK2.6 and N-terminal sequence for PaSnRK2.5 (Fig. S1C), phylogeny analyses were performed in three methods: full-length analysis for PaSnRK2.1/2.2/2.3 (Fig. S5), C-terminal analysis for PaSnRK2.1/2.2/2.3/2.4/2.6 (Fig. 1c) and N-terminal analysis for PaSnRK2.1/2.2/2.3/2.5 (Fig. 1d). As shown in Fig. 1c, d and Fig. S4, PaSnRK2.1 was classified into subclass I; PaSnRK2.2/2.3/2,6 were classified into subclass II; PaSnRK2.4 and PaSnRK2.5 were classified into subclass III via C-terminal analysis and N-terminal analysis, respectively.
Sequence alignments and phylogenetic trees of the PYL, PP2C, and SnRK2 gene families. Genes studied are marked with black diamonds. Amino acid sequences are shown only for functional residues and domains. Residue positions are marked with the numbers nearby. Conserved residues are marked with black or grey shading. More detailed alignments are shown in Supplementary Fig. S1. Phylogenetic trees are shown only with their topological structures. a Sequence alignment and phylogenetic tree of the PYL family. Residues forming the ligand-binding pocket are marked with black triangles. The gate and latch domains are indicated. Functional residues and domains are based on reports by Melcher et al. (2009) and Santiago et al. (2009). b Sequence alignment and phylogenetic tree of the PP2C. The phylogenetic tree was constructed with the N-terminal sequences. Sequences showed in break line box are C-terminal region of the amino acid. Residues interacting with ABA, PYLs, and Mn/Mg ions are marked with black triangles, asterisks, and white triangles, respectively. Phosphatase sites are marked with black circles. Functional residues and domains are based on studies by Melcher et al. (2009). c Sequence alignment and phylogenetic tree of the deduced C-terminal amino acid sequence of SnRK2 family. Potential phosphatase sites are marked with black circles according to Umezawa et al. (2009). Functional residues and domains are noted according to Yoshida et al. (2006) and Yuasa et al. (2007). d Phylogenetic tree constructed with deduced N-terminal amino acid sequences of SnRK2 family in sweet cherry and Arabidopsis. The N-terminal region, which was used to construct phylogenetic tree, was marked with red box in Fig. S1C
Growth parameters of sweet cherry fruit
By measuring changes in single fruit weight, transverse diameter and vertical diameter, a profile of berry development was established for ‘Sato Nishiki’ sweet cherry fruit (Fig. 2a). Fruit growth followed a biphasic growth pattern with time. It took about 35 days from full bloom to maturation and about 40 days to full ripeness. The first 17 DAFB was the first rapid growth stage. The following was the pit hardening stage, during which the fruit slightly enlarged and the stony endocarp underwent hardening. When the pit hardening stage ended, a second rapid growth stage occurred after 25 DAFB. Fruit growth ceased at 35 DAFB and then fruit entered into the full ripe and senescence stage.
ABA and IAA variations during sweet cherry fruit development and ripening and in response to exogenous hormone treatment
The highest ABA level during sweet cherry fruit development and ripening appeared at 13 DAFB, and then it sharply declined to lower levels at 17 DAFB. Only slight changes were detected between 17 and 33 DAFB, after which it increased and reached another peak value at 35 DAFB and after that it decreased (Fig. 2b). The highest IAA level was detected at 10 DAFB, afterwards, it gradually decreased and reached the lowest level at 29 DAFB, after which it increased and peaked at 35 DAFB (Fig. 2b). IAA content in ABA treated fruits was decreased compared to that in the control fruit (Fig. 3c). However, ABA content was increased in the IAA treated fruits (Fig. 3d).
Effects of ABA and IAA application on sweet cherry fruit ripening
Compared to that in the control fruits, soluble solids content in ABA treated fruits was slightly increased, however, it was decreased in IAA treated fruits (Fig. 3a). In addition, ABA treatment significantly increased the anthocyanin content in the peel of sweet cherry fruit, while IAA treatment slightly reduced its content (Fig. 3b).
Expressions of PaPYLs, PaPP2Cs and PaSnRK2s during sweet cherry fruit development and ripening
Within the PaPYL gene family, PaPYL1 and PaPYL2 had similar expression patterns from 10 DAFB to 25 DAFB and both of them reached the highest value at 21 DAFB, however, during the following development stages, they exhibited different expression patterns: PaPYL1 had another expression peak at 35 DAFB, while PaPYL2 had two expression peaks at 29 and 37 DAFB, respectively (Fig. 4a). PaPYL3 expression significantly decreased between 10 and 13 DAFB periods and then it gradually decreased to relatively low levels (Fig. 4a).
Within the PaPP2C gene family, PaPP2C1 expression was increased from 10 to 13 DAFB, and then it gradually decreased to the lowest level at 33 DAFB, after which it increased and peaked at 35 DAFB (Fig. 4b). PaPP2C2 expression gradually decreased from 10 DAFB to 29 DAFB, after that only slight changes were detected. Expression of PaPP2C3 was relatively high at 10 DAFB and then it significantly decreased to the lowest level at 21 DAFB, afterwards, it increased and reached the peak value at 35 DAFB (Fig. 4b). PaPP2C4 had a fluctuant expression pattern during sweet cherry fruit development and ripening (Fig. 4b). Both PaPP2C5 and PaPP2C6 expressions generally decreased during the whole development process, though there was a small peak appearing at 35 DAFB (Fig. 4b).
Within the PaSnRK2 gene family, PaSnRK2.1 expression decreased sharply from 10 DAFB to 21 DAFB, after which it increased and reached a small peak at 25 DAFB, then it decreased again and followed by another small peak at 35 DAFB (Fig. 4c). PaSnRK2.2 had a relatively high expression value at 10 DAFB, then it significantly decreased to the lowest level at 21 DAFB, afterwards, it gradually increased and peaked at 35 DAFB (Fig. 4c). PaSnRK2.3 expression was generally increased from 10 DAFB and reached the peak value at 25 DAFB, after which it declined till 35 DAFB and thereafter it increased again (Fig. 4c). PaSnRK2.4 expression was decreased from 10 DAFB to 33 DAFB, after which it gradually increased toward 40 DAFB (Fig. 4c). PaSnRK2.5 and PaSnRK2.6 expression patterns were similar during the whole development process, both of them reached the highest value and peaked at 21 DAFB (Fig. 4c).
Expressions of PaPYLs, PaPP2Cs and PaSnRK2s in response to exogenous ABA and IAA treatment in sweet cherry fruit
Among the three PaPYLs, PaPYL1 was significantly up-regulated by IAA treatment; PaPYL2 was significantly down-regulated by both ABA and IAA treatment; however, PaPYL3 was insensitive to both treatments (Fig. 5a).
a Expressions of PaPYLs in response to exogenous ABA and IAA treatments. b Expressions of PaPP2Cs in response to exogenous ABA and IAA treatments. c Expressions of PaSnRK2s in response to exogenous ABA and IAA treatments. PacACT1 (FJ560908) was used as an internal control gene. Numbers above/below the bars correspond to the last number of the genes in each gene family members. Bars in the positive quadrant indicate up-regulated gene expression, while those in the negative quadrant indicate down-regulated gene expression. Standard deviation are indicated
In terms of PaPP2C gene family, PaPP2C1, 2 and 4 expressions were not obviously changed by exogenous ABA and IAA treatment (Fig. 5b). However, PaPP2C3, 5 and 6 were all greatly up-regulated by both ABA and IAA treatment (Fig. 5b).
Within the PaSnRK2 gene family, expression of PaSnRK2.1, 2.5 and 2.6 was down-regulated in response to ABA treatment (Fig. 5c) and expression of PaSnRK2.2 and 2.3 was down-regulated by IAA treatment (Fig. 5c). However, PaSnRK2.4 was insensitive to both treatments (Fig. 5c).
Discussion
Core components of the ABA signal transduction pathway emerged with the evolution of plants from aquatic to terrestrial environment. During the evolution process, size of PYL family and group A of PP2C family increased. For example, as for PYL gene family, there are 4 members in Physcomitrella patens, 10 members in Selaginella moellendorffi, 14 members in Arabidopsis thaliana, 10 members in cucumber, 8 members in tomato and 6 members in orange; as for group A PP2C gene family, there are 2 members in P. patens, 5 members in S. moellendorffi, 9 members in Arabidopsis thaliana, 7 members in tomato and 9 members in grape (Park et al. 2009; Umezawa et al. 2010; Sun et al. 2011; Boneh et al. 2012; Romero et al. 2012; Wang et al. 2014). However, subclass III of SnRK2 family size was relatively conserved, no more than 4 members in all the reported species so far (Umezawa et al. 2010). In this study, via blasting the homologues of AtPYL, group A AtPP2C and AtSnRK2 in the sweet cherry contig database which was de novo assembled from an exocarp RNA-seq short reads pool (Alkio et al. 2012), 6 group A PP2Cs and 2 subclass III SnRK2s were identified, of which the number of family members was within the normal range; however, for the PYL family, only three members were obtained, which was far more less than the number of the homologues in other species. The reason for the small size of PaPYL family, on one hand, is that the contig database was generated only from sweet cherry fruit exocarp, which may lead to identification of genes only expressed in exocarp. On the other hand, the total number of PYL varies between different species and low total number of PaPYL may directly result in less number of genes that can be detected in fruit exocarp. Other genes in PaPYL family may be identified in the future with the progress in the sequencing of the sweet cherry whole genome.
According to previous studies, only subclass III of SnRK2 family involves in the ABA signal transduction (Umezawa et al. 2010). Among the five identified PaSnRK2s, PaSnRK2.4 and PaSnRK2.6 only have C-terminal sequences (Fig. S1C); meanwhile PaSnRK2.5 only has N-terminal sequence (Fig. S1C). For clarifying the phylogeny information and subclasses that each PaSnRK2 belonging to, three methods (full-length, N-terminal and C-terminal alignment) were used to generate the phylogenetic tree (Fig. 1; Fig. S5). Among all the three types of analysis, PaSnRK2.1 and PaSnRK2.2/2.3, which have full-length amino acid sequences, were always classified into the subclass I and II, respectively (Fig. 1; Fig. S5), from which it can be presumed that both the N-terminal and C-terminal of PaSnRK2s have the conserved sequence that can be used for the phylogeny analysis. Thus, based on the phylogeny analyses and multiple-alignments, PaSnRK2.4 and PaSnRK2.5 have a very high possibility to be the members of subclass III of SnRK2 family and transduce ABA signal.
By monitoring the growth parameters (Fig. 2a) and analyzing the endogenous ABA and IAA concentration in sweet cherry fruit during the whole development and ripening process (Fig. 2b), a similar double-peak variation pattern was observed in both kinds of phytohormones. The first peak of ABA and IAA appeared at the first rapid growth stage: for ABA, this peak was thought to involve in the activation of the sink and stimulate the nutrition accumulation in fruitlets (Talon et al. 1990); for auxin, the high concentration has been proved to involve in accelerating of cell division process (Mapelli et al. 1978). Therefore, at the first rapid growth stage of sweet cherry, ABA and IAA can be presumed to worked together to finish the cell division process by stimulating the nutrition accumulation and also accelerating the cell division. Furthermore, from the absolute and relative quantification results, PaPYL1/2, PaPP2C3/4/6 and PaSnRK2.4 may play more important roles in the transduction of ABA signal at this stage (Fig. 4).
The second peak of the two studied plant hormones were found at 35 DAFB, which is the time point between the second rapid growth stage and the ripen stage (Fig. 2). The ABA peak at this time point has been proved to increase anthocyanin biosynthesis, maturity index (SSC/TA), and thereby promoting fruit ripening (Ren et al. 2010; Luo et al. 2014). Meanwhile, the IAA dramatic variation at this stage was thought to be related to the fruit size enlargement at the second rapid stage (Liu et al. 2007). Although ABA peaked at 35 DAFB, we found that the transition of the second rapid growth stage to the ripening stage initiated 6 days earlier (28 DAFB). Therefore, the increase of ABA and the low level IAA may be critical for this transition. The functions of exogenous ABA and IAA were opposite in the regulation of sweet cherry fruit ripening: ABA accelerated ripening, while IAA delayed ripening process (Fig. 3a, b and Supplementary Fig. S7). Furthermore, at this time, ABA may play a dominant role in controlling the ripening process, because exogenous ABA treatment can down-regulate the endogenous IAA level, meanwhile applying extra IAA can lead to endogenous ABA increase, which may be caused by a negative feedback reflecting the fruit tried to overcome the ripening inhibition caused by IAA via increasing endogenous ABA level. In the aspect of ABA signal transduction, during the second rapid growth stage, PaPYL2, PaPP2C3/4/5/6, PaSnRK2.4 may be the elements that make up the signal transduction pathway, while at the ripening stage, PaPYL1, PaPP2C3/4 and PaSnRK2.4 may play critical roles in transducing the ABA signal. As the transition period from growth to ripening is very important for fruit ripening, and at the same time ABA and IAA have complex interaction during this stage, responses of ABA signal transduction core component genes to ABA and IAA are also very important to the regulation of sweet cherry fruit ripening. Through exogenous phytohormones application, we observed that among the three gene families studied in this work, only PaPP2C3/5/6 and PaSnRK2.6 significantly responded to ABA treatment (expression changes over 50 %) (Fig. 5b, c), which was similar to what had been found in tomato that only few members of the three gene families respond to exogenous ABA treatment at breaker stage (Sun et al. 2011). By comparing the expression patterns during the whole fruit development and their responses to ABA, we can presume that ABA could not be the only and main regulator in controlling the variation of its signal transduction core component genes’ expression during the whole sweet cherry fruit development. Another evidence which can support this hypothesis is that none of the studied genes’ expression patterns coincide exactly with the ABA variation during the whole period (Fig. 4). With regard to IAA, although it can delay fruit ripening and interact with ABA at the beginning of sweet cherry fruit ripening, it may not have direct effects on the expression of PaPYLs and PaPP2Cs which can be presumed from the similar responses of these two genes families to both ABA and IAA (Fig. 5a, b). The similar responses caused by IAA may through its effects on the ABA content. However, interestingly, within the PaSnRK2s gene family, some opposite responses were found between ABA and IAA treatment (Fig. 5c). Therefore, IAA may have some effects on regulating the expression of PaSnRK2s, especially PaSnRK2.4/2.5, or at least not fully through ABA.
Between the two rapid growth stages is the pit hardening stage, during which the ABA level was relatively stable. However, the expression of PaPYL2 and 3 changed dramatically at this stage. One possible explanation is the hysteresis effect of PaPYLs’ expression response to the ABA variation (Fig. 4a).
Based on what mentioned above, the expression of PaPYLs, PaPP2Cs and PaSnRK2s was related to sweet cherry fruit development and ripening and in response to exogenous ABA and IAA treatment at the initiation of fruit ripening. And this relationship is more intense than that of CHLH during sweet cherry fruit development and ripening (Ren et al. 2010). Thus, incorporation with our previous studies on sweet cherry, it can be presumed that at the onset of sweet cherry fruit ripening, ABA has an important and positive effect on promoting fruit ripening, while IAA has a negative effect on the start of fruit ripening; at transcriptional level, the endogenous ABA level was regulated by the expression of both PacNCED1, a key enzyme in the ABA biosynthetic pathway, and PaCYP707As, key enzymes in the oxidative catabolism of ABA (Ren et al. 2010; Luo et al. 2014); in the aspect of ABA signal transduction, the PYL–PP2C–SnRK2 pathway may play a more important role in regulating of sweet cherry fruit development and ripening than that of the CHLH–WRKY pathway (Ren et al. 2011), in addition, based on their sequence characteristics and expression levels, PaPYL2/3, PaPP2C3/4 and PaSnRK2.4/2.5 may play a more important role in transducing ABA signal and regulating fruit development and ripening. We hope all above information could reveal some basic information about the regulation of ABA signal transduction core component genes in sweet cherry fruit maturation and help us gain a deeper understanding about ABA and IAA’s function on sweet cherry fruit ripening at the molecular level. However, a more clear ABA signal transduction pathway during sweet cherry fruit development and ripening can not only be based on the transcriptional regulation, a protein–protein interaction analysis is very necessary and critical. Thus, direct test at protein level such as yeast-two-hybrid should be performed in the future studies.
Abbreviations
- ABA:
-
Abscisic acid
- CHLH:
-
Magnesium chelatase H subunit
- DAFB:
-
Days after full bloom
- IAA:
-
Indole acetic acid
- PP2C:
-
Type 2C protein phosphatase
- PYL:
-
Pyrabactin resistance like
- PYR1:
-
Pyrabactin resistance 1
- RCAR:
-
Regulatory components of ABA receptor
- SnRK2:
-
Subfamily 2 of SNF1-related kinases
- VIGS:
-
Virus induced gene silencing
References
Alkio M, Jonas U, Sprink T, Van Nocker S, Knoche M (2012) Identification of putative candidate genes involved in cuticle formation in Prunus avium (sweet cherry) fruit. Ann Bot 110:101–112
Boneh U, Biton I, Zheng C, Schwartz A, Ben-Ari G (2012) Characterization of potential ABA receptors in Vitis vinifera. Plant Cell Rep 31:311–321
Chai YM, Jia HF, Li CL, Dong QH, Shen YY (2011) FaPYR1 is involved in strawberry fruit ripening. J Exp Bot 62:5079–5089
Crane JC (1969) The role of hormones in fruit set and development. HortScience 4:108–111
Given NK, Venis MA, Grierson D (1988) Hormonal regulation of ripening in the strawberry, a non-climacteric fruit. Planta 174:402–406
Huai JL, Wang M, He JG, Zheng J, Dong ZG, Lv HK et al (2008) Cloning and characterization of the SnRK2 gene family from Zea mays. Plant Cell Rep 27:1861–1868
Jia HF, Chai YM, Li CL, Lu D, Luo JJ, Qin L, Shen YY (2011) Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol 157:1188–1199
Kondo S, Gemma H (1993) Relationship between abscisic acid (ABA) content and maturation of the sweet cherry. J Jpn Soc Hortic Sci 62:63–68
Kondo S, Inoue K (1997) Abscisic acid (ABA) and 1-aminocyclopropane-1-carboxylic acid (ACC) content during growth of ‘Satohnishiki’ cherry fruit, and the effect of ABA and ethephon application on fruit quality. J Hortic Sci 72:221–227
Liu BH, Jiang YM, Peng FT, Sui J, Zhao FX, Wang HY (2007) The dynamic changes of endogenous hormones in sweet cherry (Prunus avium L.) pulp. Acta Hortic Sin 34:1535–1538
Luo H, Dai SJ, Ren J, Zhang CX, Ding Y, Li Z, Sun Y, Ji K, Wang YP, Li Q, Chen P, Duan C, Wang Y, Leng P (2014) The role of aba in the maturation and postharvest life of a nonclimacteric sweet cherry fruit. J Plant Growth Regul 33:373–383
Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A et al (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324:1064–1068
Mapelli S, Frova C, Torti G, Soressi GP (1978) Relationship between set, development and activities of growth regulators in tomato fruits. Plant Cell Physiol 19:1281–1288
Melcher K, Ng LM, Zhou XE, Soon FF, Xu Y, Suino-Powell KM et al (2009) A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature 462:602–608
Nishimura N, Hitomi K, Arvai AS, Rambo RP, Hitomi C, Cutler SR et al (2009) Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science 326:1373–1379
Nitsch JP (1988) Hormonal factors in growth and development. In: Hulme AC (ed) The biochemistry of fruits and their products. UK Academic Publishers, London, pp 427–472
Ozga JA, Reinecke DM (2003) Hormonal interactions in fruit development. J Plant Growth Regul 22:73–81
Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y et al (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324:1068–1071
Rabino I, Mancinelli AL (1986) Light, temperature, and anthocyanin production. Plant Physiol 81:922–924
Ren J, Sun L, Wu JF, Zhao SL, Wang CL, Wang YP et al (2010) Cloning and expression analysis of cDNAs for ABA 8′-hydroxylase during sweet cherry fruit maturation and under stress conditions. J Plant Physiol 167:1486–1493
Ren J, Sun L, Wang CL, Zhao SL, Leng P (2011) Expression analysis of cDNA for magnesium chelatase H subunit (CHLH) during sweet cherry fruit ripening and under stress conditions. Plant Growth Regul 63:301–307
Romero P, Lafuente MT, Rodrigo MJ (2012) The Citrus ABA signalosome: identification and transcriptional regulation during sweet orange fruit ripening and leaf dehydration. J Exp Bot 63:4931–4945
Santiago J, Florine D, Adam R, Regina A, Sang-Youl P, Marc J et al (2009) The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 462:665–668
Setha S, Kondo S, Hirai N, Ohigashi H (2005) Quantification of ABA and its metabolites in sweet cherries usingdeuterium-labeled internal standards. Plant Growth Regul 45:183–188
Shen YY, Wang XF, Wu FQ, Du SY, Cao Z, Shang Y et al (2006) Nature 443:823–826
Shen CJ, Wang SK, Bai YH, Wu YR, Zhang SN, Chen M et al (2010) Functional analysis of the structural domain of ARF proteins in rice (Oryza sativa L.). J Exp Bot 14:3971–3981
Shen XJ, Zhao K, Li LL, Zhang KC, Yuan HZ, Liao X, Wang Q, Guo XW, Li F, Li TH (2014) A role for PacMYBA in ABA-regulated anthocyanin biosynthesis in red-colored sweet cherry cv. Hong Deng (Prunus avium L.). Plant Cell Physiol 55:862–880
Stern RA, Flaishman M, Applebaum S, Ben-Arie R (2007) Effect of synthetic auxins on fruit development of ‘Bing’ cherry (Prunus avium L.). Sci Hortic 114:275–280
Sun L, Wang YP, Chen P, Ren J, Ji K, Li Q et al (2011) Transcriptional regulation of SlPYL, SlPP2C, and SlSnRK2 gene families encoding ABA signal core components during tomato fruit development and drought stress. J Exp Bot 62:5659–5669
Sun L, Sun YF, Zhang M, Wang L, Ren J, Cui MM, Wang YP, Ji K, Li P, Li Q, Chen P, Dai SJ, Duan CR, Wu Y, Leng P (2012) Suppression of 9-cis-epoxycarotenoid dioxygenase, which encodes a key enzyme in abscisic acid biosynthesis, alters fruit texture in transgenic tomato. Plant Physiol 158:283–298
Talon M, Zacarias L, Primo-Millo E (1990) Hormonal changes associated with fruit set and development in mandarins differing in their parthenocarpic ability. Physiol Plant 79:400–406
Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K et al (2009) Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci USA 106:17588–17593
Umezawa T, Nakashima K, Miyakawa T, Kuromori T, Tanokura M, Shinozaki K, Yamaguchi-Shinozaki K (2010) Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol 51:1821–1839
Wan CY, Wilkins TA (1994) A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.). Anal Biochem 223:7–12
Wang YP, Wang Y, Kai W, Zhao B, Chen P, Sun L et al (2014) Transcriptional regulation of abscisic acid signal core components during cucumber seed germination and under Cu2+, Zn2+, NaCl and simulated acid rain stresses. Plant Physiol Biochem 76:67–76
Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ (2009) Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191
Wu FQ, Xin Q, Cao Z, Liu ZQ, Du SY, Mei C et al (2009) The magnesium-chelatase h subunit binds abscisic acid and functions in abscisic acid signaling: new evidence in Arabidopsis. Plant Physiol 150:1940–1954
Yoshida R, Umezawa T, Mizoguchi T, Takahashi S, Takahashi F, Shinozaki K (2006) The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in arabidopsis. J Biol Chem 281:5310–5318
Yu YM, Liu CX, Zhang WH, Wang ZF, Cao QW, Sun XL (2008) An improved HPLC method for detecting inner hormones in cucumber fruit. Shandong Agric Sci 7:97–99
Yuasa T, Tomikubo Y, Yamauchi T, Inoue A, Iwaya-Inoue M (2007) Environmental stresses activate a tomato SNF1-related protein kinase 2 homolog, SlSnRK2C. Plant Biotechnol 24:401–408
Zhang M, Yuan B, Leng P (2009) The role of ABA in triggering ethylene biosynthesis and ripening of tomato fruit. J Exp Bot 60:1579–1588
Acknowledgments
We thank Dr. Merianne Alkio from University of Hannover for providing the sweet cherry contig database and for the helpful advice in the revision of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, Y., Chen, P., Sun, L. et al. Transcriptional regulation of PaPYLs, PaPP2Cs and PaSnRK2s during sweet cherry fruit development and in response to abscisic acid and auxin at onset of fruit ripening. Plant Growth Regul 75, 455–464 (2015). https://doi.org/10.1007/s10725-014-0006-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-014-0006-x