Abstract
The interactions of major topographic features with climate fluctuations over time are key factors that influence the spatial genetic structure of trees species. As an important characteristic of China’s terrain, the boundary between the mountainous West China and the lowland East China has been found to contribute to the divergence of many species. In this study, we examined whether the stepped geomorphology of China has shaped the genetic structure of Quercus acutissima, a widespread oak species native to East Asia, at a much larger geographic scale. Thirty natural populations of the species were sampled across the entire distribution in China and genotyped using ten nuclear microsatellite markers. To determine the genetic structure and demographic history, we conducted Bayesian cluster analysis and approximate Bayesian computation. A significant east-west divergence was detected at the species level, with western populations presenting much stronger differentiation and lower genetic diversity than eastern populations. The divergence between the groups of Southwest China and East China was estimated to have occurred 3.55–2.37 million years ago (Ma), closely associated with the recent uplift of the Qinghai-Tibetan Plateau (QTP) and climate changes during the late Pliocene to the early Pleistocene. The Central China group may arise from the admixture of these two groups, which was dated to 0.83–0.55 Ma, a period covering several interglacial intervals with unusually warm and wet climate in Central China. The spatial genetic structure of Q. acutissima indicates the possibility of central-marginal dispersal dynamics along latitude and existence of multiple refugia at distribution edges. Our findings highlight that the stepped geomorphology in China and climatic changes since the Pliocene play an important role in shaping the intraspecific genetic structure of widespread species in East Asia.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The geological history of China has shaped the evolutionary dynamics of endemic species in a way that differs from other well-studied regions of the world, such as Europe and North America (e.g., Kou et al. 2015; Meng et al. 2017). Topographic changes, such as the uplift of the Qinghai-Tibetan Plateau (QTP), and climatic shifts, such as the intensification of East Asian monsoons, and aridification in Central Asia, have exerted a significant influence on the evolution of the flora of China (Chen et al. 2017; Yu et al. 2017; Lu et al. 2018). Several critical periods marked by a close relationship between geological/palaeoclimatic events and intraspecific/interspecific evolutionary events have been identified, e.g., the middle Miocene (Kou et al. 2015; Bai et al. 2016; Meng et al. 2017), the late Pliocene to the early Pleistocene (Chen et al. 2015; Wang et al. 2017; Ye et al. 2018; Zhao et al. 2018), shortly after the mid-Pleistocene transition (Tian et al. 2015; Ye et al. 2018), the Last Interglacial and the Last Glacial Maximum (Tian et al. 2015; Xu et al. 2016; Fan et al. 2018). However, most recent phylogeographic investigations were carried out based on species that inhabit a separate climate region or geographic unit of East Asia, e.g., subtropical China, Qinghai-Tibetan Plateau, and northern China. Only a few focused on the impact of palaeoenvironmental changes on the intraspecific divergence and range dynamics of widespread species across multiple climatic zones or floristic regions (e.g., Chen et al. 2012; Guo et al. 2014; Ye et al. 2017). Hence, more studies at a larger geographic scale are expected to provide new insight into the diversity of evolutionary patterns amid the complex topography and climatic heterogeneity in East Asia.
Oaks (Quercus spp.) are one of the most widely distributed forest-tree genera in the Northern Hemisphere (Hubert et al. 2014) and are considered to be models to integrate ecology, evolution, and genomics (Petit et al. 2013). The evolutionary histories of oaks are strongly influenced by palaeoenvironmental changes in different ways, such as historical introgression/hybridization triggered by range fluctuations (Bagnoli et al. 2015; Ortego et al. 2017), interspecific divergence along with formation of geographical barriers (Cavender-Bares et al. 2015; Rodríguez-Correa et al. 2017), and lineage diversification caused by local adaptation to diverging climates (Meng et al. 2017; Hipp et al. 2018). Climatic forces and environmental heterogeneity have been found to be key factors driving the intraspecific genetic divergence of oaks (Ortego et al. 2012; Gugger et al. 2013; Riordan et al. 2016). Meanwhile, intraspecific admixture pattern of oaks is also associated with the stability of the local habitat. Recent studies have shown that higher stability may contribute to genetic connectivity across different climatic periods, whereas lower habitat stability may promote the genetic admixture among regions (Ortego et al. 2015).
Quercus acutissima Carruthers, one of the members of section Cerris, is a dominant species in warm temperate deciduous forests of East Asia (Fujiwara and Harada 2015). As an ecologically important and economically valuable tree species, it has been listed as one of the precious wood species by the government. In China, its modern distribution range extends from 18°N to 41°N latitude and from 91°E to 123°E longitude, across seven temperature zones and three moisture regions of China (Zhang et al. 2014a, 2015). The historical distribution of Q. acutissima remains unknown, but leaf and fruit fossils of morphologically closely related species from section Cerris were observed in the Neogene floras of southwestern China and northern China (Zhou 1993; Song et al. 2000), suggesting a probable widespread palaeodistribution of Q. acutissima in this region.
A previous study based on chloroplast markers indicated that Q. acutissima may have two localized glacial refugia in Central China and Southwest China, but the phylogeographic structure was weak (Zhang et al. 2015). Population genetic analysis of 28 provenances implied that a nuclear genetic differentiation may occur between East China and West China (Zhang et al. 2013a). Other researchers also described the genetic variation of Q. acutissima in China, South Korea, and Japan (Chung et al. 2002; Choi et al. 2005; Ye and Zeng. 2009; Zhang et al. 2013b; Saito et al. 2017). However, the information on the evolutionary history of the species was limited by small sample size and a narrow sampling range.
Due to the widespread distribution and the dominant position in warm temperate deciduous forests of East Asia, Quercus acutissima provides a useful model to evaluate the impact of the stepped geomorphology in China on the intraspecific genetic structure and demographic history of a tree species at a larger geographic scale. Here, we used ten nuclear microsatellite (nSSR) markers to assess the genetic variation of Q. acutissima based on 30 natural populations sampled throughout the distribution in China. The specific goals of this work were to (1) examine range-wide patterns of intraspecific genetic diversity, (2) investigate whether a distinct genetic structure exists, (3) infer the intraspecific evolutionary history and evaluate the effects of geological/palaeoclimatic events, and (4) reveal possible locations of glacial refugia.
Materials and methods
Population sampling
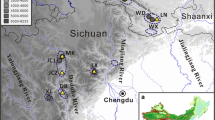
A total of 707 individuals were sampled from 30 natural populations across the entire distribution of Q. acutissima in China during the spring and summer of 2014 (Fig. 1). In each population, eight to ten fresh leaves per tree were collected from 10 to 30 adult individuals at least 30 m apart from each other. Leaf tissues were quickly dried with silica gel and stored at room temperature in the laboratory. Spatially explicit information of each sampled population, including latitude, longitude, and elevation, was recorded using a handheld GPS unit (Magellan, USA) (Tables 1 and S1).
Bayesian cluster analysis for 30 Quercus acutissima populations. Area of the pie is proportional to the probability of membership to the West China group (green) or the East China group (red). Small dots indicate the subgroup each population belongs to when we divided all the populations into seven subgroups. The bottom right map shows ranges of Southwest China (SW), Central China (C), and East China (E) groups. Light blue dashed line represents the boundary between the second step region and the third step region of China’s terrain. Population codes are described in Tables 1 and S1
DNA extraction and microsatellite genotyping
Total genomic DNA was extracted from 30 mg silica gel-dried leaf tissue of each individual using a Plant Genomic DNA Kit (Tiangen, Beijing, China). The quality and concentration of the genomic DNA were evaluated by electrophoresis with 1% agarose gels and one drop spectrophotometer (OD-1000, Shanghai Cytoeasy Biotech Co., Ltd., China), respectively. DNA samples were diluted to 20 ng/μL and stored at − 20 °C for PCR amplification.
All the 707 DNA samples were genotyped at 12 nSSR loci using primers developed for Q. acutissima or other oak tree species, including quru-GA-0M05 and quru-GA-0M07 (Aldrich et al. 2002); MSQ16 (Dow and Ashley 1996); QM67-3M1 (Isagi and Suhandono 1997); ssrQrZAG4, ssrQrZAG7, ssrQrZAG11, ssrQrZAG20, and ssrQrZAG112 (Kampfer et al. 1998); ssrQpZAG9 and ssrQpZAG110 (Steinkellner et al. 1997); and EE743802 (Zhang et al. 2013a). The forward primers were labeled with fluorescent dye, 6-FAM, or HEX (Applied Biosystems, USA). Polymerase chain reactions (PCRs) were performed using a Mastercycler pro Thermal Cycler (Eppendorf, Germany) in 20-μL volumes containing 2× Taq PCR MasterMix (Tiangen, Beijing, China), 10 μM of each primer, 20–40 ng of template DNA and ddH2O. The PCR protocols were designed as an initial denaturing of 4 min at 94 °C, followed by 30 cycles of 45 s at 94 °C, 45 s of annealing at a primer-specific temperature (Table 2), 45 s of elongation at 72 °C, and ending with a final extension of 8 min at 72 °C. PCR products were separated on an ABI3730xl automated Genetic Analyzer using ROX-500 as an internal standard (Applied Biosystems, USA). Allele sizes were determined manually using Genemarker version 2.2.0 (Applied Biosystems, USA).
Data analysis
Null alleles
The frequency of null alleles at each locus in each population was estimated using INEST version 2.2 (Chybicki and Burczyk 2009) based on an individual inbreeding model (IIM) simultaneously including three parameters, i.e., genotyping failures (b), inbreeding coefficients (f), and null alleles (n). This approach was implemented with 500,000 iterations, discounting 50,000 iterations as burn-in, and sampling every 50th update. We compared deviance information criterion (DIC) values between models with null alleles (nfb) and models without null alleles (fb) to evaluate the significance of null alleles in each population. A smaller DICnfb than DICfb indicated that the model with null alleles was favored; in other words, we can verify the significance of null alleles in the model. Using this method, we found that the mean null allele frequency of quru-GA-0M05 and EE743802 across all the 30 populations were 0.21 and 0.13, respectively, greater than the threshold of 0.10. These two loci were then excluded from all subsequent analyses.
Genetic diversity
Genetic diversity statistics were estimated using POPGENE version 1.32 (number of alleles, NA; effective number of alleles, NE; observed heterozygosity, HO; Yeh et al. 1999) and FSTAT version 2.9.3.2 (allelic richness, AR; genetic diversity within populations, HS; inbreeding coefficient, FIS; Goudet 2001) for each locus and each population. Linkage disequilibrium (LD) for all locus pairs in each population and significant deviations from Hardy-Weinberg equilibrium (HWE), as indicated by deviations of FIS from zero, were tested by randomization using FSTAT. The obtained P values were adjusted using a sequential Bonferroni correction (Rice 1989). Since the presence of null alleles may affect the estimation of FIS, a corrected inbreeding coefficient (FIS′) was also evaluated for each population with INEST version 2.2 using the full model (nfb). The significance of FIS′ was assessed by comparing DIC values between the full model and the model without inbreeding (nb).
To estimate the effect of reduction in population size on genetic diversity due to bottlenecks, the Wilcoxon sign-rank test was implemented in Bottleneck version 1.2.02 (Cornuet and Luikart 1996) under two mutation models: the stepwise mutation model (SMM) and the two-phase mutation model (TPM) with 70% stepwise mutations and 30% multistep mutations. We performed 10,000 simulations for each population.
Genetic differentiation
We estimated both global FST (Weir and Cockerham 1984) and the standardized measure G′ST (Hedrick 2005) for each locus and over all loci in MSA version 4.05 (Dieringer and Schlotterer 2003), with the significance tested by 10,000 permutations. To evaluate the impact of null alleles on estimation of genetic differentiation among populations, we also calculated FST with a correction using the ‘exclusion null alleles’ (ENA) method (cFST) in FreeNA (Chapuis and Estoup 2007). The 95% confidence intervals (CIs) of both global FST and cFST across loci were obtained through bootstrap re-sampling in the same program. In addition, we applied the allele size permutation test (Hardy et al. 2003) in SPAGeDi version 1.5 (Hardy and Vekemans 2002) to detect the significant influence of stepwise-like mutations on genetic structuring, as indicated by RST > FST, where RST is an FST analog based on allele size variance (Slatkin 1995).
We tested the significance of differences in genetic statistics AR, HO, HS, and FST among geographic regions identified by STRUCTURE using FSTAT version 2.9.3.2. The two-sided P values were obtained after 10,000 permutations. A hierarchical analysis of molecular variance (AMOVA) was performed in ARLEQUIN version 3.5 (Excoffier and Lischer 2010) to examine the pattern of genetic variance partitioned among regions (sub-regions), among populations within regions (sub-regions), and within populations. The significance of fixation indices (FCT, FSC, and FST) was tested with 10,000 permutations. To test isolation by distance (IBD), a Mantel test with 10,000 random permutations was also carried out using GENALEX 6.5 (Peakall and Smouse 2012) between the matrix of pairwise FST/(1-FST) and the matrix of logarithm of the geographic distances.
Genetic structure
We used Bayesian cluster analysis to detect the genetic structure of Q. acutissima using STRUCTURE version 2.3.4 (Pritchard et al. 2000). A LOCPRIOR model was applied to use sampling locations as prior information (Hubisz et al. 2009). Twenty independent runs for each K (from 1 to 12) were performed with a 500,000 burn-in period and 500,000 repetitions. The optimal number of clusters (K) was determined through the delta K method as described in Evanno et al. (2005). Values of membership coefficient (q) were post-processed over the 20 runs using CLUMPP version 1.1.2 (Jakobsson and Rosenberg 2007) and displayed with DISTRUCT version 1.1 (Rosenberg 2004). Complementary to Bayesian cluster analysis, we also performed principal coordinate analysis (PCoA) based on pairwise population FST values using GENALEX 6.5 (Peakall and Smouse 2012) and conducted UPGMA cluster analysis in NTSYS-pc version 2.1 (Rohlf 1999) based on Nei’s unbiased genetic distances (Nei 1978) calculated by POPGENE version 1.32 (Yeh et al. 1999).
Gene flow among regions
We used a maximum likelihood procedure and a Brownian motion microsatellite model as implemented in MIGRATE-N version 3.6.11 (Beerli 2006) to estimate the mutation-scaled population size (θ) and immigration rates (M) among the three geographic regions identified by STRUCTURE. Three independent replicates were run with ten short chains (10,000 sampled trees for each chain), five long chains (100,000 sampled trees for each chain), and the first 10,000 sampled trees discarded as burn-in. The adaptive heating scheme at four temperatures (1.0, 1.5, 3.0, and 6.0) was used to efficiently search the genealogy space.
Demographic history
The approximate Bayesian computation (ABC) procedure was carried out in DIYABC version 2.1.0 (Cornuet et al. 2014) to infer the probable demographic history of the three genetic groups identified in this study. Specifically, the Southwest China group (SW) comprises 61 individuals from Southwest China and the Hainan Island, the Central China group (C) comprises 258 individuals from Central China and the Loess Plateau, and the East China group (E) comprises 388 individuals from East China and the Tianmu Mountain. Eight scenarios were compared to test different hypotheses about glacial refugia: three refugia in Southwest China, Central China, and East China (scenario 1); two refugia in Southwest China and Central China (scenario 2); and one refugium in East China (scenario 3), Central China (scenario 4), or Southwest China (scenarios 5–8) (Fig. 2 and Table S2). The analysis was run five times independently, with one million simulations for each scenario in each replicate. The prior distributions of historical parameters are shown in Table S3. Eighteen summary statistics including mean number of alleles, mean genic diversity for three regions, FST, classification index, and shared allele distance between pairs of regions were defined in the ABC analysis. The 1% simulated data closest to the observed data was used to estimate the posterior distributions of historical parameters and the relative posterior probabilities of each scenario via a logistic regression approach. The model checking process was performed to evaluate the goodness of fit for the preferred scenario.
Eight demographic scenarios tested by approximate Bayesian computation (ABC). NA, NSW, NC, and NE are effective population sizes of the common ancestor (A), the Southwest China group (SW), the Central China group (C), and the East China group (E). Parameters t1, t2, and t3 are times of occurrence of various events including population size variation, divergence, and admixture, which are counted in generations and described in Table S2. Parameters rA and 1-rA are the admixture rates from the Southwest China group to the Central China group and from the East China group to the Central China group
Results
Genetic diversity of microsatellite loci and populations
Mean null allele frequency at each of the ten loci was estimated to be lower than the threshold of 0.10 across all the 30 populations (Table 2). Comparison of DIC values showed that the model with null alleles was slightly favored in 22 populations of Q. acutissima (Tables 1 and S4), while estimated frequency averaged across loci was relatively low (range 0.02–0.07; Table 1). Ten populations (TC, LN, TE, MX, AK, TB, NJ, LYG, QHD, and WTM) had an inbreeding coefficient that significantly deviated from zero (P < 0.05 after Bonferroni correction), but these signs of departure from HWE were not shown after excluding the bias introduced by the existence of null alleles (Tables 1 and S4). No evidence of significant LD was observed for each pair of loci in each population (P < 0.05 after Bonferroni correction), indicating that all loci were inherited independently and thus can be used for investigating the genetic variation of Q. acutissima.
At the population level, range of mean effective number of alleles (NE) was 2.7–4.7 (mean 4.0), allelic richness (AR) was 4.1–6.5 (mean 5.7), and genetic diversity within populations (HS) was 0.572–0.763 (mean 0.709; Table 1). Higher intrapopulation genetic diversity was observed in East China than in West China (P = 0.007 for AR; P < 0.001 for HS; Table 3). Central populations between 26°N and 35°N also presented higher genetic diversity than southern populations (P = 0.014 for AR; P = 0.022 for HS) and northern populations (P = 0.044 for AR; except for P = 0.117 for HS; Table S5). Additionally, a significant quadratic regression relationship was detected between latitude and genetic diversity statistics for all populations (R = 0.500, P = 0.020 for AR; R = 0.515, P = 0.016 for HS), and in both Southwest-Central China (R = 0.659, P = 0.043 for AR; R = 0.789, P = 0.005 for HS) and East China (R = 0.686, P = 0.016 for AR; except for P = 0.082 for HS; Fig. S1), suggesting that central populations have higher genetic diversity than southern and northern populations. No signal of recent bottleneck was detected in each population based on the stepwise mutation model (SMM) and the two-phase mutation model (TPM), except for in YF under the stepwise mutation model (P = 0.016; Table S6).
Genetic differentiation among populations and regions
The impact of null alleles on the estimation of population differentiation was very mild, as indicated by the almost equal values and 95% CIs for both global FST (0.056, 95% CI: 0.049–0.063) and cFST (0.056, 95% CI: 0.049–0.062). Genetic differentiation at each locus was significant (P < 0.001) but relatively low (Table 2). The observed value of multilocus RST (0.058) laid between the upper and lower limits of the 95% CI of the null distribution of the permuted RST (0.034–0.075), and the difference was not statistically significant (P = 0.710). None of the global RST values for each locus was significantly larger than the permuted RST (P > 0.05; Table 2), suggesting that RST was equal to FST and the stepwise-like mutations have not contributed to the genetic divergence of Q. acutissima.
Results of AMOVA indicated significant genetic differentiation between West China and East China (FCT = 0.019, P < 0.0001), whereas only 1.85% of the genetic variation was partitioned among groups (Table 4). At the population level, a lower FST was detected in East China than in West China (P = 0.005) or in Southwest China (P = 0.022; Table 3), implying stronger genetic differentiation among populations within the West China region. Additionally, a significant pattern of isolation by geographic distance was observed for all the 30 populations (R = 0.346, P < 0.001), West China populations (R = 0.548, P < 0.001), and Central China populations (R = 0.292, P = 0.039), but not for East China populations (P > 0.05; Fig. S2).
Genetic structure and gene flow
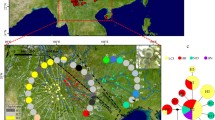
Bayesian cluster analysis showed that the optimal number of genetic groups (K) was two (Fig. 3a). All the 30 populations of Q. acutissima were divided into the West China group and the East China group, across a well-known boundary of the stepped geomorphology of China, which separates the second step region characterized by plateaus with higher elevation of 1000–2000 m, and the third step region characterized by plains with lower elevation < 500 m (Fig. 1 and Table 1). A significant decline in the probability of membership to the West China cluster (qW) in each population was detected with increasing longitude, latitude, and decreasing elevation (all P values < 0.001; Fig. 3d–f). According to this trend, we then subdivided all the populations into three geographic regions: (1) Southwest China with qW > 0.75, (2) East China with qW < 0.25, and (3) Central China, an admixed cluster of the former two regions, with 0.25 ≤ qW ≤ 0.75 (Figs. 1 and 3g; Table 1). The Central China region can further be subdivided into the West Central China sub-region with 0.50 < qW ≤ 0.75 and the East Central China sub-region with 0.25 ≤ qW ≤ 0.50. The STRUCTURE result of K = 4 is shown in Figs. 1 and 3g. Three of the four genetic clusters were found to be dominant only in a single population at the distribution edge, i.e., cluster I in the southern-most population CJ (qI = 0.948), cluster II in the northwestern-most population HL (qII = 0.856), and cluster III in the southeastern-most population WTM (qIII = 0.826) (Table S7). Cluster IV was not only dominant in the northeastern-most populations such as QHD (qIV = 0.922), DL (qIV = 0.892), and ZH (qIV = 0.891), but also in other populations in East China (Table S7). Significant linear correlations were observed between latitude and the probability of membership to the genetic cluster I (qI) and II (qII) in each population from West China (all P values < 0.01; Fig. 3b, c). Based on the STRUCTURE result of K = 4, we further divided all the 30 populations into seven sub-regions (Tables 1 and 5, Fig. 1). Both PCoA and UPGMA cluster analysis showed good consistency with STRUCTURE, except for populations TB and NS nested within the East China cluster in the UPGMA dendrogram (Figs. 4 and S3).
STRUCTURE results for 707 Quercus acutissima individuals from 30 populations. a Delta K statistics calculated according to Evanno et al. (2005). b, c Linear correlations between latitude and the probability of membership (q) to the cluster I (qI) and the cluster II (qII) in each West China population when K = 4. d–f Linear correlations between geographic information and the probability of membership to the West China cluster (qW) in each population when K = 2. g Histogram of individual assignments when K = 2, 3, and 4. Each vertical bar represents one individual. Roman numerals show genetic clusters when K = 4. Colors of dots in b–f and of horizontal bars in g represent different geographic regions (sub-regions) as shown in Fig. 1. Population and region codes are shown in Tables 1 and S1
Taking the three main regions identified by STRUCTURE into account, all six pairwise maximum likelihood estimates of mutation-scaled immigration rate were significant with no 95% CIs overlapping zero (Table 6). The evidence of distinctly asymmetric historical gene flow was detected among three region-pairs, with migration direction from Southwest China to Central China (27.31 vs. 11.58), from East China to Central China (39.88 vs. 27.00), as well as from Southwest China to East China (20.15 vs. 13.86).
Demographic history
ABC analysis showed that scenario 8 (ancient divergence with recent admixture model, Fig. 2) had the highest posterior probability (0.491, 95% CI: 0.473–0.510) across all the five runs. No overlap of 95% CIs was observed with the other seven scenarios (Table S2). A good fit to the observed data was assessed by 16 of 18 summary statistics not significantly different from the observed values. The mean median values of t1 (the admixture time between SW and E), t2 (the divergence time between SW and E), and t3 (the time of ancestral effective population size variation) across the five runs were 5508 (95% CI: 309–23,300), 23,680 (95% CI: 4400–47,700), and 106,400 (95% CI: 24,900–195,000) generations, respectively (Table S8). Similar admixture rates, rA, were detected between SW/E and C (SW → C = 0.511; E → C = 0.489; Table S8). The mean median values of the effective population sizes NSW, NC, NE, and NA were 254,400, 535,600, 617,400, and 300,000, respectively (Table S8).
Discussion
Genetic east-west differentiation revealed by nuclear markers and discordant pattern at chloroplast genome
Topographic differences between the mountainous West China and the lowland East China may have affected the intraspecific differentiation patterns of extant plant species in this region (Shi et al. 2014). Previous studies have observed an obvious phylogeographic break between West China and East China, such as in Juglans cathayensis (Bai et al. 2014), Castanopsis eyrei (Shi et al. 2014), Castanopsis fargesii (Li et al. 2014; Sun et al. 2014), and Castanopsis carlesii (Sun et al. 2016). However, most recent studies are restricted in subtropical China. Our investigation of Q. acutissima confirms that a significant nuclear east-west divergence occurs at a much larger geographic scale, coincident with Zhang et al. (2013a), which examined the genetic structure of 28 provenances of Q. acutissima at a regional scale. Such a finding strongly supports the hypothesis that the boundary between the second step region and the third step region of China’s terrain plays an important role in shaping the genetic structure of widespread tree species in East Asia. Furthermore, a distinct genetic pattern across the boundary was also detected for Q. acutissima. The West China group, characterized by habitats with more complex topography, exhibited a lower level of genetic diversity, a higher level of population differentiation, and a stronger pattern of isolation-by-distance (Table 3 and Fig. S2), implying that topography heterogeneity in West China has exerted a significant influence on the intraspecific genetic structure of Q. acutissima, probably through long-term isolation with restricted gene flow among populations.
In contrast to the above, Zhang et al. (2015) did not detect a clear longitudinal divergence of Q. acutissima at the chloroplast genome level. Instead, the most common chlorotype (H1) was identified in 23 of 30 populations in China, occurring at six of seven sub-regions defined in this study. The diagnostic power of intraspecific structure was thus reduced for chloroplast markers. Similar cases were also reported in plants of subtropical China, such as Castanopsis eyrei (Shi et al. 2014) and Sargentodoxa cuneata (Tian et al. 2015). Indeed, the cytoplasmic-nuclear discordance is more common across closely related species (e.g., Qi et al. 2012; Jose-Maldia et al. 2017; Zhou et al. 2017). Markers associated with the farthest dispersing sex may better delimit species (Petit and Excoffier 2009). Previous studies have shown that in conifers, variation in paternally inherited chloroplast DNA was more species-specific than variation in maternally inherited mitochondrial DNA, and biparentally inherited nuclear DNA exhibited an intermediate pattern (Zhou et al. 2010; Zhou et al. 2017). In oaks, two hypotheses are usually used to explain the origin of shared chlorotypes among species, i.e., chloroplast capture (Acosta and Premoli 2010; Premoli et al. 2012), and incomplete lineage sorting (ILS; Simeone et al. 2016; Vitelli et al. 2017).
In our case at the intraspecific level, we integrate multiple clues to argue for the range expansions and incomplete lineage sorting. First, the star-like pattern of the chlorotype lineages implied that Q. acutissima may have experienced ancient range expansions during the interglacial periods (Tian et al. 2015). This process would have provided an opportunity for ancestral haplotypes, such as the most common haplotype (H1) that occupied the interior position in the network, to disperse to different parts of the distribution, which was a necessary condition for the formation of shared variation between groups. Second, recent studies pointed out that high intraspecific gene flow and high mutation rates may accelerate lineage sorting between species (Zhou et al. 2010). In oaks, gene flow through pollen was estimated to be 200 times higher than that through seeds (Ennos 1994; Mohammad-Panah et al. 2017). Thus, biparentally inherited nuclear markers may have experienced a faster lineage sorting between two geographic groups, whereas maternally inherited chloroplast markers likely retained ancestral polymorphisms for a long time. Finally, the current random distribution pattern of ancestral haplotypes throughout the whole range exactly coincides with the expectation of the ILS scenario (Zhou et al. 2017). If the extensive shared haplotypes arose from the genetic admixture between groups or chloroplast capture between species, it would be expected to be concentrated geographically in contact zones or adjacent areas (Zhou et al. 2010; Yang et al. 2016). Therefore, we conclude that the widespread distribution of ancestral chlorotypes was more likely a consequence of ancient range expansions combined with incomplete lineage sorting. Furthermore, recent repeated local range expansions (Tian et al. 2015; Fan et al. 2018; Ye et al. 2018) may also promote the haplotype sharing in Q. acutissima. These factors will give chloroplast markers limited efficiency in detecting intraspecific genetic structure.
Ancient divergence and recent admixture triggered by topographic changes and climatic fluctuations
Based on the STRUCTURE result, we subdivided all the populations of Q. acutissima into three genetic groups, namely Southwest China, Central China, and East China. The scenario of ancient divergence and recent admixture was strongly supported by ABC analysis. Assuming an average generation time of 100–150 years for Q. acutissima (Cavender-Bares et al. 2011; Zeng et al. 2015), the split between the East China group and the Southwest China group was estimated to have occurred during the late Pliocene to the early Pleistocene (3.55–2.37 Ma, mean 2.96 Ma). Compared to other oak species in China, this time of the initial divergence between major clades was slightly earlier than that of deciduous oaks, e.g., Q.variabilis (1.45 Ma, Chen et al. 2012) and Q. mongolica (1.38 Ma, Zeng et al. 2015), but much later than that of evergreen oaks, e.g., Q. glauca (9.07 Ma, Xu et al. 2015), Q. arbutifolia (10.25 Ma, Xu et al. 2016), and Q. aquifolioides (8.60 Ma, Du et al. 2017). This finding supports the hypothesis that the occurrence of deciduous oaks in China may be later than that of evergreen oaks, which was also inferred from the fossil history of Chinese oaks (Zhou 1993).
Our findings also indicated that the intraspecific divergence of Q. acutissima was likely associated with the recent uplift of the QTP and climate changes during the late Pliocene to the early Pleistocene in East Asia. Several forms of evidence have shown that the accelerating uplift of the QTP during the period of 3.4–2.6 Ma has increased the tectonic activities within the adjacent mountains and regions, e.g., uplift of the West Qinling Mountain and the formation of fault basins on the Yunnan-Guizhou Plateau (An et al. 1999). The increasing topographic differences between the second step region and the third step region may deepen the divergence between the eastern and western populations. Furthermore, East Asia experienced a significant change in climate around the Pliocene/Pleistocene boundary. Previous studies have shown that both the East Asian summer monsoon and the Indian summer monsoon displayed a trend of intensification during the period of 3.6–2.6 Ma (An et al. 2001; Chang et al. 2010), whereas the Asian interior was under the control of enhanced aridification (Zhang et al. 2014b), with the most easterly record observed at the eastern edge of the Loess Plateau (Li et al. 2015). Although the link between the rapid uplift of the QTP and climate changes is considered controversial (Wang et al. 2005), it is possible that the increased climate heterogeneity and topographic differences may drive the intraspecific divergence of Q. acutissima simultaneously.
Our analysis detected a recent intraspecific admixture event of Q. acutissima between the Southwest China group and the East China group, which was estimated to have occurred 0.83–0.55 Ma (mean 0.69 Ma) under the assumption of an average generation time of 100–150 years. This period comprises several interglacial intervals in the middle Pleistocene, including Marine isotope stages (MISs) 13, 15, 17, and 19. An extra-long interglacial may have persisted throughout MISs 15–13 (0.62–0.48 Ma), associated with the limited extent of Arctic ice sheets in glacial MIS 14 (Hao et al. 2015). Notably, an unusually warm and wet climate accompanied by the extremely strong East Asian summer monsoon may have existed in China during this time. Specifically, in the southern Loess Plateau, Guo et al. (1998) have recognized a period of greatest warmth since the last 1.2 million years, corresponding to MIS 13 and 15. According to their estimations, this area was likely under a subtropical semi-humid climate in MIS 13, with the annual mean temperature at least 4–6 °C higher and the annual rainfall 200–300 mm higher than at present. In the eastern Tibetan Plateau, the warmest interglacial over the last 0.8 million years was also related to MIS 13, and the appearance of Quercus pollen distinguished this time from other interglacials (Chen et al. 1999). More similar cases were also recorded in the paleosol sediments of southern China (Yin and Guo 2007). Interestingly, all the studied sites mentioned here were around the distribution of the Central China group, implying that a regional warm and humid climate may have occurred, at least in Central China. Under such a favorable condition, we infer that Q. acutissima may have expanded from Southwest China and East China refugia into the mountainous area of Central China, which then triggered the occurrence of genetic admixture along with the duration of recolonization. Our findings highlight that Central China was not only a potential glacial refugium (Zhang et al. 2015), but also likely a ‘melting pot’ of genetic diversity for Q. acutissima.
Genetic evidence for central-marginal hypothesis and multiple cryptic refugia at the distribution edges
For Q. acutissima, a gradual decline of intraspecific genetic diversity from center to margin along latitudinal gradients was observed at both the whole-distribution level and regional level (Fig. S1). Populations between 35°N and 26°N presented higher allelic richness and within-population genetic diversity than southern populations (< 26°N) and northern populations (> 35°N) (Table S5). This result exactly coincides with the expectation of the central-marginal (C-M) hypothesis, which predicts marginal populations have lower genetic diversity within populations due to small population size and increased spatial isolation (Micheletti and Storfer 2015). In East Asia, this prediction has been successfully verified in Euptelea pleiospermum (Wei et al. 2016). However, our study did not detect a C-M pattern along longitudinal gradient. Instead, a significantly higher genetic diversity was observed in East China than in West China, indicating again the profound influence of the topographic difference between the second step region and third step region of China’s terrain.
According to the STRUCTURE result of K = 4, our study also suggests the existence of multiple cryptic refugia in situ at distribution edges of Q. acutissima. Populations at the southern-most point (CJ), northwestern-most point (HL), and southeastern-most point (WTM) constituted three distinct single-population clusters (I–III). Another three populations (TC, KM, and FN) in the Southwest China sub-region were geographically close to CJ, with relatively higher qI (0.6–0.8). Finally, although cluster IV is common in both East China and Central China regions, populations dominated by this cluster with qIV larger than or approximately equal to 0.9 were only found in the northwestern-most part, e.g., TS, QHD, ZH, and DL (Table S7). Such a finding shows a strong signal for the existence of multiple refugia at the southwestern, southeastern, northwestern, and northeastern edges for Q. acutissima.
Based on maternally inherited chloroplast markers, Zhang et al. (2015) also uncovered a close genetic relationship among these four refugia. In their study, four haplotypes from the same lineage were found specific to Southwest China refugium (H2 in FN and TC), Southeast China refugium (H4 in QY), Northwest China refugium (H14 in HL), and Northeast China refugium (H9 in DL), presenting a distribution pattern similar to the genetic structure revealed by nSSR markers. Moreover, H2, the ancestor haplotype occupying the interior position of the network, generated H4, H9, and H14 through one to two single-site mutations, implying an ancient common origin in Southwest China and subsequent range expansions or long-distance dispersal events from Southwest China to the other three refugia during interglacials. This inference was consistent with the result of ABC analysis, which also provides evidence of the ancient common origin of the three main genetic groups in Southwest China.
Another explanation, although unlikely, for the close genetic relationship among these four refugia is human-mediated historical seed dispersal. A recent study has found that human activities resulted in a homogeneous genetic structure and lower genetic diversity of Q. acutissima in Japan (Saito et al. 2017). However, our study detected a distinct genetic structure (Fig. 1) and a central-marginal pattern of genetic diversity along latitude (Fig. S1). Additionally, the gradient change of probability of membership to the genetic cluster I and II along the latitude was obvious in West China (Fig. 3b, c). Significant patterns of isolation by geographic distance were also observed for populations across the entire distribution and within West China (Fig. S2). The co-occurrence of these genetic imprints more likely supports the hypothesis of natural processes such as range expansions, and not human-mediated seed flow. Even if the formation of putative refugia populations was associated with human-mediated gene flow, a subsequent gradual recolonization was required to shape the genetic structure of Q. acutissima. This process may need at least 10,000 years according to the estimation method in Saito et al. (2017), which is much longer than the history of artificial cultivation of oaks in China (You et al. 2017). Therefore, the existence of multiple refugia was most likely a consequence of naturally occurring range expansions or long-distance seed dispersal events, not human-mediated historical seed transfer.
Implications for conservation
Q. acutissima is able to regenerate naturally; however, long-term exploitation and habitat fragmentation have caused a considerable decrease in its distribution across China. Field surveys found that in some provinces such as Sichuan, Chongqing, Guizhou, and Hainan, the habitats of several wild populations have been devastated for farmland reclamation, tourism development, and commercial forest afforestation (e.g., Hevea brasiliensis and Eucalyptus robusta). Therefore, it is essential to take effective conservation measures for this species.
One of the important goals of species conservation is maintaining its genetic diversity and evolutionary potential (Mingeot et al. 2016). Our study reveals that Q. acutissima has higher genetic diversity at the species level, with genetic variation mainly partitioned within populations. Considering that natural populations of the species have not yet reached the endangered level, we suggest that in situ conservation should be the primary management activity in the future. Populations for priority conservation should be those with higher genetic diversity (e.g., populations in Central China) or with unique alleles (e.g., populations in putative refugia). Under the condition of limited funds, populations with the highest genetic diversity among those with close genetic relationships should be selected first for protection. Zhang et al. (2013a) observed a similar level of genetic diversity in provenances of Q. acutissima, indicating that it is feasible to preserve the genetic diversity of this species by constructing germplasm banks. According to the genetic structure identified in this study, we suggest that in practical situations, at least two germplasm banks should be set up, corresponding to the West China group and the East China group.
References
Acosta MC, Premoli AC (2010) Evidence of chloroplast capture in south American Nothofagus (subgenus Nothofagus, Nothofagaceae). Mol Phylogenet Evol 54:235–242
Aldrich PR, Michler CH, Sun W, Romero-Severson J (2002) Microsatellite markers for northern red oak (Fagaceae: Quercus rubra). Mol Ecol Notes 2:472–474
An ZS, Wang SM, Wu XH, Chen MY, Sun DH, Liu XM, Wang FB, Li L, Sun YB, Zhou WJ, Zhou J, Liu XD, Lu HY, Zhang YX, Dong GR, Qiang XK (1999) Eolian evidence from the Chinese loess plateau: the onset of the late Cenozoic great glaciation in the northern hemisphere and Qinghai-Xizang plateau uplift forcing. Sci China Ser D Earth Sci 42:258–271
An ZS, Kutzbach JE, Prell WL, Porter SC (2001) Evolution of Asian monsoons and phased uplift of the Himalaya-Tibetan plateau since Late Miocene times. Nature 411:62–66
Bagnoli F, Tsuda Y, Fineschi S, Bruschi P, Magri D, Zhelev P, Paule L, Simeone MS, González-Martínez SC, Vendramin GG (2015) Combining molecular and fossil data to infer demographic history of Quercus cerris: insights on European eastern glacial refugia. J Biogeogr 43:679–690
Bai WN, Wang WT, Zhang DY (2014) Contrasts between the phylogeographic patterns of chloroplast and nuclear DNA highlight a role for pollen-mediated gene flow in preventing population divergence in an east Asian temperate tree. Mol Phylogenet Evol 81:37–48
Bai WN, Wang WT, Zhang DY (2016) Phylogeographic breaks within Asian butternuts indicate the existence of a phylogeographic divide in East Asia. New Phytol 209:1757–1772
Beerli P (2006) Comparison of Bayesian and maximum likelihood inference of population genetic parameters. Bioinformatics 22:341–345
Cavender-Bares J, Gonzalez-Rodriguez A, Pahlich A, Koehler K, Deacon N (2011) Phylogeography and climatic niche evolution in live oaks (Quercus series Virentes) from the tropics to the temperate zone. J Biogeogr 38:962–981
Cavender-Bares J, González-Rodríguez A, Eaton DA, Hipp AA, Beulke A, Manos PS (2015) Phylogeny and biogeography of the American live oaks (Quercus subsection Virentes): a genomic and population genetics approach. Mol Ecol 24:3668–3687
Chang ZG, Xiao JL, Lü LQ, Yao HT (2010) Abrupt shifts in the Indian monsoon during the Pliocene marked by high-resolution terrestrial records from the Yuanmou Basin in Southwest China. J Asian Earth Sci 37:166–175
Chapuis M, Estoup A (2007) Microsatellite null alleles and estimation of population differentiation. Mol Biol Evol 24:621–631
Chen FH, Bloemendal J, Zhang PZ, Liu GX (1999) An 800 ky proxy record of climate from lake sediments of the Zoige Basin, eastern Tibetan Plateau. Palaeogeogr Palaeoclimatol Palaeoecol 151:307–320
Chen DM, Zhang XX, Kang HZ, Sun X, Yin S, Du HM, Yamanaka N, Gapare W, Wu HX, Liu CJ (2012) Phylogeography of Quercus variabilis based on chloroplast DNA sequence in East Asia: multiple glacial refugia and mainland-migrated island populations. PLoS One 7:e47268
Chen JM, Zhao SY, Liao YY, Gichira AW, Gituru RW, Wang QF (2015) Chloroplast DNA phylogeographic analysis reveals significant spatial genetic structure of the relictual tree Davidia involucrate (Davidiaceae). Conserv Genet 16:583–593
Chen YS, Deng T, Zhou Z, Sun H (2017) Is the east Asian flora ancient or not? Nat Sci Rev. https://doi.org/10.1093/nsr/nwx156
Choi HS, Kim YY, Hong KN, Hong YP, Hyun JO (2005) Genetic structure of a population of Quercus acutissima in Korea revealed by microsatellite markers. Korean J Genet 27:267–271
Chung MY, Nason J, Chung MG, Kim KJ, Park CW, Sun BY, Pak JH (2002) Landscape-level spatial genetic structure in Quercus acutissima (Fagaceae). Am J Bot 89:1229–1236
Chybicki IJ, Burczyk J (2009) Simultaneous estimation of null alleles and inbreeding coefficients. J Hered 100:106–113
Cornuet JM, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014
Cornuet JM, Pudlo P, Veyssier J, Dehne-Garcia A, Gautier M, Leblois R, Marin JM, Estoup A (2014) DIYABC v2.0: a software to make approximate Bayesian computation inferences about population history using single nucleotide polymorphism, DNA sequence and microsatellite data. Bioinformatics 30:1187–1189
Dieringer D, Schlotterer C (2003) MICROSATELLITE ANALYSER (MSA): a platform independent analysis tool for large microsatellite data sets. Mol Ecol Notes 3:167–169
Dow BD, Ashley MV (1996) Microsatellite analysis of seed dispersal and parentage of saplings in bur oak, Quercus macrocarpa. Mol Ecol 5:615–627
Du FK, Hou M, Wang WT, Mao KS, Arndt H (2017) Phylogeography of Quercus aquifolioides provides novel insights into the Neogene history of a major global hotspot of plant diversity in south-West China. J Biogeogr 44:294–307
Ennos RA (1994) Estimating the relative rates of pollen and seed migration among plant populations. Heredity 72:250–259
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and windows. Mol Ecol Resour 10:564–567
Fan L, Zheng H, Milne RI, Zhang L, Mao K (2018) Strong population bottleneck and repeated demographic expansions of Populus adenopoda (Salicaceae) in subtropical China. Ann Bot 121:665–679. https://doi.org/10.1093/aob/mcx198
Fujiwara K, Harada A (2015) Character of warm-temperate Quercus forests in Asia. In: Box E, Fujiwara K (eds) Warm-temperate deciduous forests around the northern hemisphere. Geobotany studies (basics, methods and case studies). Springer, Cham, pp 27–80
Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). http://www2.unil.ch/popgen/softwares/fstat.htm
Gugger PF, Ikegami M, Sork VL (2013) Influence of late Quaternary climate change on present patterns of genetic variation in valley oak, Quercus lobata Née. Mol Ecol 22:3598–3612
Guo ZT, Liu TS, Fédoroff N, Wei LY, Ding ZL, Wu NQ, Lu HY, Jiang WY, An ZS (1998) Climate extremes in loess of China coupled with the strength of deep-water formation in the North Atlantic. Glob Planet Chang 18:113–128
Guo XD, Wang HF, Bao L, Wang TM, Bai WN, Ye JW, Ge JP (2014) Evolutionary history of a widespread tree species Acer mono in East Asia. Ecol Evol 4:4332–4345
Hao QZ, Wang L, Oldfeld F, Guo ZT (2015) Extra-long interglacial in northern hemisphere during MISs 15-13 arising from limited extent of Arctic ice sheets in glacial MIS 14. Sci Rep 5:12103
Hardy OJ, Vekemans X (2002) SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620
Hardy OJ, Charbonnel N, Freville H, Heuertz M (2003) Microsatellite allele sizes: a simple test to assess their significance on genetic differentiation. Genetics 163:1467–1482
Hedrick PW (2005) A standardized genetic differentiation measure. Evolution 59:1633–1638
Hipp AL, Manos PS, González-Rodríguez A, Hahn M, Kaproth M, McVay JD, Avalos SV, Cavender-Bares J (2018) Sympatric parallel diversification of major oak clades in the Americas and the origins of Mexican species diversity. New Phytol 217:439–452
Hubert F, Grimm GW, Jousselin E, Berry V, Franc A, Kremer A (2014) Multiple nuclear genes stabilize the phylogenetic backbone of the genus Quercus. Syst Biodivers 12:405–423
Hubisz MJ, Falush D, Stephens M, Pritchard JK (2009) Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour 9:1322–1332
Isagi Y, Suhandono S (1997) PCR primers amplifying microsatellite loci of Quercus myrsinifolia Blume and their conservation between oak species. Mol Ecol 6:897–899
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Jose-Maldia LS, Matsumoto A, Ueno S, Kanazashi A, Kanno M, Namikawa K, Yoshimaru H, Tsumura Y (2017) Geographic patterns of genetic variation in nuclear and chloroplast genomes of two related oaks (Quercus aliena and Q. serrata) in Japan: implications for seed and seedling transfer. Tree Genet Genome 13:121
Kampfer S, Lexer C, Glössl J, Steinkellner H (1998) Characterization of (GA)n microsatellite loci from Quercus robur. Hereditas 129:183–186
Kou YX, Cheng SM, Tian S, Li B, Fan DM, Chen YJ, Soltis DE, Soltis PS, Zhang ZY (2015) The antiquity of Cyclocarya paliurus (Juglandaceae) provides new insights into the evolution of relict plants in subtropical China since the late Early Miocene. J Biogeogr 43:351–360
Li C, Sun Y, Huang HW, Cannon CH (2014) Footprints of divergent selection in natural populations of Castanopsis fargesii (Fagaceae). Heredity 113:533–541
Li YM, Ferguson DK, Zhao Q, Wang YF, Wang RX, Li CS (2015) Diatom-inferred salinity changes from the Yushe paleolake indicate an aridification during the Pliocene-Pleistocene transition in North China. Palaeogeogr Palaeoclimatol Palaeoecol 417:544–553
Lu LM, Mao LF, Yang T, Ye JF, Liu B, Li HL, Sun M, Miller JT, Mathews S, Hu HH, Niu YT, Peng DX, Chen YH, Smith SA, Chen M, Xiang KL, Le CT, Dang VC, Lu AM, Soltis PS, Soltis DE, Li JH, Chen ZD (2018) Evolutionary history of the angiosperm flora of China. Nature 554:234–238
Meng HH, Su T, Gao XY, Li J, Jiang XL, Sun H, Zhou ZK (2017) Warm-cold colonization: response of oaks to uplift of the Himalaya-Hengduan Mountains. Mol Ecol 26:3276–3294
Micheletti SJ, Storfer A (2015) A test of the central-marginal hypothesis using population genetics and ecological niche modelling in an endemic salamander (Ambystoma barbouri). Mol Ecol 24:967–979
Mingeot D, Husson C, Mertens P, Watillon B, Bertin P, Druart P (2016) Genetic diversity and genetic structure of black alder (Alnus glutinosa [L.] Gaertn) in the Belgium-Luxembourg-France cross-border area. Tree Genet Genome 12:24
Mohammad-Panah N, Shabanian N, Khadivi A, Rahmani MS, Emami A (2017) Genetic structure of gall oak (Quercus infectoria) characterized by nuclear and chloroplast SSR markers. Tree Genet Genome 13:70
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Ortego J, Riordan EC, Gugger PF, Sork VL (2012) Influence of environmental heterogeneity on genetic diversity and structure in an endemic southern Californian oak. Mol Ecol 21:3210–3223
Ortego J, Gugger PF, Sork VL (2015) Climatically stable landscapes predict patterns of genetic structure and admixture in the Californian canyon live oak. J Biogeogr 42:328–338
Ortego J, Gugger PF, Sork VL (2017) Genomic data reveal cryptic lineage diversification and introgression in Californian golden cup oaks (section Protobalanus). New Phytol 218:804–818. https://doi.org/10.1111/nph.14951
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Petit RJ, Excoffier L (2009) Gene flow and species delimitation. Trends Ecol Evol 24:386–393
Petit RJ, Carlson J, Curtu AL, Loustau ML, Plomion C, González-Rodríguez A, Sork V, Ducousso A (2013) Fagaceae trees as models to integrate ecology, evolution and genomics. New Phytol 197:369–371
Premoli AC, Mathiasen P, Acosta MC, Ramos VA (2012) Phylogeographically concordant chloroplast DNA divergence in sympatric Nothofagus s.s. how deep can it be? New Phytol 193:261–275
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Qi XS, Chen C, Comes HP, Sakaguchi S, Liu YH, Tanaka N, Sakio H, Qiu YX (2012) Molecular data and ecological niche modelling reveal a highly dynamic evolutionary history of the east Asian tertiary relict Cercidiphyllum (Cercidiphyllaceae). New Phytol 196:617–630
Rice WP (1989) Analyzing tables of statistical tests. Evolution 43:223–249
Riordan EC, Gugger PF, Ortego J, Smith C, Gaddis K, Thompson P, Sork VL (2016) Association of genetic and phenotypic variability with geography and climate in three southern California oaks. Am J Bot 103:73–85
Rodríguez-Correa H, Oyama K, Quesada M, Fuchs EJ, Quezada M, Ferrufino L, Valencia-Ávalos S, Cascante-Marín A, González-Rodríguez A (2017) Complex phylogeographic patterns indicate central American origin of two widespread Mesoamerican Quercus (Fagaceae) species. Tree Genet Genome 13:62
Rohlf FJ (1999) NTSYS-pc. Numerical taxonomy and multivariate analysis system, version 2.1. Exeter Software, Setauket
Rosenberg NA (2004) Distruct: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138
Saito Y, Tsuda Y, Uchiyama K, Fukuda T, Seto S, Kim P, Shen H, Ide Y (2017) Genetic variation in Quercus acutissima Carruth., in traditional Japanese rural forests and agricultural landscapes, revealed by chloroplast microsatellite markers. Forests 8:451
Shi MM, Michalski SG, Welk E, Chen XY, Durka W (2014) Phylogeography of a widespread Asian subtropical tree: genetic east-west differentiation and climate envelope modelling suggest multiple glacial refugia. J Biogeogr 41:1710–1720
Simeone MC, Grimm GW, Papini A, Vessella F, Cardoni S, Tordoni E, Piredda R, Franc A, Denk T (2016) Plastome data reveal multiple geographic origins of Quercus group Ilex. PeerJ 4:e1897
Slatkin M (1995) A measure of population subdivision based on microsatellite allele frequencies. Genetics 139:457–462
Song SY, Krajewska K, Wang YF (2000) The first occurrence of the Quercus section Cerris Spach fruits in the Miocene of China. Acta Palaeobot 40:153–163
Steinkellner H, Fluch S, Turetschek E, Lexer C, Streiff R, Kremer A, Burg K, Glössl J (1997) Identification and characterization of (GA/CT)n-microsatellite loci from Quercus petraea. Plant Mol Biol 33:1093–1096
Sun Y, Hu HQ, Huang HW, Vargas-Mendoza CF (2014) Chloroplast diversity and population differentiation of Castanopsis fargesii (Fagaceae): a dominant tree species in evergreen broad-leaved forest of subtropical China. Tree Genet Genome 10:1531–1539
Sun Y, Surget-Groba Y, Gao S (2016) Divergence maintained by climatic selection despite recurrent gene flow: a case study of Castanopsis carlesii (Fagaceae). Mol Ecol 18:4580–4592
Tian S, Lei SQ, Hu W, Deng LL, Li B, Meng QL, Soltis DE, Soltis PS, Fan DM, Zhang ZY (2015) Repeated range expansions and inter−/postglacial recolonization routes of Sargentodoxa cuneata (Oliv.) Rehd. Et Wils. (Lardizabalaceae) in subtropical China revealed by chloroplast phylogeography. Mol Phylogenet Evol 85:238–246
Vitelli M, Vessella F, Cardoni S, Pollegioni P, Denk T, Grimm GW, Simeone MC (2017) Phylogeographic structuring of plastome diversity in Mediterranean oaks (Quercus group Ilex, Fagaceae). Tree Genet Genome 13:3
Wang PX, Clemens S, Beaufort L, Braconnot P, Ganssen G, Jian ZM, Kershaw P, Sarnthein M (2005) Evolution and variability of the Asian monsoon system: state of the art and outstanding issues. Quat Sci Rev 24:595–629
Wang YH, Comes HP, Cao YN, Guo R, Mao YR, Qiu YX (2017) Quaternary climate change drives Allo-peripatric speciation and refugial divergence in the Dysosma versipellis-pleiantha complex from different forest types in China. Sci Rep 7:40261
Wei XZ, Sork VL, Meng HJ, Jiang MX (2016) Genetic evidence for central-marginal hypothesis in a Cenozoic relict tree species across its distribution in China. J Biogeogr 43:2173–2185
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Xu J, Deng M, Jiang XL, Westwood M, Song YG, Turkington R (2015) Phylogeography of Quercus glauca (Fagaceae), a dominant tree of east Asian subtropical evergreen forests, based on three chloroplast DNA interspace sequences. Tree Genet Genome 11:805
Xu J, Jiang XL, Deng M, Westwood M, Song YG, Zheng SS (2016) Conservation genetics of rare trees restricted to subtropical montane cloud forests in southern China: a case study from Quercus arbutifolia (Fagaceae). Tree Genet Genome 12:90
Yang J, Di XY, Meng X, Feng L, Liu ZL, Zhao GF (2016) Phylogeography and evolution of two closely related oak species (Quercus) from north and Northeast China. Tree Genet Genome 12:89
Ye QL, Zeng XY (2009) Optimization of SRAP-PCR system and genetic diversity analysis in Quercus acutissima Carr. Biotechnology 19:25–27
Ye JW, Bai WN, Bao L, Wang TM, Wang HF, Ge JP (2017) Sharp genetic discontinuity in the aridity-sensitive Lindera obtusiloba (Lauraceae): solid evidence supporting the tertiary floral subdivision in East Asia. J Biogeogr 44:2082–2095
Ye JW, Jiang T, Wang HF, Wang TM, Bao L, Ge JP (2018) Repeated expansions and fragmentations linked to Pleistocene climate changes shaped the genetic structure of a woody climber, Actinidia arguta (Actinidiaceae). Botany 96:19–31
Yeh FC, Yang R, Boyle T (1999) POPGENE, version 1.31: Microsoft Windows-based free software for population genetic analysis. https://sites.ualberta.ca/~fyeh/popgene.html
Yin QZ, Guo ZT (2007) Strong summer monsoon during the cool MIS-13. Clim Past Discuss 3:1119–1132
You LX, Li Y, Yin ZF, Fang YM (2017) The study of sawtooth oak in China: provenance test, afforestation and forest management. World For Res 30:75–79 (in Chinese with English abstract)
Yu XQ, Gao LM, Soltis DE, Soltis PS, Yang JB, Fang L, Yang SX, Li DZ (2017) Insights into the historical assembly of east Asian subtropical evergreen broadleaved forests revealed by the temporal history of the tea family. New Phytol 215:1235–1248
Zeng YF, Wang WT, Liao WJ, Wang HF, Zhang DY (2015) Multiple glacial refugia for cool-temperate deciduous trees in northern East Asia: the Mongolian oak as a case study. Mol Ecol 24:5676–5691
Zhang YY, Fang YM, Yu MK, Li XX, Xia T (2013a) Molecular characterization and genetic structure of Quercus acutissima germplasm in China using microsatellites. Mol Biol Rep 40:4083–4090
Zhang YY, Fang YM, Yu MK, Zhou XL (2013b) Molecular characterization of sawtooth oak (Quercus acutissima) germplasm based on randomly amplified polymorphic DNA. Plant Syst Evol 299:1829–1837
Zhang XW, Li Y, Fang YM (2014a) Geographical distribution and prediction of potential ranges of Quercus acutissima in China. Acta Botan Boreal-Occiden Sin 34:1685–1692 (in Chinese with English abstract)
Zhang YB, Sun DH, Li ZJ, Wang F, Wang X, Li BF, Guo F, Wu S (2014b) Cenozoic record of aeolian sediment accumulation and aridification from Lanzhou, China, driven by Tibetan Plateau uplift and global climate. Glob Planet Chang 120:1–15
Zhang XW, Li Y, Liu CY, Xia T, Zhang Q, Fang YM (2015) Phylogeography of the temperate tree species Quercus acutissima in China: inferences from chloroplast DNA variations. Biochem Syst Ecol 63:190–197
Zhao YJ, Yin GS, Pan YZ, Gong X (2018) Ecological and genetic divergences with gene flow of two sister species (Leucomeris decora and Nouelia insignis) driving by climatic transition in Southwest China. Front Plant Sci 9:31
Zhou ZK (1993) The fossil history of Quercus. Acta Bot Sin 15:21–33 (in Chinese with English abstract)
Zhou YF, Abbott RJ, Jiang ZY, Du FK, Milne RI, Liu JQ (2010) Gene flow and species delimitation: a case study of two pine species with overlapping distributions in Southeast China. Evolution 64:2342–2352
Zhou Y, Duvaux L, Ren G, Zhang L, Savolainen O, Liu J (2017) Importance of incomplete lineage sorting and introgression in the origin of shared genetic variation between two closely related pines with overlapping distributions. Heredity 118:211–220
Acknowledgments
We thank the editor and three anonymous reviewers for their critical comments; all members from Sork’s lab at UCLA: Victoria L. Sork, Alayna Mead, Claudia Henriquez, Dylan Burge, Luke Browne, Rachel Meyer, Scott O’Donnell, and Sorel Fitz-Gibbon for their insightful comments and detailed edits; Qing-Feng Huang, Shao-Bin Ma, Ming Cao, Zhi-Gao Liu, Mao Wang, Su-Fang Yu, Heng Jia, Su-Jing Fu, Hua-Chen Wang, and Luo-Zhong Tang for their help with fieldwork; Gao-Ming Wei, Xiao-Meng Shi, and Yu-Xiang Liu for their assistant with data analysis.
Data archiving statement
Microsatellite genotype dataset is available from the Zenodo: https://doi.org/10.5281/zenodo.1412808.
Funding
This research was supported by the National Natural Science Foundation of China (31770699, 31370666), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD) and the Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYLX15_0922).
Author information
Authors and Affiliations
Contributions
Xing-Wang Zhang conducted molecular laboratory work. Xing-Wang Zhang and Yao Li collected the samples. Xing-Wang Zhang, Yao Li, Qiang Zhang, and Yan-Ming Fang conceived the study, analyzed the data, and wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by J. Wright
Electronic supplementary material
ESM 1
(DOCX 876 kb)
Rights and permissions
About this article
Cite this article
Zhang, XW., Li, Y., Zhang, Q. et al. Ancient east-west divergence, recent admixture, and multiple marginal refugia shape genetic structure of a widespread oak species (Quercus acutissima) in China. Tree Genetics & Genomes 14, 88 (2018). https://doi.org/10.1007/s11295-018-1302-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-018-1302-9