Abstract
Long noncoding RNAs (lncRNAs) play significant roles in the growth and development of herbaceous plants by regulating target genes; however, the significance of lncRNA-messenger RNA (mRNA) interactions needs to be investigated in perennial trees. Here, we combined transcript profiling and multi-single-nucleotide polymorphism (SNP) association mapping to analyze the genetic variation and putative interactions of the lncRNA UDP-glucosyltransferase-related lncRNA (UGTRL) and its predicted target PtoUGT88A1 in a natural population of 435 unrelated Populus tomentosa individuals. We detected 41 and 67 common SNPs (minor allele frequency >0.05) in UGTRL and PtoUGT88A1, respectively, in the association population. Using additive and dominant association models, we identified 86 associations with 12 traits measuring tree growth, wood properties, and photosynthetic parameters. These associations represent 36 significant SNPs (P < 0.01) from UGTRL and its putative target and explained 0.06 to 7.28 % of the phenotypic variance, indicating that UGTRL and its putative target affect wood formation. An epistasis model uncovered 84 SNP-SNP association pairs representing 38.89 % of the significant SNPs in UGTRL and PtoUGT88A1 with information gain of −8.01 to 5.57 %, revealing the strong interactions between UGTRL and its putative target. Tissue-specific expression analysis in eight tissues, including xylem and cambium, showed that UGTRL and PtoUGT88A1 displayed similar expression patterns (r = 0.77), which implied the putative lncRNA-mRNA interaction and the potential roles of the lncRNA and its target in wood formation. Our study provides a novel method integrating association studies and expression profiling for functional annotation of lncRNAs and dissection of lncRNA-mRNA interactions in trees.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Unlike most herbaceous species, trees have long growth cycles, large growth sizes, and highly lignified tissues (Barbazuk et al. 2008). Trees can adapt to complex and changeable environmental conditions, provide renewable materials for biofuels, and also perform various ecological services, such as preventing erosion (Neale and Kremer 2011). Hence, many tree research programs aim to enhance wood quality and quantity and improve utilization of elite germplasm. Wood formation and tree growth are regulated by elaborate, coordinated networks and involve various dynamic processes and diverse metabolic pathways that include cell division, secondary cell wall deposition, cell expansion, and programmed cell death (Samuels et al. 2006). Dissection of the polygenic genetic regulatory mechanism that affects wood properties may enable breeding programs to harness the genetic variation in natural tree populations.
In Populus, several key genes involved in wood formation have been identified, such as cellulose synthase A (CesA) (Suzuki et al. 2006), which catalyzes the biosynthesis of cellulose, a major component of secondary cell walls, UDP-glucuronate decarboxylase (UXS) (Du et al. 2013), which affects fiber formation, growth and development, and wood quality of Populus tomentosa, and UDP-glucosyltransferases (UGTs), which participate in the metabolism of cell wall polysaccharides in poplar (Aspeborg et al. 2005). UGTs are indispensable for ensuring conjugation of sugar moieties to secondary metabolites. UGTs have been identified in many plants as involved in various pathways, such as stress resistance and carbohydrate metabolism (Sun et al. 2013), with many functions in biosynthesis and modification of natural products, such as flavonoids and cytokinins (Jones and Vogt 2001). In addition, some putative UGT genes were also predicted to function under high nitrogen availability in cell wall polysaccharide synthesis in poplar (Plavcová et al. 2013). Even though UGTs have pivotal roles in the pathway for long-distance transport of photosynthetic products and the accumulation of fixed carbon in secondary xylem, no studies have yet reported the roles of UGTs in photosynthesis and wood formation in plants.
Long noncoding RNAs (lncRNAs), whose length varies from 200 nt to dozens of kilobases with little or no protein-coding capacity, are essential noncoding RNAs (ncRNAs) that function at all biological regulatory levels (Quan et al. 2015). The numerous lncRNAs identified in animals participate in many biological processes (Ponting et al. 2009), such as Xist lncRNA in X-chromosome inactivation (Lee and Bartolomei 2013; Zhao et al. 2008). In addition, lncRNAs regulate the expression of their target genes by various modes (Quan et al. 2015). For example, in Saccharomyces cerevisiae, the lncRNA SER regulatory gene 1 (SRG1) lies in the promoter region and depresses the transcription of SER3, which participates in biosynthesis of serine (Martens et al. 2004). In plants, transcriptome sequencing identified numerous lncRNAs in maize (Li et al. 2014b), Arabidopsis (Liu et al. 2012) and Populus (Chen et al. 2015; Shuai et al. 2014), and increasing numbers of lncRNAs have been shown to regulate the development of plants. For example, many classes of lncRNAs influence flowering time of Arabidopsis in multiple pathways; these lncRNAs include COOLAIR, COLDAIR, and ASL (Heo and Sung 2011; Shin and Chekanova 2014; Swiezewski et al. 2009). Previous studies have demonstrated that ncRNAs interact with their target genes to regulate the growth and development of plants (Yang et al. 2015), and transgenic and reverse genetics methods have been used to uncover many lncRNA-messenger RNA (mRNA) interactions in plants, such as lncRNA HIDDEN TREASURE 1 (HID1), which negatively regulates PHYTOCHROME-INTERACTING FACTOR 3 (PIF3) in modulating red-light-mediated photomorphogenesis in Arabidopsis seedlings (Wang et al. 2014a). Nevertheless, no reports have clarified the roles of lncRNAs in wood formation in trees.
Given the experimental limitations of trees, with their long generation times and complex biological characteristics, association genetics approaches, also called linkage disequilibrium (LD) mapping, can be used to identify the interactions of lncRNAs and their target genes. In particular, application of association-based molecular marker-assisted selection (MAS) in several agricultural crops demonstrated that MAS can greatly shorten the breeding cycles and increase the efficiency of selection (Neale and Kremer 2011). In addition, MAS can also explain a large proportion of the variation in breeding populations and dissect the complex associations of markers and traits (Guerra et al. 2013). Also, single-nucleotide polymorphism (SNP)-based association mapping for forest trees can detect the link between natural allelic variation and a specific phenotype, thus enabling MAS for that phenotype (Sexton et al. 2012). For example, previous studies identified polymorphisms in cinnamoyl CoA reductase (CCR), a key gene for lignin biosynthesis, which were associated with microfibril angle in Eucalyptus nitens (Thumma et al. 2005). Similarly, research concentrating on candidate gene-based association studies clarified several major SNPs within candidate genes associated with tree growth and wood property traits (Tian et al. 2012). Recently, SNP-based association has also been applied to evaluate microRNA (miRNA)-mRNA interactions in specific traits and examine the multi-gene networks affecting tree growth and wood properties (Du et al. 2015; Yang et al. 2015), which takes association genetics a step further than single-gene association studies. However, to date, no studies have examined lncRNA-mRNA interactions by SNP-based association methods in trees.
P. tomentosa, with its widely distributed natural population, adaptive evolution to complex environments, and abundant allelic variation, is an important plantation wood species and commercial trees for pulp and timber production in northern China (Zhang et al. 2007). Therefore, a better understanding of the molecular genetic mechanisms underlying wood formation in P. tomentosa will assist modern tree breeding programs. Here, we decipher the genetic variations and putative interactions of lncRNA UGT-related lncRNA (UGTRL) and its target PtoUGT88A1 in a population of 435 unrelated individuals from P. tomentosa. Specially, combined with examination of the expression patterns of UGTRL and PtoUGT88A1, we used joint genetic effects, including additive, dominant, and epistatic models, to dissect multiple SNP-based associations within UGTRL and PtoUGT88A1 for growth, wood properties, and photosynthetic traits. Thus, we elucidated the putative roles of the lncRNA UGTRL in wood formation and provided an alternative method to address the interactions of lncRNAs and mRNAs associated with multiple traits in trees.
Materials and methods
Association population and phenotypic data
Association population
The association population (natural population) consisted of 435 unrelated individuals of P. tomentosa, which were randomly sampled from the collection of 1047 individuals, which collected from 100 provenances of P. tomentosa with each individual within one provenance was collected at least 1 km apart, representing the entire natural distribution of P. tomentosa, which covers three climate regions (southern, northeastern, and northwestern China) and an area of 1 million km2 along the Yellow River (30–40° N, 105–125° E) (Zhang et al. 2010b). A clonal arboretum of this collection was grown in Guan Xian County, Shandong Province, China (36° 23′ N, 115° 47′ E), in a randomized complete block design approach with three clonal replications (Du et al. 2012). Additionally, a set of 43 unrelated individuals randomly selected from the association population were used for SNP discovery.
Phenotypic data
According to the potential functions of the predicted target PtoUGT88A1, such as the transfer and accumulation photosynthesis products, it is indicated that UGTRL and PtoUGT88A1 may contribute to phenotypic variation of growth, wood property, and photosynthesis traits of trees. Thus, three growth traits, seven wood property traits, and four photosynthetic traits, which were measured in the 435 individuals of P. tomentosa with at least three replications per parameter, were used for further analysis. The growth traits were as follows: diameter at breast height (DBH, cm), tree height (H, m), and stem volume (V, m3), and the sampling and measurement were conducted as previously described (Zhang et al 2006). The wood property traits were follows: fiber length (FL, mm), fiber width (FW, μm), and microfibril angle (MFA, °), holocellulose content (HC, %), α-cellulose content (CC, %), lignin content (LC, %), and hemicellulose content (HEC, %), and the measurement of these traits was conducted as previously described (Tian et al 2014). In addition, the photosynthetic traits were as follows: photosynthetic rate (Pn, μmol m−2 s−1), conductance to H2O (Cond, mol m−2 s−1), intercellular CO2 (Ci, μmol mol−2), and transpiration rate (Trmmol, g m−2 h−1), and the measurements were conducted as previously described (Wang et al. 2014b). Analysis of correlations for these phenotypic data in the association population has been reported previously (Wang et al. 2014a).
Identification of UGTRL and its target gene PtoUGT88A1
Identification and sequence analysis of UGTRL
Total RNA was extracted from the stem cambium, developing xylem, and mature xylem of a 1-year-old P. tomentosa clone “LM50” planted in Guan Xian County, using the Plant Qiagen RNeasy kit as described in the manufacturer’s instructions. To remove DNA contamination, additional on-column DNase digestions were performed three times during RNA purification using the RNase-Free DNase Set (Qiagen). Library construction and Illumina sequencing for three RNA samples were conducted by Shanghai Biotechnology Corporation (Shanghai, China), using Illumina HiSeq 2500. High-quality reads, filtered by FASTX-Toolkit version 0.0.13 (http://hannonlab.cshl.edu/fastx_toolkit/index.html), were mapped to the Populus trichocarpa genome (Tuskan et al. 2006), using Tophat v2.0.9 (Trapnell et al. 2012), and Cufflinks v2.1.1 (Trapnell et al. 2012) was used to assemble the transcripts and calculated the expression levels using fragments per kilobase of transcript per million fragments (FPKM) for normalized (SRA accession number: SRP060593) (data unpublished). The prediction of lncRNAs from RNA-seq was conducted as previously described (Chen et al. 2015). The differentially expressed lncRNAs were filtered by fold change (FC) ≥2 or ≤0.5 (P < 0.01, FC = FPKM of A/FPKM of B, where A and B represent any two tissues of cambium, developing xylem, and mature xylem). We selected UGTRL as a candidate lncRNA, as it detected in the three tissues and showed different expression levels among cambium, developing xylem, and mature xylem (P < 0.01), indicating that it may be involved in wood formation. To evaluate the conservation of UGTRL, we conducted BLAST in National Center for Biotechnology Information (NCBI, www.ncbi.nih.gov) to obtain sequences homologous to UGTRL in P. tomentosa, P. trichocarpa, Populus euphratica, Populus deltoides, Ricinus communis, Eucalyptus grandis, Fragaria vesca subsp. vesca, Prunus mume, and Vitis vinifera, and the alignment and phylogenetic tree were constructed with MEGA 5.0 (Tamura et al. 2011) following the description of Du et al (2013).
Prediction of the UGTRL target gene PtoUGT88A1
To ensure the reliability and correctness of target prediction, we applied two methods to predict the trans-regulatory target genes of lncRNAs. The first option was based on sequence complementarity (Fig. S1) and the prediction of RNA duplex energy (Han et al. 2012). We used sequence complementarity of UGTRL for first-round screening with a BLAST cutoff E-value <1E−5; then, RNAplex (Tafer and Hofacker 2008) was used for the second-round screening considering the RNA duplex energy with parameters of E-value <−30. The other tool used to predict the putative target was LncTar (Li et al. 2014a), which identified the potential interaction of lncRNAs and mRNAs by finding the minimum free energy of the joint structure with a highest cutoff of normalized delta free energy (ndG) <−0.2. After filtering by the two methods, we predicted four target genes in the complementary DNA (cDNA) library of P. tomentosa; however, only one of these genes showed differential expression in cambium, developing xylem, and mature xylem and showed strong expression in mature xylem. Thus, this gene was selected as the candidate target gene, and its full-length cDNA was isolated from the mature xylem of P. tomentosa by reverse transcription (RT)-PCR (accession number: KR492613). Then, we identified the cDNA with high sequence similarity to Potri.017G150000. BLAST searches showed that Potri.017G150000 is a homolog of At-UGT88A1 (AT3G16520.3); thus, we named this putative target PtoUGT88A1.
Analysis of expression of UGTRL and PtoUGT88A1
To measure the expression of UGTRL and PtoUGT88A1, RT quantitative PCR (RT-qPCR) was performed on RNA isolated from eight tissues: root, young leaves, old leaves, and stem, which included shoot apex, cambium, developing xylem, mature xylem, and phloem, on the 7500 Fast Real-Time PCR System with SYBR Premix EX Taq (TaKaRa, Dalian, China). RNA was isolated from fresh tissues with at least three individual samples from 1-year-old LM50, using the Plant Qiagen RNeasy kit, and RNA integrity was confirmed on an agarose gel. The cDNA templates for the eight tissues were reverse transcribed with the Plant Qiagen RNeasy kit from the total RNA. We used gene-specific and poplar internal control (Actin, accession number EF145577) primers designed by Primer Express 3.0 software (Applied Biosystems) (Table S1). The PCR program contained an initial denaturation at 94 °C for 5 min; 40 cycles of 94 °C for 30 s, 58 °C for 30 s, and 72 °C for 30 s; and a final with the melting curve of 70–95 °C. All the reactions were conducted with triplicate technical and biological repetitions, and the melting curves were used to confirm the specificity of the amplifications. Also, the results for all tissues were standardized to an internal control. Opticon Monitor Software 3.1 was used for data analysis following the protocol described in the manual.
SNP discovery and genotyping
Total genomic DNA was extracted from fresh leaves using the DNeasy Plant Mini Kit (Qiagen China, Shanghai). PCR amplification was conducted as described by Zhang et al (2010a). The genomic DNAs of UGTRL and PtoUGT88A1 were sequenced using the BigDye Terminator Cycle Sequencing kit (version 3.1, Applied Biosystems, Beijing, China) and the 4300 DNA Analyzer (Li-Cor Biosciences, Lincoln, NE, USA). The full-length genomic DNAs of UGTRL and PtoUGT88A1 were sequenced and analyzed in 43 unrelated individuals randomly selected from the association population of P. tomentosa to identify SNPs, not including Indels (insertions/deletions). A total of 86 full-length sequences of UGTRL and PtoUGT88A1 were deposited in NCBI with accession numbers KR492527–KR492569 and KR492570–KR492612, respectively. MEGA 5.0 was used for manually removing primer sequences and sequence alignment, and π (Nei 1987) and θ w (Watterson 1975), calculated by DnaSP 5.10 (Librado and Rozas 2009), were used to evaluate the nucleotide diversity. All the common SNPs (minor allele frequency >0.05) identified were genotyped in 435 individuals of the association population using the Beckman Coulter (Franklin Lakes, NJ, USA) sequencing system.
Data analysis
LD analysis
The squared correlation of allele frequencies (R 2) between every pair of common SNPs (minor allele frequency >0.05) (Hill and Robertson 1968) within lncRNA UGTRL and PtoUGT88A1 was used to measure LD using TASSEL v.2.0.1 (http://www.maizegenetics.net/) (Bradbury et al. 2007). To evaluate the extent of LD within the sequenced regions of UGTRL and PtoUGT88A1, the decay of LD with physical distance (base pairs) of each pair of common SNP sites within a gene was calculated with 105 permutations by nonlinear regression (Remington et al. 2001). Singletons were excluded in LD analyses. This analysis was done for both UGTRL and PtoUGT88A1.
Single-SNP-based associations
We used a mixed linear model (MLM) with 105 permutations in TASSEL v.2.0.1 (Bradbury et al. 2007) for single-SNP associations between 108 common SNPs from UGTRL and its putative target and 14 traits. The MLM can be described as follows: y = μ + Qv + Zu + e, in which y is a vector of phenotypic trait values, μ is the intercept vector, v is a vector for population effects, u represents the vector of random polygenic background effects, e represents random experimental errors, and the Q and Z matrixes denote population structure and relate y to u, respectively. In addition, for Var(u) = G = σ 2 a K, σ 2 a is defined as unknown additive genetic variance and K represents the relative kinship matrix (Yu et al. 2006). In this Q + K model, based on 20 species-specific SSR markers described previously (Du et al. 2012), K was evaluated by SPAGeDi 1.3 (Hardy and Vekemans 2002), and the association population structure (Q) was obtained based on significant subpopulations (k = 3) with the statistical model described by Evanno et al (2005), using STRUCTURE 2.3.4 (Hubisz et al. 2009). The positive false discovery rate (FDR) was calculated to correct for errors related to multiple testing, using QVALUE software (Storey and Tibshirani 2003).
Haplotype-based associations
Haplotype frequencies from genotype data were estimated and haplotype-based association with growth, wood properties, and photosynthetic traits of P. tomentosa were tested by the haplotype trend regression (HTR) software (Zaykin et al. 2002). An overlapping three-marker sliding window was used to estimate possibilities of the haplotypes for associations with traits, and the significance of haplotype associations was based on 1000 permutation tests.
Multiple SNP-based associations under additive and dominance models
The fGWAS 2.0 (http://statgen.psu.edu/software/fgwas-soft.html) running in R (http://www.r-project.org/) was used to identify the significantly associated SNPs (P < 0.01) for each trait and estimate the possible additive and dominant effects of these SNPs on the associated traits (Li et al. 2011). The phenotypic and genotypic data in this model were analyzed through a two-stage procedure, by first conducting a preconditioning step using principal component analysis and then formulating a Bayesian Lasso model, which is implemented with a hierarchical model processing by the Markov chain Monte Carlo algorithm. The details of this model were described by Li et al (2011). In addition, the phenotypic variance explained by each SNP was also estimated in this model.
Multi-SNP epistasis models
We used epistasis models to characterize the genetic architecture and dissect the nonlinear SNP-SNP interactions with associated traits. To detect and characterize the interactions among multiple factors, a multi-factor dimensionality reduction method was developed for processing high-dimensionality genetic data into a single dimension, thus permitting the interactions to be detected in a relatively small set (Ritchie et al. 2001). Here, MDR 3.0.2 was applied to assess the SNP-SNP interactions with associated traits and investigate the epistatic interactions of these SNP combinations for the associated traits (Hahn et al. 2003). The Relief F algorithm in the MDR software filtered all unlinked SNPs (R 2 < 0.1 or different gene) and provided the five most-significant SNP loci for each trait after improving the reliability of probability approximation. The information gain (IG) calculated by an entropy-based model was used to assess the epistatic effect of significant SNP-SNP combinations with associated traits (Moore et al. 2006).
Transcript analysis of SNP genotypes
The SNP genotypes affect the transcript levels, which, in turn, affect the phenotypes. For the three significant SNP-SNP pairs (Q < 0.1) with highest epistatic effect in three categories of phenotypic traits identified by MDR, RT-qPCR was performed on 36 trees (9 trees for each pair of genotypes) for each SNP-SNP pair to detect the different transcript levels produced by different genotypic pairs, and then, the effects of SNP-SNP pairs on associated traits were tested. The RT-qPCR with gene-specific primers was performed as described above (Table S1). Additionally, total RNA isolated from the mature xylem of 36 individuals and RT were performed as described above. ANOVA was used to test the differential expression across different genotypic combinations of SNPs in lncRNA UGTRL and PtoUGT88A1 as described by Tian et al (2014).
Results
Identification of UGTRL and prediction of its target gene PtoUGT88A1
Transcriptome sequencing in three tissues showed that the lncRNA UGTRL is most abundant in mature xylem, followed by developing xylem and cambium, indicating the potential role of UGTRL in regulatory networks associated with wood formation (Fig. 1a). Sequencing yielded the 574-nt sequence of UGTRL (accession number: KR492527); we conducted BLAST searches of known miRNA sequence databases, which did not show any miRNA derived from UGTRL, suggesting that UGTRL likely does not function as a precursor of miRNAs. To elucidate the evolutionary conservation of UGTRL, alignment results revealed homologous sequences mostly in woody plants, like P. trichocarpa and E. grandis, and a few in liana and herbaceous plants, like V. vinifera and F. vesca. The phylogenetic tree revealed that UGTRL in P. tomentosa has close similarity to UGTRL in P. trichocarpa, P. euphratica, and P. deltoides (Fig. 1b), indicating that UGTRL is highly conserved in Populus, and has weak conservation in distantly related species.
Expression levels of UGTRL and PtoUGT88A1, phylogenetic tree of UGTRL, and gene structure of UGTRL and PtoUGT88A1. a The expression levels of UGTRL and PtoUGT88A1 in three tissues were detected via RNA-seq revealed by FPKM (left). The relative expression levels were measured by RT-qPCR in eight tissues with Actin as the internal control (right). b The phylogenetic tree of homologs of P. tomentosa UGTRL, including P. tomentosa, P. trichocarpa, P. euphratica, P. deltoides, Ricinus communis, Eucalyptus grandis, Fragaria vesca subsp. vesca, Prunus mume, and Vitis vinifera. c Gene structure and SNP diversity of UGTRL and PtoUGT88A1. The white dots in UGTRL and its putative target PtoUGT88A1 represent their common SNPs
To further explore the roles of UGTRL in plant development, we predicted PtoUGT88A1, a differentially expressed gene in mature xylem, developing xylem, and cambium of P. tomentosa, as its putative trans-regulatory target genes. We used two methods, one based on the sequence complementary of overlapping regions (identity = 71.73 %; E-value = 7.00E−82) and the RNA duplex energy (E-value = −70) predicted by RNAplex and the other based on ndG (−0.32) calculated by LncTar. With these pipelines, we identified that PtoUGT88A1 had a complementary region with UGTRL at 580–1156 nt of PtoUGT88A1 (Figs. 1c and S1). We isolated the cDNA sequence of PtoUGT88A1 by RT-PCR from a cDNA library prepared from mature xylem from P. tomentosa stem and sequenced the full-length genomic sequence of PtoUGT88A1, which identified PtoUGT88A1 as 2290 bp in length, lacking introns, and having an ORF (1434 bp) encoding a polypeptide of 477 amino acids with an estimated molecular weight of 52.8 kD and theoretical pI of 5.96, flanked by a 571 bp 5′-untranslated region (UTR) and a 285 bp 3′-UTR (Fig. 1c). Sequence analysis of PtoUGT88A1 revealed that it contains the essential domain of UDPGT, which is a conserved feature of the Arabidopsis homolog UGT88A1 (AT3G16520.3).
High correlation of lncRNA UGTRL and PtoUGT88A1 revealed by expression patterns
To provide insights into the functions and interactions of UGTRL and PtoUGT88A1, RT-qPCR was conducted to measure transcript abundances of UGTRL and PtoUGT88A1 in various organs and tissues of P. tomentosa, with Actin as the internal control. As shown in Fig. 1a, in roots and leaves, UGTRL transcript levels peaked in old leaves (12.04 ± 0.35) and were low in young leaves (3.63 ± 0.05). In stem, UGTRL showed the highest abundance in mature xylem (5.99 ± 0.25) and developing xylem (5.74 ± 0.12), followed by phloem (4.73 ± 0.07), with the lowest expression in cambium (1.07 ± 0.05) and moderate abundance in the shoot apex (2.79 ± 0.08). For PtoUGT88A1, the highest transcript level was observed in old leaves (51.98 ± 0.47), root (41.45 ± 0.18), mature xylem (40.69 ± 0.19), and phloem (29.43 ± 0.07). The developing xylem (11.16 ± 0.04), shoot apex (9.00 ± 0.16), and cambium (8.13 ± 0.07) had moderate abundance, with the lowest abundance in young leaves (2.67 ± 0.13). Comparison analysis showed that UGTRL and its target had similar expression trends in mature xylem, developing xylem, and cambium, which was observed by both RT-qPCR and RNA-seq. Generally, UGTRL showed lower transcript abundance compared with PtoUGT88A1, and UGTRL and PtoUGT88A1 showed nearly consistent expression patterns with a Pearson’s product-moment correlation of 0.77 (P < 0.05) revealed by RT-qPCR in eight tissues, indicating strong correlation of transcript levels between UGTRL and PtoUGT88A1.
Single-nucleotide diversity and LD in UGTRL and its target PtoUGT88A1
To characterize the polymorphisms for association mapping, we obtained the full-length genomic sequences for UGTRL and PtoUGT88A1 from 43 unrelated individuals of the association population in P. tomentosa. We detected 54 SNPs in UGTRL and 102 SNPs in PtoUGT88A1, with a high density of 1/11 and 1/22 bp, respectively. The nucleotide diversity (excluding indels) in UGTRL (π = 0.02607 and θ w = 0.02174) was about twofold higher than in PtoUGT88A1 (π = 0.01200 and θ w = 0.01029). The most-conserved region was in the exon of PtoUGT88A1, and the lncRNA UGTRL had the highest density of nucleotide polymorphisms, indicating that the coding region shows more conservation than the noncoding regions under selection pressure (Table 1). Within the coding region of PtoUGT88A1, the average nucleotide diversity of synonymous polymorphisms (d S , π = 0.01051) was higher than nonsynonymous polymorphisms (d N , π = 0.00783). The ratio of d N /d S (0.745) was <1 for exon of PtoUGT88A1, indicating purifying selection for nonsynonymous sites in the exon.
To perform LD analysis, 41 and 67 common SNPs (minor allele frequency >0.05) from UGTRL and PtoUGT88A1, respectively, were successfully genotyped in 435 unrelated individuals of the natural population of P. tomentosa (Fig. 1c). The R 2 values, combined with physical distances, were merged to evaluate the overall patterns of LD (Fig. 2). LD declined rapidly within ∼50 bp in UGTRL (R 2 < 0.1, P < 0.001) (Fig. 2a) and within ∼500 bp in PtoUGT88A1 (R 2 < 0.1, P < 0.001) (Fig. 2b), indicating that the LDs of UGTRL and PtoUGT88A1 do not extend to the entire genes. In addition, the degree of LD for both UGTRL and PtoUGT88A1 was low, as a large portion of SNPs were in linkage equilibrium (R 2 < 0.1) across the whole region. Thus, UGTRL and PtoUGT88A1 are appropriate for association analysis of candidate genes.
lncRNA UGTRL and its target gene share the pathway for phenotypic variation revealed by association studies
Single-SNP- and haplotype-based association
To explore the effect of lncRNA UGTRL and its putative target PtoUGT88A1 on growth and development of P. tomentosa, we used MLM in TASSEL, a method that considers population structure and kinship, to evaluate the genetic association between 108 SNPs of UGTRL and PtoUGT88A1 and 14 quantitative traits. This identified nine significant (P < 0.01, Q < 0.1) associations of nine SNPs in UGTRL and PtoUGT88A1 (Table S2) with five traits, and the phenotype variation explained (R 2) by each association ranged from 0.25 to 11.02 %. Four SNPs in UGTRL associated with four growth, wood property, and photosynthetic traits of P. tomentosa, with an average R 2 of 5.16 %. For the significant associations detected in PtoUGT88A1, five SNPs associated with four traits of three categories with the R 2 from 4.94 to 11.02 %, of which, SNP46 in the exon of PtoUGT88A1 associated with DBH with R 2 of 6.83 %. In general, single-SNP-based associations of lncRNA UGTRL and its target indicate their potential role in tree growth and wood properties.
Next, we conducted haplotype-based association to identify significant haplotypes associated with growth, wood properties, and photosynthetic traits by using HTR. We found nine common haplotypes (frequency ≥0.05) within UGTRL and PtoUGT88A1 that associated with five traits (DBH, V, FW, Pn, and Cond), explaining 1.58–9.03 % of the phenotypic variance (Table S3). Only one haplotype (A-T) from SNP28–30 within UGTRL was associated with FW. Among the haplotypes identified in PtoUGT88A1, one haplotype (C-G) from SNP1–2, two haplotypes (G-T and T-G) from SNP21–22, and two haplotypes (C-C and T-G) from SNP46–47 were associated with Cond, Pn, and DBH, respectively, which were strongly supported by three single-SNP-based associations (SNP2, SNP22, and SNP46) for the same traits (Tables S2 and S3).
Multi-SNP association under additive and dominant effects
To further decipher the roles and interactions of UGTRL and PtoUGT88A1 in tree growth and wood formation, 108 common SNPs were used to conduct multi-SNP association analysis with 14 quantitative traits in the association population, using a Bayesian Lasso model, which reveals multi-SNP additive and dominant genetic effects and uncovers some minor effects of SNPs on quantitative traits. In total, we detected 115 significant associations (P < 0.01) between 37 unique SNPs from UGTRL and its target and 12 quantitative traits of P. tomentosa with average R 2 of 4.33 % (Fig. 3a, b), indicating the common role of UGTRL and its putative target in the phenotypic variation. Of these, 16 and 21 SNPs were from UGTRL and PtoUGT88A1, respectively, and 57.14 % of the 21 associated loci in PtoUGT88A1 were in the coding region.
The SNP-trait associations with additive and dominant effects in the association population of P. tomentosa. a The associations with additive effect between 36 SNPs from UGTRL and its target and 12 traits. The pale and deep blue circles indicate the SNPs from UGTRL and PtoUGT88A1, respectively, and green circles indicate the associated traits. b The associations with dominant effect between 39 SNPs from UGTRL and its target and 12 traits. The jacinth and pink circles indicate the SNPs from UGTRL and PtoUGT88A1, respectively, and red circles in the central indicate the associated traits. For a, b, the solid lines indicate the associations detected in both additive and dominant models, and dotted lines in a, b indicate the associations only identified in the additive and dominant models, respectively. In addition, the circles with L markers (such as L7) and T markers (such as T10) in the outer lane indicate the associated SNPs from UGTRL and PtoUGT88A1, respectively. c Each number of associated trait corresponding to the numbers of SNP from UGTRL and PtoUGT88A1. For example, a total of 11 SNPs, five in UGTRL and seven in PtoUGT88A1, associated with two traits. d The number of associated SNPs in UGTRL and PtoUGT88A1 with joint additive and dominant effects for each trait
Additive effects
We detected 101 significant associations with additive effects involving 12 traits and 36 unique SNPs within UGTRL and PtoUGT88A1. The additive values for these associations ranged from 0.30 to 29.89 % (Fig. 3a, Tables 2 and S4). Forty-three associations were with SNPs in UGTRL and 58 associations with SNPs in PtoUGT88A1; 65.52 % of the associations in PtoUGT88A1 were in the coding region. The numbers of identified associations varied for the 12 traits, with the most SNP-trait associations affecting wood property traits, including 11 for wood physical properties and 40 for wood chemical composition, of which 43.14 % associations were with SNPs in UGTRL. In particular, SNP2 and SNP7 in UGTRL associated with HEC had the largest (27.18 %) and smallest (0.49 %) effect, respectively.
Dominant effects
Under the dominant model, we identified 100 SNP-trait associations among 12 traits and 37 unique SNPs from UGTRL and PtoUGT88A1 (Fig. 3b). The dominant effect of these associations ranged from −27.80 to 28.19 %, and 52 % were positive dominant values (Tables 3 and S5). Each trait had 2–17 associations with different SNPs of UGTRL and PtoUGT88A1, and the trait for wood chemical composition had the most associations, with 17 associated SNPs in UGTRL. Both lncRNA UGTRL and PtoUGT88A1 associated with 12 traits of three categories, and the majority of SNPs within UGTRL and its target were associated with more than one trait, exhibiting opposite and different dominant values for different traits, like SNP7 in UGTRL, which associated with eight traits with different dominant effects from −22.59 to 24.94 %, of which 62.5 % were negative values.
Among the 115 associations, 86 had a joint additive and dominant effect between 12 traits and 36 significant SNPs from UGTRL and PtoUGT88A1 with R 2 of 0.06 to 7.28 % (Tables 4 and S6), which is revealing the strong effects led by the common role of the lncRNA UGTRL and its putative target for the phenotype and uncovering their potential role in regulating growth and development of trees. Of these associated SNPs, 66.67 % were associated with more than one trait with disparate contributions to each trait (Fig. 3c). In addition, each trait associated with 2–15 significant SNPs in both UGTRL and PtoUGT88A1 and contributed differently to each trait (Fig. 3d), indicating that lncRNA UGTRL and its target may act in the same pathway in affecting the phenotypic variation.
Epistatic modeling demonstrated the strong genetic interactions of lncRNA UGTRL and its target
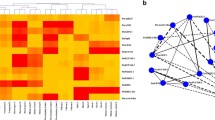
The additive and dominant effects uncovered the joint effect of unique SNPs from UGTRL and PtoUGT88A1 in phenotypic variation. Epistasis effects between SNP pairs from different genes revealed the strong correlation of different genes to phenotypic variation. We used a novel algorithm to detect the epistasis of SNP-SNP pairs between UGTRL and PtoUGT88A1, in pairs associated with 14 traits across different categories. After correcting for multiple testing, 84 significant pairwise associations (Q < 0.1) were identified involving 13 traits and 42 unique SNPs from UGTRL (18) and PtoUGT88A1 (24) with main effects from 0 to 10.04 % and pairwise effect from 0 to 20.20 % (Tables 5 and S7). Of these associated SNPs identified in MDR, 21.43 % of unique SNPs discovered in the epistasis model were also significant in the additive and dominant models. The epistasis network consisting of SNPs from UGTRL and its putative target provided powerful evidence for the interactions of lncRNA-mRNA and the potential role of this gene pair in wood formation and tree growth (Fig. 4a).
The epistatic network within the SNPs from UGTRL and PtoUGT88A1 and phenotypic variation produced by the genotypic combinations of different SNP pairs. a A structural network reveals the epistatic interactions of different loci in UGTRL and PtoUGT88A1. The gray lines represent the SNP pairs of UGTRL and its putative target identified with epistatic effects on different traits, which are showed in the gray lines. The blue and red circles represent the SNPs from UGTRL and its target, respectively. The yellow dotted lines indicate the SNPs from UGTRL or PtoUGT88A1. The numbers with L and T markers represent the SNPs from UGTRL and PtoUGT88A1, respectively. b–d The phenotypic variation of DBH (cm), FW (μm), and Cond (mol m−2 s−1) produced by different genotypic combinations for SNP pairs of L41-T43 (b), L9-T59 (c), and L32-T45 (d), respectively. The blue bars represent the transcript levels of UGTRL with different genotypes in three SNP pairs, and the red bars represent the transcripts levels of PtoUGT88A1 with different genotypes of three SNP pairs. The green nodes connected by green lines in each class represent the mean values of DBH (b), FW (c), and Cond (d) for four genotypic combinations
In the epistasis model, we used IG to evaluate the epistatic effect of SNP-SNP pairs and IG ranged from −8.01 to 5.57 % (Tables 5 and S7). Of the IGs for these associated SNP-SNP pairs, 73.81 % showed negative effects. By contrast, 21 SNP-SNP pairs showed positive effects across 11 traits and 25 unique SNPs (11 from UGTRL and 14 from PtoUGT88A1) with IGs of 0.04 to 5.57 %; of these, the SNP pairs of SNP41 in UGTRL (L41) and SNP43 in PtoUGT88A1 (T43) showed the highest IG for DBH (5.57 %) (Table S6). Interestingly, nine SNPs in UGTRL and nine in PtoUGT88A1 associated with more than one trait, such as SNP24 in UGTRL, which had 17 associations with 13 unique SNPs from its target for six traits (H, V, CC, FL, MFA, and Ci) harboring different main effects (0.11–3.44 %) to each trait. Notably, five SNP-SNP pairs, including L3-T34, L24-T13, L24-T21, L24-T67, and L24-T8, were associated with two traits with different IGs, representing the crucial roles of the SNP pairs in tree growth and development.
The effects of genotypic combinations on phenotypic variation
To further investigate the effects of genotypic combinations of different SNPs in UGTRL and its putative target on variation in phenotype, we measured the transcript levels of L41-T43, L9-T59, and L32-T45. These pairs possessed the highest positive IGs for three categories of traits, indicating that the interactions of these SNP pairs explained more genetic variation than the sum of the individual effects of the two SNPs (Fig. 4b–d). Taking the L41-T43 pair as an example, the plants where L41 had the genotype GT had higher UGTRL transcript levels than plants with the TT genotype; plants with GT had transcript levels of 6.03 (arbitrary units normalized to control) compared with 5.10 for TT plants. Similarly, plants where T43 had the AA genotype had higher PtoUGT88A1 transcript levels than plants with the AG genotype (42.44 compared with 39.88) (Fig. 4b). Then, we calculated the average values of DBH for the four genotypic combinations of L41-T43 (GT-AA, GT-AG, TT-AA, and TT-AG) in the association population. Interestingly, we found that in plants with the GT genotype of L41, the average DBH varied depending on the genotype of T43; the average DBH was 27.09 cm for GT-AA and 22.02 cm for GT-AT. We detected no significant differences in the average values of DBH when the genotype of L41 was TT (Fig. 4b), for the genotypic pairs of TT-AA (20.74 cm) and TT-AG (20.31 cm). The SNP-SNP pair of L9-T59 exhibited similar genotypic effects for the FW trait and L32-T45 similarly affected Cond (Fig. 4c, d).
Discussion
Extensive studies on lncRNAs in animals and plants in recent years have produced some remarkable achievements, such as in the treatment of cancer (Wapinski and Chang 2011). Also, a variety of approaches have demonstrated the vital roles of lncRNAs in plants. For example, the lncRNA long-day-specific male-fertility-associated RNA (LDMAR) in rice affects photoperiod-sensitive male sterility and provides a practical tool for generating hybrid rice (Ding et al. 2012). lncRNAs perform their various roles through all kinds of mechanisms, such as interacting with RNAs and proteins (Quan et al. 2015), and lncRNA-mRNA interaction based on base pairing is a common, significant mechanism. For example, the lncRNA BACE1-AS, which is a biomarker for Alzheimer’s disease, binds its target BACE1 mRNA, increases the stability of BACE1 mRNA, and thus regulates the expression of BACE1 (Faghihi et al. 2008). The HID1 lncRNA associates with the promoter region of PIF3 and represses the expression of PIF3 to positively regulate red-light-mediated seeding photomorphogenesis in Arabidopsis (Wang et al. 2014a). Thus, to identify the putative roles of the UGTRL lncRNA in tree growth and development, we used methods based on sequence complementarity and thermodynamic parameters, feasible and efficient bioinformatics methods for predicting the target of lncRNAs. However, the putative functions of lncRNA UGTRL revealed by its putative target need to be further analyzed, as it seems insufficient to certify the effect of the lncRNA-mRNA interaction on phenotypic variation in trees, based only on the bioinformatics prediction.
The characteristics of perennial trees, such as their long generation times and the lack of characterized mutants, make transgenic and reverse genetic strategies difficult for studying gene function. Association genetics conquers these deficiencies and provides an efficient, feasible method for detecting significant SNP markers in natural populations of trees (Thumma et al. 2005). Single-SNP-based associations have been widely used in studies for identifying the causal SNPs within gene attribute to quantitative traits (Tian et al. 2012; Wegrzyn et al. 2010). In our study, four SNPs in UGTRL and five SNPs in PtoUGT88A1 associated with DBH, HC, FW, Pn, and Cond and five haplotype-based associations identified in this study were strongly supported by three single-marker associations (Tables S2 and S3), suggesting that UGTRL and its target may have roles in tree growth and wood formation. In addition, four haplotypes simultaneously associated with more than one trait, such as C-G haplotype in SNP1–2 of PtoUGT88A1 associated with three traits (DBH, V, and Cond), which indicated the pleiotropic effects and significance of the loci (Du et al. 2015) (Table S3).
Multi-SNP association with additive and dominant effects, which covers the effects of multiple, weaker SNPs, was used to decipher the effects of an lncRNA-mRNA pair on phenotypic variation underlying growth, wood properties, and photosynthetic traits. Here, 86 associations with joint additive and dominant effects were detected to associate with 12 traits, representing 36 significant SNPs from UGTRL and PtoUGT88A1 (Fig. 3), indicating a common effect of the lncRNA-mRNA pair on a certain trait, and supporting the putative roles of UGTRL in affecting phenotypic variation in P. tomentosa, likely through the same pathway as its target. For example, seven SNPs from UGTRL and PtoUGT88A1 were found to associate with LC under additive and dominant effects with R 2 from 0.56 to 5.60 %, indicating that lncRNA UGTRL and its target may affect LCs through a shared pathway. In addition, the phenotypic variance explained by SNPs in UGTRL (11.88 %) was higher than that explained by SNPs in PtoUGT88A1 (9.42 %), indicating that UGTRL may have a major effect on variation in LC. Previous studies showed that UGT72E2 in Arabidopsis thaliana was responsible for accumulation of coniferyl and sinapyl alcohol 4-O-glucosides, the basic monolignols for lignin biosynthesis in the phenylpropanoid pathway (Boerjan et al. 2003; Lanot et al. 2006). Based on the associations under additive and dominant effects, our findings indicate that UGTRL and PtoUGT88A1 may co-regulate LCs through affecting the biosynthesis of the precursors of lignin and UGTRL may have a similar role as its target. Furthermore, a majority of SNPs associated with multiple traits under additive and dominant models with different contributions, such as SNP7 in UGTRL associated with six traits with varied R 2 from 1.31 to 5.97 %, suggesting the pleiotropy of specific SNPs, consistent with results of previous studies (Yang et al. 2015).
Furthermore, a high correlation in expression of UGTRL and its target (r = 0.77) was also observed, together with association studies with additive and dominance effects, which provide strong evidence that UGTRL and its target may affect phenotypic variation in the same pathway. In our study, comparing the expression levels of mRNAs revealed that UGTRL expressed at a relative low level (Chen et al. 2015). UGTRL and PtoUGT88A1 showed higher expression in secondary tissues (xylem) than in primary tissues (cambium) except leaves, indicating their potential effect on accumulation of secondary metabolites. In addition, UGTs participate in secondary metabolism in poplar, such as metabolism of cell wall polysaccharides (Aspeborg et al. 2005), thus suggesting that UGTRL and its target may function in secondary cell wall biosynthesis. UGTs are encoded by a multi-gene family and catalyze the transfer of glycosyl from UDP-glucose (activated nucleotide sugars) to aglycones (acceptor molecules) (Ross et al. 2001), and the high abundance of the two transcripts in old leaves and 29 associations with photosynthetic traits detected under multi-SNP associations indicate that both UGTRL and PtoUGT88A1 may commonly function in transferring the glucose synthetized by photosynthesis to acceptor molecules in all tree organs, such as xylem. The results of multi-SNP associations and transcriptome profiling demonstrate that lncRNA UGTRL and its putative target, PtoUGT88A1, may have common roles in wood formation through transferring sugar moieties synthetized by photosynthesis to organs of trees, secondary cell wall biosynthesis, and lignin biosynthesis.
Epistasis, which profoundly influences evolution, can be described as nonadditive interactions among separate mutations (Huang et al. 2012). Previous studies on disease susceptibility showed that complex gene interactions have a much stronger effect than single genes (Chou et al. 2011). Also, epistasis analysis addresses the limitation of examining only a single gene and provides an additional and reasonable pathway for further investigating lncRNA-mRNA interactions and their effects on quantitative traits. Our study characterized 84 significant SNP-SNP epistatic associations involved in 13 traits and 42 unique SNPs from UGTRL and PtoUGT88A1 (Fig. 4a), which provides necessary genetic evidence for dissecting the interactions of lncRNA-mRNA pairs for phenotypic variation. Of the SNPs identified in the epistasis model, 21.43 % were also significant in the additive and dominant models, mainly because the SNPs detected in epistasis have minor effects; thus, some of these SNPs could not be detected under additive and dominant models (Xu and Jia 2007). Interestingly, 18 SNPs from UGTRL and PtoUGT88A1 associated with more than one trait, which improved our understanding of pleiotropy for certain genes. In addition, SNP24 in UGTRL formed four SNP pairs with SNP8, SNP13, SNP21, and SNP67 in PtoUGT88A1 under the epistasis model, and each pair associated with two traits with different IGs. The observation further demonstrated the vital roles of SNP24 in UGTRL and clearly showed the significance of these SNP pairs detected under the epistasis model for phenotypic variation, thus indicating the putative regulatory roles of interactions of UGTRL and PtoUGT88A1 in wood formation. In addition, 73.81 % of these SNP-SNP pairs showed negative epistatic effects, indicating the redundant information carried by the pairs for each specific trait (Collins et al. 2013), and indicate that UGTRL and its target may contribute to the phenotypic variation through the same pathway with similar roles. Twenty-one SNP-SNP pairs showed positive epistatic effects on 11 traits, indicating that the SNP pairs showed a greater contribution to phenotypic variation than the sum of the individual effects of the two SNPs. Both negative and positive IGs revealed the close interactions of the lncRNA-mRNA pairs.
The relationship between epistatic effects produced by different genotypic combinations and phenotypic traits also requires further discussion. For example, the SNP pair L41-T43 had high IG and associated with DBH (Fig. 4b). The DBH was controlled by the genotype of T43 when the genotype of L41 is GT, but the effect of T43 on DBH is masked when the genotype of L41 is TT. More broadly, the different genotypes of L41 and T43 represented different expression levels of UGTRL and PtoUGT88A1, and the mean DBH depends on the transcript levels of PtoUGT88A1 when lncRNA UGTRL had a high abundance. However, the low abundance of UGTRL transcript decreased the effect of PtoUGT88A1 on DBH. A similar phenomenon was also identified in L9-T59 for FW and L32-T45 for Cond. In other words, the genotype of one SNP or genotype combination of several SNPs may affect the expression of genes, and epistasis among the genotype combinations of different genes in the same pathway could affect phenotypic variation. Epistasis provides an additional and effective path for application of MAS, which is needed in future studies to understand the functional roles of genes in the networks for a specific trait or pathway.
Conclusions
Here, we first predicted PtoUGT88A1 as the putative target of lncRNA UGTRL using bioinformatics methods and clarified the genetic variations and putative interactions of UGTRL and PtoUGT88A1 in wood formation with a combined approach using multiple SNP-based association analysis and examination of the expression patterns of them. Multiple SNP-based association under additive and dominant models and expression pattern analysis provided the genetic evidence that both UGTRL and PtoUGT88A1 commonly act on the phenotypic variation of P. tomentosa, indicating the potential joint roles of UGTRL and its target in wood formation. Furthermore, the epistasis model provided a feasible approach and uncovered the interactions of UGTRL and PtoUGT88A1 for each trait, indicating that the epistatic interactions of lncRNA UGTRL and its putative target affected certain traits. Thus, this genetic analysis annotated the putative function of UGTRL and revealed the potential roles of interactions of UGTRL and its target PtoUGT88A1 in wood formation by affecting the transfer of sugar moieties synthetized by photosynthesis to organs of trees, secondary cell wall biosynthesis, and lignin biosynthesis. Overall, the strategy of integrating expression analysis and multi-SNP-based association analysis with additive, dominant, and epistasis models can be used in population studies for dissecting the interactions of candidate genes and the effect of their genetic variation on certain phenotypes. Importantly, some SNPs detected in our study have significant associations with phenotypic characteristics with joint genetic effects, a crucial clue for MAS breeding to screen key traits.
References
Aspeborg H, Schrader J, Coutinho PM, Stam M, Kallas A, Djerbi S, Nilsson P, Denman S, Amini B, Sterky F, Master E, Sandberg G, Mellerowicz E, Sundberg B, Henrissat B, Teeri TT (2005) Carbohydrate-active enzymes involved in the secondary cell wall biogenesis in hybrid aspen. Plant Physiol 137:983–997
Barbazuk WB, Fu Y, McGinnis KM (2008) Genome-wide analyses of alternative splicing in plants: opportunities and challenges. Genome Res 18:1381–1392
Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54:519–546
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Chen J, Quan M, Zhang D (2015) Genome-wide identification of novel long non-coding RNAs in Populus tomentosa tension wood, opposite wood and normal wood xylem by RNA-seq. Planta 241:125–143
Chou HH, Chiu HC, Delaney NF, Segre D, Marx CJ (2011) Diminishing returns epistasis among beneficial mutations decelerates adaptation. Science 332:1190–1192
Collins RL, Hu T, Wejse C, Sirugo G, Williams SM, Moore JH (2013) Multifactor dimensionality reduction reveals a three-locus epistatic interaction associated with susceptibility to pulmonary tuberculosis. BioData Min 6:4
Ding J, Lu Q, Ouyang Y, Mao H, Zhang P, Yao J, Xu C, Li X, Xiao J, Zhang Q (2012) A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proc Natl Acad Sci U S A 109:2654–2659
Du Q, Wang B, Wei Z, Zhang D, Li B (2012) Genetic diversity and population structure of Chinese White poplar (Populus tomentosa) revealed by SSR markers. J Hered 103:853–862
Du Q, Pan W, Tian J, Li B, Zhang D (2013) The UDP-glucuronate decarboxylase gene family in Populus: structure, expression, and association genetics. PLoS One 8:e60880
Du Q, Tian J, Yang X, Pan W, Xu B, Li B, Ingvarsson PK, Zhang D (2015) Identification of additive, dominant, and epistatic variation conferred by key genes in cellulose biosynthesis pathway in Populus tomentosa. DNA Res 22:53–67
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St LGR, Kenny PJ, Wahlestedt C (2008) Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med 14:723–730
Guerra FP, Wegrzyn JL, Sykes R, Davis MF, Stanton BJ, Neale DB (2013) Association genetics of chemical wood properties in black poplar (Populus nigra). New Phytol 197:162–176
Hahn LW, Ritchie MD, Moore JH (2003) Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics 19:376–382
Han L, Zhang K, Shi Z, Zhang J, Zhu J, Zhu S, Zhang A, Jia Z, Wang G, Yu S, Pu P, Dong L, Kang C (2012) LncRNA profile of glioblastoma reveals the potential role of lncRNAs in contributing to glioblastoma pathogenesis. Int J Oncol 40:2004–2012
Hardy O, Vekemans X (2002) SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes: 618-620
Heo JB, Sung S (2011) Encoding memory of winter by noncoding RNAs. Epigenetics 6:544–547
Hill WG, Robertson A (1968) Linkage disequilibrium in finite populations. Theor Appl Genet 38:226–231
Huang W, Richards S, Carbone MA, Zhu D, Anholt RR, Ayroles JF, Duncan L, Jordan KW, Lawrence F, Magwire MM et al (2012) Epistasis dominates the genetic architecture of Drosophila quantitative traits. Proc Natl Acad Sci U S A 109:15553–15559
Hubisz MJ, Falush D, Stephens M, Pritchard JK (2009) Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour 9:1322–1332
Jones P, Vogt T (2001) Glycosyltransferases in secondary plant metabolism: tranquilizers and stimulant controllers. Planta 213:164–174
Lanot A, Hodge D, Jackson RG, George GL, Elias L, Lim EK, Vaistij FE, Bowles DJ (2006) The glucosyltransferase UGT72E2 is responsible for monolignol 4-O-glucoside production in Arabidopsis thaliana. Plant J 48:286–295
Lee JT, Bartolomei MS (2013) X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell 152:1308–1323
Li J, Das K, Fu G, Li R, Wu R (2011) The Bayesian lasso for genome-wide association studies. Bioinformatics 27:516–523
Li J, Ma W, Zeng P, Wang J, Geng B, Yang J, Cui Q (2014a) LncTar: a tool for predicting the RNA targets of long noncoding RNAs. Brief Bioinform
Li L, Eichten SR, Shimizu R, Petsch K, Yeh CT, Wu W, Chettoor AM, Givan SA, Cole RA, Fowler JE, Evans MM, Scanlon MJ, Yu J, Schnable PS, Timmermans MC, Springer NM, Muehlbauer GJ (2014b) Genome-wide discovery and characterization of maize long non-coding RNAs. Genome Biol 15:R40
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Liu J, Jung C, Xu J, Wang H, Deng S, Bernad L, Arenas-Huertero C, Chua NH (2012) Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 24:4333–4345
Martens JA, Laprade L, Winston F (2004) Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429:571–574
Moore JH, Gilbert JC, Tsai CT, Chiang FT, Holden T, Barney N, White BC (2006) A flexible computational framework for detecting, characterizing, and interpreting statistical patterns of epistasis in genetic studies of human disease susceptibility. J Theor Biol 241:252–261
Neale DB, Kremer A (2011) Forest tree genomics: growing resources and applications. Nat Rev Genet 12:111–122
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Plavcová L, Hacke UG, Almeida-Rodriguez AM, LI E, DOUGLAS CJ (2013) Gene expression patterns underlying changes in xylem structure and function in response to increased nitrogen availability in hybrid poplar. Plant Cell Environ 36:186–199
Ponting CP, Oliver PL, Reik W (2009) Evolution and functions of long noncoding RNAs. Cell 136:629–641
Quan M, Chen J, Zhang D (2015) Exploring the secrets of long noncoding RNAs. Int J Mol Sci 16:5467–5496
Remington DL, Thornsberry JM, Matsuoka Y, Wilson LM, Whitt SR, Doebley J, Kresovich S, Goodman MM, Buckler ET (2001) Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc Natl Acad Sci U S A 98:11479–11484
Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF, Moore JH (2001) Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am J Hum Genet 69:138–147
Ross J, Li Y, Lim E, Bowles DJ (2001) Higher plant glycosyltransferases. Genome Biol 2:S3004
Samuels AL, Kaneda M, Rensing KH (2006) The cell biology of wood formation: from cambial divisions to mature secondary xylem. Can J Bot 84:631–639
Sexton TR, Henry RJ, Harwood CE, Thomas DS, McManus LJ, Raymond C, Henson M, Shepherd M (2012) Pectin methylesterase genes influence solid wood properties of Eucalyptus pilularis. Plant Physiol 158:531–541
Shin JH, Chekanova JA (2014) Arabidopsis RRP6L1 and RRP6L2 function in FLOWERING LOCUS C silencing via regulation of antisense RNA synthesis. PLoS Genet 10:e1004612
Shuai P, Liang D, Tang S, Zhang Z, Ye CY, Su Y, Xia X, Yin W (2014) Genome-wide identification and functional prediction of novel and drought-responsive lincRNAs in Populus trichocarpa. J Exp Bot 65:4975–4983
Storey JD, Tibshirani R (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci U S A 100:9440–9445
Sun YG, Wang B, Jin SH, Qu XX, Li YJ, Hou BK (2013) Ectopic expression of Arabidopsis glycosyltransferase UGT85A5 enhances salt stress tolerance in tobacco. PLoS One 8:e59924
Suzuki S, Li L, Sun YH, Chiang VL (2006) The cellulose synthase gene superfamily and biochemical functions of xylem-specific cellulose synthase-like genes in Populus trichocarpa. Plant Physiol 142:1233–1245
Swiezewski S, Liu F, Magusin A, Dean C (2009) Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462:799–802
Tafer H, Hofacker IL (2008) RNAplex: a fast tool for RNA-RNA interaction search. Bioinformatics 24:2657–2663
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thumma BR, Nolan MF, Evans R, Moran GF (2005) Polymorphisms in cinnamoyl CoA reductase (CCR) are associated with variation in microfibril angle in Eucalyptus spp. Genetics 171:1257–1265
Tian J, Du Q, Chang M, Zhang D (2012) Allelic variation in PtGA20Ox associates with growth and wood properties in Populus spp. PLoS One 7:e53116
Tian J, Du Q, Li B, Zhang D (2014) Single-nucleotide polymorphisms in the 5′ UTR of UDP-glucose dehydrogenase (PtUGDH) associate with wood properties in Populus tomentosa. Tree Genet Genomes 10:339–354
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578
Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A et al (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313:1596–1604
Wang B, Du Q, Yang X, Zhang D (2014a) Identification and characterization of nuclear genes involved in photosynthesis in Populus. BMC Plant Biol 14:81
Wang Y, Fan X, Lin F, He G, Terzaghi W, Zhu D, Deng XW (2014b) Arabidopsis noncoding RNA mediates control of photomorphogenesis by red light. Proc Natl Acad Sci U S A 111:10359–10364
Wapinski O, Chang HY (2011) Long noncoding RNAs and human disease. Trends Cell Biol 21:354–361
Watterson GA (1975) On the number of segregating sites in genetical models without recombination. Theor Popul Biol 7:256–276
Wegrzyn JL, Eckert AJ, Choi M, Lee JM, Stanton BJ, Sykes R, Davis MF, Tsai CJ, Neale DB (2010) Association genetics of traits controlling lignin and cellulose biosynthesis in black cottonwood (Populus trichocarpa, Salicaceae) secondary xylem. New Phytol 188:515–532
Xu S, Jia Z (2007) Genome-wide analysis of epistatic effects for quantitative traits in barley. Genetics 175:1955–1963
Yang X, Du Q, Chen J, Wang B, Zhang D (2015) Association mapping in Populus reveals the interaction between Pto-miR530a and its target Pto-KNAT1. Planta 242:77–95
Yu J, Pressoir G, Briggs WH, Vroh BI, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, Holland JB, Kresovich S, Buckler ES (2006) A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet 38:203–208
Zaykin DV, Westfall PH, Young SS, Karnoub MA, Wagner MJ, Ehm MG (2002) Testing association of statistically inferred haplotypes with discrete and continuous traits in samples of unrelated individuals. Hum Hered 53:79–91
Zhang D, Zhang Z, Yang K (2006) QTL analysis of growth and wood chemical content traits in an interspecific backcross family of white poplar. Can J Forest Res 36:2015–2023
Zhang D, Zhang Z, Yang K (2007) Identification of AFLP markers associated with embryonic root development in Populus tomentosa Carr. Silvae Genet: 27-32
Zhang D, Du Q, Xu B, Zhang Z, Li B (2010a) The actin multigene family in Populus: organization, expression and phylogenetic analysis. Mol Genet Genomics 284:105–119
Zhang D, Yang X, Zhang Z, Li B (2010b) Expression and nucleotide diversity of the poplar COBL gene. Tree Genet Genomes 6:331–344
Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT (2008) Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322:750–756
Acknowledgments
This work was supported by the National “863” Plan Project (No. 2013AA102702), the State Key Basic Research Program of China (No. 2012CB114506), and the Fundamental Research Funds for the Central Universities (No. BLYJ201408).
Data archiving statement
Sequence data from this article have been deposited with the GenBank Data Library under the accession nos. KR492527–KR492612.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by P. Ingvarsson
Mingyang Quan and Jiaxing Tian contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1
The real-time PCR primers used in this study.(DOC 29 kb)
Table S2
Significant SNPs associated with all traits calculated by MLM in the association population of Populus tomentosa.(DOC 41 kb)
Table S3
Significant haplotypes from UGTRL and its putative target PtoUGT88A1 associated with growth, wood properties, and photosynthetic traits in the association population of Populus tomentosa.(DOC 50 kb)
Table S4
Additive effect of all significant SNPs from UGTRL and PtoUGT88A1 associated with each trait in the association population of Populus tomentosa.(DOC 142 kb)
Table S5
Dominant effect of all significant SNPs from UGTRL and PtoUGT88A1 associated with each trait in the association population of Populus tomentosa(DOC 113 kb)
Table S6
The additive, dominant and R 2 (phenotypic contributions) of significant SNPs associated with all the traits under both additive and dominant effects in the association population of Populus tomentosa.(DOC 157 kb)
Table S7
The SNP pairs and their main effects detected from UGTRL and PtoUGT88A1 under epistasis model in the association population of Populus tomentosa.(DOC 123 kb)
Fig. S1
Sequence complementarity of UGTRL and its putative target PtoUGT88A1.(DOC 82 kb)
Rights and permissions
About this article
Cite this article
Quan, M., Tian, J., Yang, X. et al. Association studies reveal the effect of genetic variation in lncRNA UGTRL and its putative target PtoUGT88A1 on wood formation in Populus tomentosa . Tree Genetics & Genomes 12, 8 (2016). https://doi.org/10.1007/s11295-015-0967-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-015-0967-6