Abstract
Postglacial migration paths for most of the tree species of Eastern North America remain unknown. The presence of no-analogue forest communities prior to the last glacial advance suggests that individual trees species in Eastern North America may respond differently as climate changes and human impacts increase. In this study, we examined chloroplast haplotypes from natural populations of Juglans cinerea L., a North American forest tree, to infer postglacial migration patterns. Sequences from eight different regions of the chloroplast genome in 197 trees distributed across the range revealed 10 haplotypes. A minimum spanning network, phylogenetic analysis and haplotype distributions revealed that the three most common haplotypes were geographically disjunct and not closely related. Haplotype 6 (73 trees) occurred only in western populations, haplotype 10 (83 trees) occurred only in eastern populations and haplotype 7 (21 trees) occurred only at the southern edge of the native range. The southernmost population contained the most haplotype diversity but included no eastern haplotypes. Haplotype phylogeography suggested geographical differentiation prior to the last glacial advance in eastern populations and separate postglacial migration paths for eastern and western populations. As migration of J. cinerea to Atlantic Canada from southern refugia does not appear possible given known seed dispersal mechanisms, the possibility of northern refugia or dispersal by extinct megafauna merits serious consideration. Differences among species in preglacial history, ecological niche preferences and seed dispersal mechanisms suggest that response to long-term climate change and acute human disturbance may be highly species specific.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The presence of no-analogue forest communities prior to the last glacial advance suggests that individual trees species in Eastern North America may respond differently as climate changes and human impacts increase. The two Juglans species native to Eastern North America, Juglans nigra L. (eastern black walnut) and Juglans cinerea L. (butternut), are of particular concern, given the economic value of the former and the threatened status of the latter. Both species bear hard-shelled nuts encased in thick, indehiscent, toxic husks. As birds cannot crack either nut unaided and do not cache them (Stapanian and Smith 1986), the primary animal agents of seed dispersal are scatter hoarding squirrels (Wall 2001). Given the apparent lack of a long distance seed dispersal mechanism, the disparity between where the Juglans were, given the Last Glacial Maximum (LGM) pollen distributions and where both species went, appears to be a particularly acute case of Reid’s paradox (Clark et al. 1998). Chloroplast phylogeography, not previously available for either species, could provide some insight into how both Eastern North American Juglans species migrated in response to the last major climate shift in this region.
As J. cinerea L. (butternut) faces extinction from the combined threats of disease and climate change, we have focussed on the chloroplast phylogeography of this species while the data can still be gathered. J. cinerea is listed as an endangered tree in Canada and a threatened tree or species of special concern in the USA (Schultz 2003) due to extensive mortality from butternut canker, a disease caused by the non-native fungus Ophiognomonia clavigignenti-juglandacearum (Broders and Boland 2011). Predictive models of twentieth century J. cinerea distribution based on Paleoclimate modelling and pollen records are in moderate agreement with the observed distributions suggesting ecological niche conservation over this period (Martínez-Meyer and Peterson 2006). While ecological niche preferences provide guidance for the choice of future ex situ sites for conservation, inference of the patterns of postglacial migrations could provide essential information on the rate of unassisted migration and the locations of glacial refugia in a large-seeded, cold-adapted angiosperm forest tree.
The ecological niche preferences of J. cinerea result in low population densities and a patchy distribution (Bryan 1965). Wind pollination suggests a capacity for long distance gene flow, but rate of seed dispersal determines the rate of migration. Birds and small rodents do not cache the nuts of this species, while tree squirrels (Tamiasciurus hudsonicus, Sciurus carolinensis and Sciurus niger) cache the nuts a maximum distance of 40–60 m from the tree (Goheen and Swihart 2003; Ivan and Swihart 2000; Moore et al. 2007). As the nut falls while still encased in a buoyant husk, downstream water dispersal is possible but the seed must strand on a suitable regeneration site before the husk becomes waterlogged (the nut itself will sink if viable). These limitations on seed dispersal and the limited number of suitable regeneration sites once dispersed suggest a limited capacity for rapid natural migration. The natural range of J. cinerea prior to the introduction of butternut canker extended as far as ~1,000 km northeast of the LGM, indicating an average migration rate of ~100 m/year if refugial populations were located close to the LGM. This is the upper limit of the rate estimated for other Eastern North American forest trees under the same assumption (McLachlan et al. 2005). The postglacial pollen record for J. cinerea in central and Eastern North America suggests refugia located north of the contemporary southern range edge, but the record is sparse. The pollen of J. cinerea tends to appear in the early stages of postglacial transition from spruce-dominated forests to mesic deciduous hardwoods (Finkelstein et al. 2006; Watts 1975, 1979). In studies in which J. cinerea pollen was distinguished from that of J. nigra, J. cinerea pollen occurs in sediments dated 25,000 years ago (25 ka) at Anderson Pond, Tennessee (Delcourt 1979), 19.5 ka at Lookout Mountain in northern Georgia (Watts 1975), 14.5 ka at Powers Fort Swale in southwestern Missouri (Royall et al. 1991) and 13 ka at Crystal Lake, IL (Gonzales et al. 2009). However, J. cinerea tends to have a patchy distribution that could leave little trace in sediment cores, and thus the sparse sediment core evidence does not rule out small northern refugia (Cruzan and Templeton 2000). Climate reconstructions for Eastern North America during the LGM suggest that cool season conditions suitable for J. cinerea could have existed as far north as 35° N (Jackson et al. 2000).
Bayesian cluster analysis of the first range wide genetic diversity and differentiation study in J. cinerea showed higher diversity in nuclear microsatellite polymorphisms and more extensive gene flow in the southern parts of the range when compared to the northern range edge, despite severe population losses in the south in the last 50 years due to the fungal disease (Hoban et al. 2010). Evaluation of possible mechanisms for this pattern indicated that lower genetic diversity and higher differentiation along the northern edges of the range was most likely due to founder effects as the range shifted northward during glacial retreat. Although the nuclear data did not clearly show differentiation between eastern and western populations, gene flow in the central and southern parts of the native range may have obscured the original patterns of postglacial migration since the LGM. The J. cinerea native range crosses two continental divides, the Eastern and the St. Lawrence (Fig. 1), and includes three major watersheds: the Mississippi, the Atlantic and the St. Lawrence. Migration into the valleys that traverse the continental divide could be slowed by the lack of suitable regeneration sites and seasonal flooding. The nuts of J. cinerea fall while tightly encased in a thick, buoyant oblate husk, permitting hydrochorous transport (Middleton 2000; Schultz 2003; Victory et al. 2006). Even if water dispersal is relatively infrequent, flooding could cause a pulse of long-distance migration down river, sending the seeds of upland migrants back into source populations. A study of fine-scale J. cinerea differentiation in the Great Smoky Mountains National Park did indicate a small but significant effect of watershed between some but not all populations (Parks et al. 2014).
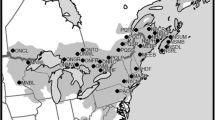
Locations of butternut sites, haplotypes and relative proportions of haplotypes per site. Grey area is the native range for butternut. Dashed line is the approximate ice margin at the LGM. Inset: haplotype network with the black lines showing the inferred connections between the haplotypes based on parsimonious mutation models. An intersection of lines without a circle represents an unobserved haplotype. Size of circles for haplotypes 2, 6, 7 and 10 is proportional to number of individuals. The remaining haplotypes represent one tree each
The fossil and molecular evidence indicates that J. cinerea and the other Juglans species evolved north of the Arctic circle (66° N) and migrated south, differentiating into separate sections and species during the last 40 million years (Aradhya et al. 2007). Fossils and pollen of a J. cinerea ancestor, Juglans eocinerea, occur in sediments dated to the early to mid-Miocene (Hills et al. 1974; Whitlock and Dawson 1990; Wolfe 1980) in the western Arctic Canadian islands. As the species migrated into Eastern North America, water dispersal and an ecological preference for lower altitudes (<500 m) (Parks et al. 2014) could have resulted in differentiation across the Eastern and St. Lawrence continental divides. In the Atlantic watershed, the Appalachian mesophytic ecoregion could have provided refugia for eastern populations during the Pleistocene glaciations. In the Mississippi watershed, west of the Appalachians, areas amenable to northern refugia are not as evident. Ecologically suitable regions in the Ozark valleys in southern Missouri and northern Arkansas and foothills in southern Tennessee, the contemporary southern edge of the J. cinerea range, are already located within 34-35° N. Pollen and macrofossil data from this region indicates the presence of other cool temperate hardwoods during the LGM (Jackson et al. 2000).
In most angiosperms, the chloroplast is inherited only through the egg cell (Zhang et al. 2003) and thus is only dispersed through the seed. When seed dispersal is more limited than pollen dispersal, as is the case with J. cinerea, chloroplast haplotypes can provide a more reliable indication of past migrations than nuclear genotypes (Petit et al. 2003). The inference of postglacial migration paths based on geographical distribution of chloroplast haplotypes assumes that founder effects should decrease haplotype richness and increase haplotype differentiation, in the direction of migration (Le Corre and Kremer 1998). Based on this assumption, chloroplast haplotypes from several species of oaks widely distributed across Europe suggest refugia in Iberian, Italian and eastern Balkan refugia with evidence of haplotype exchange during the last or previous glacial cycles (Petit et al. 2002). In Eastern North America, chloroplast haplotype distribution in American beech (Fagus grandifolia Ehrh.) and red maple (Acer rubrum L.) suggests that some populations colonized northward from refugia closer to the LGM (McLachlan et al. 2005) than the contemporary southern edge of the range. As chloroplast haplotype studies in angiosperm North American trees accumulate (Birchenko et al. 2009; Breen et al. 2012; Magni et al. 2005; Marsico et al. 2009), a complex picture of postglacial migration emerges, in which ecological niche requirements, seed dispersal dynamics, local geography and the intensity of climate instability interact to produce large variations in where tree species find refuge and how long they stay there (Breen et al. 2012; Finkelstein et al. 2006; Soltis et al. 2006; Stewart et al. 2010; Watts 1979). In the wide ranging North American conifer species Picea mariana (Mill.) Britton, Sterns & Poggenb. (black spruce) and Pinus banksiana Lamb. (jack pine), mitochondrial haplotypes have revealed evidence of multiple refugia, cryptic refugia, population mixing at suture zones and complex patterns of postglacial migration (Godbout et al. 2005; Jaramillo-Correa et al. 2004), suggesting that individual species histories, as well as continental geography, influence forest tree response to long-term climate change. Evidence for forest communities in North America with no modern analogue during the LGM (Gill et al. 2009; Jackson et al. 1997; Williams and Jackson 2007; Williams et al. 2001) and evidence for the extinction of at least one tree species in North America (Jackson and Weng 1999; Liu et al. 2013) during glacial retreat indicate that prediction of the location and species composition of future forests as the climate changes requires more information on the postglacial history of a wider range of North American forest tree species.
As butternut canker has now spread throughout the entire native range of J. cinerea (Furnier et al. 1999), the opportunity for studies on the postglacial history of this species based on living descendants is rapidly fading. Although range wide fine-scale geographical sampling in this species is no longer possible due to the local extinctions of southern and central populations, the naturally occurring populations that still remain make a chloroplast haplotype study feasible. Based on evolutionary history, ecological niche preferences and sediment core evidence, we hypothesized that haplotype richness and distribution would be consistent with migration from the contemporary southern range edge in the Mississippi watershed and from Appalachian cryptic refugia close to the last glacial margin in the Atlantic watershed.
Materials and methods
Human and animal subjects
This study did not include or involve the use of human or animal subjects.
Species characteristics and population sampling scheme
The most cold-tolerant species in the Juglans genus, J. cinerea, occurs in scattered, low density populations in the Eastern United States and in parts of Ontario, Quebec and New Brunswick. J. cinerea nuts fall while still fully encased in a thick husk. Tree squirrels are the only animal known to disperse the nuts for winter caching but the oblate green fruits float briefly in water, providing downstream transport in waterways and overland dispersal in seasonal floods (Schultz 2003). Populations for this investigation were selected according to the following criteria: (1) location on a latitudinal gradient to ensure that surviving northern, southern and middle populations were represented; (2) adequate number of trees per population to allow accurate estimation of haplotype richness and frequency (Feng et al. 2008); and (3) inclusion of populations showing a wide range of differentiation for nuclear markers (Hoban et al. 2010). We chose 14 populations with 6–21 individuals from each population. Butternut canker, large-scale agriculture and urban development in the central part of the J. cinerea range have resulted in the disappearance of adequately sized naturally occurring populations (>5 trees) and thus this region is unavoidably underrepresented. Many of the trees we sampled in our previous studies (Hoban et al. 2009, 2010) are now dead.
Hybrid exclusion
Japanese walnut (Juglans ailantifolia Carr.), imported throughout the J. cinerea range in the nineteenth century, hybridizes with J. cinerea, producing fertile and vigorous hybrids that can backcross into local populations (Hoban et al. 2012b). All trees included in this study were positively identified as J. cinerea (as opposed to J. ailantifolia or hybrids) based on 12 nuclear microsatellites (Hoban et al. 2008) and 3 J. cinerea chloroplast markers (McCleary et al. 2009). Although sympatric in many locations, black walnut and butternut arise from different sections of Juglans genus and do not hybridize, eliminating this complication.
DNA extraction, amplification and sequencing
DNA extraction and amplification were performed as previously described (McCleary et al. 2013), using annealing temperatures optimized for the primers used in this study (Table 1). Amplicons were purified and then sequenced on a ABI 3730xl DNA Analyser as previously reported (McCleary et al. 2009). As our preliminary data suggested that sequence polymorphism in the J. cinerea chloroplast was low, we sequenced 16 regions of the chloroplast genome, including 10 regions in which our previous study detected polymorphism in northern red oak (Quercus rubra L.). Primers first reported in our previous study for six of these regions, atp1-rps2, rpoC2-rpoC1, rpoB-rpoC1, ndhK-ndhC, psbH-petB, petB-petD, were identified as universal in that study based on successful amplification across 11 tracheophyte clades (Borkowski et al. 2014). The primers we used for two of these regions, trnC-trnD (Demesure et al. 1995) and trnV-rbcL (Dumolin-Lapegue et al. 1997), were as reported by the original authors (Table 1). The primers we used for the psaA-trnS region were different than those previously reported for the same region (Demesure et al. 1995). The reverse primer we used for trnH-trnK was the same as previously reported (Demesure et al. 1995) but the forward primer was different. We also included petG-trnP and trnT-trnL (Huang et al. 2002), psbB-psbF and rpl20-rps12 (Hamilton 1999), and trnL(UAG)-rpl32 and trnQ(UUG)-5’rps16 (Shaw et al. 2007). We screened these 16 regions in eight individuals each from populations in Waupaca, WI; Keswick, NB; Gilbert Island, NB; Alley Spring State Park, MO; the Allegheny National Forest, PA; and the St. Francis National Forest, AR. Only 8 of the 16 regions we sequenced in the population subsets were polymorphic in J. cinerea (Table 1). We then sequenced 197 trees in 14 populations for all eight regions.
Sequence analysis
We used Sequencher ver. 4.10.1 (Gene Codes Corporation, Ann Arbor, MI) to detect single nucleotide polymorphisms (SNPs) and indels. We generated the geographical distributions of haplotypes in ArcGis v 9.3 (Environmental Systems Research Institute Inc., Redlands, CA). A phylogenetic tree was generated using MrBayes 3.2 (Ronquist et al. 2012) with HKY model of DNA evolution, gamma-distributed rate variation among sites, some sites invariant and the number of generations = 410,000. The number of generations was empirically determined by adding generations to the run until the standard deviation of the split frequencies stabilized at values below 0.01 and the potential scale reduction factors were 1 ± 0.002 for all parameters. The best fit DNA evolution model was chosen based using Modeltest (Posada and Crandall 1998), as implemented in MEGA5 (Tamura et al. 2011). We used network v 4.6.1 (Fluxus Technology Ltd) to generate a minimum spanning tree using median-joining (Bandelt et al. 1999). For calculating the minimum spanning tree, epsilon (a weighting factor for the depth of the search) was set to 0 (default value) and deletions were weighted twice (non-default value) as recommended in the manual for the software. The Connection Cost and Greedy FHP algorithms yielded identical networks.
As our sample numbers varied from 6 to 21 individuals, we used rarefaction to six individuals to calculate haplotype richness as implemented in contrib (Petit et al. 1998). We tested for phylogeographic structure using the method implemented in permut&cpssr (Bordacs et al. 2002; Pons and Petit 1996). Differentiation was calculated for unordered (G ST) and ordered (N ST) alleles then 10,000 haplotype permutations among populations were generated to test if N ST was significantly greater than G ST. When more closely related haplotypes occur more often in the same population than distant populations, N ST will be greater than G ST (Pons and Petit 1996). contrib and permut&CPSSR are available at http://www.pierroton.inra.fr/genetics/labo/Software/.
Results
Out of the 16 chloroplast intergenic regions sequenced, only 8 showed polymorphisms. Seven of the polymorphic regions (atpI-rps2, ndhK-ndhC, psaA-trnS, rpoB-rpoC1, trnC-trnD and trnH-trnK) are also polymorphic in Q. rubra (Fagaceae, Fagales) and have demonstrated utility as universal primers. The region trnL(UAG)-rpl32, originally selected by Shaw and colleagues (2007) for investigations at low taxonomic levels, proved informative in this one. Sequences for all eight regions in 197 individuals across 14 populations revealed 11 polymorphic sites: 10 SNPs and 1 indel. Two of the SNPs were located in trnL(UAG)-rpl32, in the small single copy region of the chloroplast, a region not queried in most chloroplast haplotype studies. Eight of the 10 SNPs detected were transversions (Table 2).
The 10 haplotypes detected (Table 2) were highly differentiated among sites (G st = 0.842 se = 0.069) and haplotypes richness was low across the range (Table 3). The value of N st (0.91, se = 0.053), a measure of differentiation for ordered alleles, was significantly greater (P = 0.01) than the measure of differentiation for unordered alleles (G st = 0.842 se = 0.069), based on 10,000 permutations (P = 0.01), indicating high phylogeographic structure. Over 80 % of the trees genotyped (158) had one of two haplotypes (6 or 10), with haplotype 6 present only in western populations and haplotype 10 present only in eastern populations (Table 3, Fig. 1). Haplotypes 6 and 10 differed at 7 of the 11 polymorphic sites detected (Table 2): four A-C transversions, two G-T transversions and one C-T transition.
The southernmost population (St. Francis National Forest, AK) contained the highest haplotype diversity. Eight populations were monomorphic. Populations in the northeastern part of the range contained haplotypes 8 and 9, in addition to 10; none of which were found in the southern or western populations. Haplotype pairs 1-2, 5-6, 9-10 and 3-4 differed from each other by a single SNP. The pairs 1-2, 5-6 and 9-10 occurred together but haplotypes 3 and 4, each represented by a single tree, occurred at opposite sides of range (Fig. 1, Table 3).
The minimum spanning tree (Fig. 1), based on a parsimony model, and the maximum likelihood phylogeny (Fig. 2), based on a DNA evolution model of different rates of transitions and tranversions, reveal at least two lineages. Under the simple two-lineage interpretation, lineage A (haplotypes 1–6) and lineage B (haplotypes 7–10) are largely but not completely geographically disjunct. However, the minimum spanning tree and the phylogenetic analysis indicate that this interpretation is too simple. Haplotype 10 is the predominant haplotype in all of the populations in the Atlantic and the St. Lawrence watersheds. Haplotype 6 occurred only in populations in the Mississippi watershed. The five westernmost locations, distributed across a latitudinal gradient spanning nearly 11°, share related haplotypes and display a pattern of haplotype loss northward as predicted by the stepping stone model for postglacial migration. The seven northeast locations span ~8.5° of latitude but are nearly monomorphic. Haplotype 8, not closely related to haplotype 10, occurred in one tree at one of the New Brunswick sites, at the extreme northeast corner of the range, where haplotype 10 predominates. Haplotype 7, not directly related to either haplotype 6 or haplotype 10, was the predominant haplotype in the Tennessee population.
Discussion
We queried 16 different intergenic regions of the J. cinerea chloroplast (>7,000 bp) and found polymorphism in only eight of these regions. The SNPs detected included more transversions than transitions, a result also noted in the sequence of the trnT − trnF and atpB − rbcL chloroplast regions in 17 taxa of the Juglans genus (Aradhya et al. 2007), including J. cinerea. Ultimately, we detected 11 polymorphisms comprising 10 haplotypes in populations preselected to represent a wide range of geographical and genetic diversity within the limitations imposed by local extinctions. These 10 haplotypes show high geographical differentiation, with little or no haplotype richness within northern populations. Interpretation of this pattern requires consideration of Pleistocene glacial dynamics, geography and seed dispersal mechanisms.
Assuming that the three most common haplotypes existed prior to the Holocene and taking the phylogeography into consideration, we argue that our data are consistent with multiple glacial refugia. Haplotype phylogeography in the west suggests source populations close to or south of the current southwestern edge of the contemporary range. Sediment cores reveal black walnut pollen (J. nigra) at Nonconnah Creek, Mississippi (35.083° N, −89.916° W) during the LGM (Delcourt et al. 1980), suggesting that J. cinerea, the most cold-tolerant species in the Juglans genus, could have existed in this region and further north. The St. Francis National Forest site, the only site in which we detected four haplotypes, occurs almost at the same latitude, in the Central Mississippi Alluvial Valley. Juglans pollen (species not specified) is just detectable in sediment cores dated to ~16 ka at the Fort Powers Swale, MO, site (36.58° N, −90.56° W), but in the interval 14.5 to 9.5 ka, the authors report both J. cinerea and J. nigra pollen at this site.
The eastern haplotypes could be interpreted as the result of a migration out of a southern refugium east of the Mississippi embayment, in southern Georgia and Florida. Juglans pollen is never abundant but it is present in sediment cores in southern Georgia and Florida during the LGM (Jackson et al. 1997, 2000). Although most pollen records do not distinguish J. nigra from J. cinerea pollen, the latter is more cold tolerant and thus it is reasonable to suppose that J. cinerea was present. The major difficulty with one southeastern refugium is the lack of seed dispersal mechanisms that could have moved J. cinerea from southern Georgia to New Brunswick, a distance of over 2,300 km, in 18–20,000 years. Tree squirrels are the only known animal dispersal agent. Johnson and Webb, in their discussion of bird-dispersed tree seeds, do not consider the extinct passenger pigeon (Ectopistes migratorius) a viable seed dispersal agent, as the birds ate beechnuts and acorns but did not cache them (Johnson and Thompson Webb 1989). Even assuming that delayed digestion could have resulted in occasional transport of viable, intact seed, J. cinerea nuts are large (~3 × 5 cm) and have deeply ridged shells much harder and thicker than the shells of pecan (Carya illinoensis (Wangenh.) K.Koch) and Juglans regia L., the cultivated Persian walnut. Birds cannot crack the nuts unaided. Crows (Corvus brachyrhynchos) crack the nuts of J. regia and Northern California walnut (Juglans hindsii (Jeps.) Jeps. ex R.E. Sm.) by dropping the nuts from a height (Cristol and Switzer 1999), but there are no reports of any bird attempting to do this with the large, sharp-edged nuts of J. cinerea.
Glacial refugia further north could have made the migration to Atlantic Canada possible. Possibilities include eastern refugia further north and a northeastern coastal refugium on the now submerged continental shelf. Many investigators have proposed cryptic northern refugia as a possible solution to Reid’s paradox, the disparity between the predicated rate of tree migration assuming known seed dispersal mechanisms and the apparent rate, as inferred from pollen and fossil evidence (Clark et al. 1998). Although J. cinerea can persist in southern sites where water is abundant, this species as a rule prefers cool, moist conditions and could have survived even further north than the 34-35° N suggested for the northern limit of cold-tolerant temperate hardwoods during the LGM (Jackson et al. 2000) if the periglacial climate was more moderate than previously thought (Loehle 2007). Although our data give little indication of the location of possible northern refugia in the east due to lack of polymorphism, the geological, pollen and fossil evidence suggests a warm thermal enclave between the Southern Appalachians and the Atlantic Ocean, extending as far north as Cape Hatteras (35.2053° N, −75.552° W) and east onto what is now submerged continental shelf (Royall et al. 1991). However, even if butternut has persisted in this region, the distance to Atlantic Canada still exceeds 1,700 km.
Nuclear microsatellite genotypes have revealed that J. cinerea populations in New Brunswick (Hoban et al. 2010) and the red pine (Pinus resinosa Ait.) populations of Newfoundland and New Brunswick (Boys et al. 2005) are genetically distinct from conspecifics further south and west. P. banksiana populations in New Brunswick, Nova Scotia and Prince Edward Island are nearly monomorphic for a mitochondrial haplotype not found anywhere else in a native range that extends to Alberta (Godbout et al. 2005). These results, as well as other data from coastal plain flora and fauna in this region, suggest the possibility of a now submerged coastal glacial refugium in northeastern North America (Godbout et al. 2010; Jaramillo-Correa et al. 2004). As speculative as this interpretation is, given the absence of fossil evidence, it is difficult to explain the data otherwise.
J. cinerea has migrated long distances before, but over much longer time scales. Fossil remains of the species inferred as the ancestor of J. cinerea, J. eocinerea n.sp., occur at many sites in Arctic Canada (Fyles et al. 1994; Hickey et al. 1988; Hills et al. 1974; Whitlock and Dawson 1990) dated to the early Miocene epoch (~22 Ma). As the climate cooled and the deciduous forest migrated south, J. cinerea appeared and eventually populated Eastern North America well before the Pleistocene glaciations. We have shown in other studies that seed dispersal distances in J. cinerea are small (<100 m) and population sizes are limited by the limited availability of favourable regeneration sites (Hoban et al. 2012a). Assuming the ecological preferences of butternut have not changed and that no dispersal agents existed other than squirrels, water remains the least speculative option for long distance seed dispersal. Seed from J. cinerea populations located along rivers could easily be transported further than 100 m downstream or overland during seasonal floods. During the northward retreat of the glacial margin 18–20 ka, butternut could have migrated slowly west and north from eastern coastal refugia. Once northern migrants got across the Laurentian divide, water transport to open sites northward could have provided an opportunity for bursts of seedling regeneration, unlike the resource restricted sites provided by gaps in upland forests (Hoban et al. 2014). Further south, migration westward and north would have been slow, but water transport would have hastened migration in the south east direction. This patchy, uncertain pattern of regeneration would have resulted in repeated founder effects during recolonization.
If haplotype diversity in eastern glacial refugia, regardless of where they were, was high, this proposed regeneration pattern should have resulted in inland populations nearly fixed for different haplotypes. Instead, we see a near monomorphism for haplotype 10, with two divergent haplotypes (3 and 8) in different populations in the northeast. This pattern suggests that the haplotype 10 was already predominant prior to the last glacial advance. The highly divergent nuclear genotypes of the New Brunswick butternuts in the absence of unique chloroplast haplotypes also suggests that this lack of chloroplast diversity existed before the last glacial advance. The individuals having haplotypes 3 and 8 were sequenced twice with the same result, so cannot be dismissed as sequencing errors. Haplotypes 3 and 8 may represent secondary contact during previous glacial episodes but now are nearly lost through genetic drift.
Still assuming that the only dispersal mechanisms for this species are squirrels and water, haplotype 7, detected in one southern site and not closely related to haplotype 6 or haplotype 10, may represent a local chloroplast lineage that became separated during the original colonization of the Eastern United States. This population is the result of a documented burst of natural regeneration within the last 50–70 years (Hoban et al. 2012a). A previous study (Hoban et al. 2010) using nuclear microsatellites indicated that this population is divergent from the other populations (although not as divergent as the butternuts in New Brunswick) and consists of related groups of half sib families, a classic case of genetic drift. The haplotype 7 butternuts in Butternut Valley may represent a remnant of the southern glacial refugium east of the Mississippi embayment. The expectation that additional haplotypes not present in the north should be present here cannot be tested now, due to the extensive local extinction of butternut populations in the South.
Furthermore, the assumption that squirrels were the only mammalian animal dispersal agent for butternut should be reconsidered, given the number of herbivorous Pleistocene megafauna that became extinct in Eastern North America as the ice margins receded and the first people arrived. The contemporary distribution of butternut (before butternut canker) shares similarities to another disappearing tree, the Kentucky coffee tree (Gymnocladus dioicus (L.) K. Koch), native to the central USA. The only contemporary long-distance seed dispersal mechanism for both species and indeed the only seed dispersal mechanism for Kentucky coffee tree is water (Zaya and Howe 2009). Despite the toxicity of the pods and seeds, Zaya and Howe hypothesize that G. dioicus relied on megafaunal fruit dispersal, a mechanism that requires consumption of the fruit, fruit pulp or husk. Based on their argument, three indications of possible megafaunal seed dispersal are large, indehiscent fruits toxic to smaller animals, well-protected seeds that could have withstood megafaunal teeth and fruits that now attract few or no native mammals. Butternut have large, indehiscent toxic fruits (the thick greenish husk) not eaten by extant wild animals or domesticated livestock and a hard, thick nut shell with high, sharp ridges. Eastern North American forest dwelling megafauna existed until 10–12 ka, overlapping with the first people to arrive as the glacier retreated (Boulanger and Lyman 2014). Even if the megafauna usually discarded the nuts, the occasional long distance transport of a few may have sufficed to move the species from midcountry (~35° N) to New England before the extinction. Finally, the first people may have played a role thereafter, as butternut was highly valued for the oily nut and for making dye and medicines (Omar et al. 2000; Schlarbaum et al. 1997).
The decimation and local extinction of southern and central J. cinerea populations (Anderson and LaMadelaine 1978; Schlarbaum et al. 1997) due to butternut canker, intensive agriculture and other anthropogenic disturbances creates an unavoidable ascertainment bias in our data. A more complete geographical sampling may have enabled us to speculate less on the migration patterns of this large-seeded species adapted to cool mesic conditions. This ecological preference makes J. cinerea an excellent candidate for a tree that could have persisted in the cryptic northern refugia or the eastern coastal refugia proposed by other investigators (McLachlan et al. 2005; Stewart and Lister 2001; Stewart et al. 2010). The evidence in living trees is rapidly fading but J. cinerea macrofossils, particularly nut shell fragments and pollen, are distinctive and may lie undiscovered in caves and sediments not yet investigated.
Although our study did not provide a definitive answer to the question of how J. cinerea responded to the last glacial retreat in eastern populations, our data does suggest that in Eastern North America, tree migration patterns are the result of complex interactions of climate shifts, regional geography, ecological preferences and seed dispersal mechanisms. The role of watersheds in butternut migration must remain speculative as the species is dying out but this concept could be tested in black walnut, the only other Juglans species native in Eastern North America. Black walnut is not threatened, has the same seed dispersal mechanisms and is sympatric with butternut in the southern half of the range.
Finally, chloroplast or mitochondrial haplotyping, paleobotanical sampling and pollen counts in sediment cores may all be insufficient, when used alone, to reconstruct past tree migrations. Pollen records do not provide evidence of absence or define either the southern or northern edge range boundaries under some conditions (Jackson et al. 1997). Present day chloroplast haplotype diversity may not coincide with the paleobotanical evidence. Breen and colleagues (2012) found in their study of chloroplast haplotypes in Populus balsamifera a shallow chloroplast genealogy not at all suggestive of the LGM Beringian refuge indicated in the paleobotanical record. The last major climate shift in Eastern North America was accompanied by the appearance of humans, two events whose combined impact resulted in the extinction of Pleistocene megafauna, a consequence that may have had a not fully recognized impact on forest community composition that endures to this day.
References
Anderson RL, LaMadelaine LA (1978) The distribution of butternut decline in the eastern United States. Northeastern Area State and Private Forestry, USDA Forest Service, Forest Survey Report S-3-78. Northeastern Area State and Private Forestry, Broomall, PA. 5 p
Aradhya M, Potter D, Gao F, Simon C (2007) Molecular phylogeny of Juglans (Juglandaceae): a biogeographic perspective. Tree Gen Genom 3:363–378. doi:10.1007/s11295-006-0078-5
Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48
Birchenko I, Feng Y, Romero-Severson J (2009) Biogeographical distribution of chloroplast diversity in northern red oak (Quercus rubra L.). Am Midl Nat 161:134–145
Bordacs S et al (2002) Chloroplast DNA variation of white oaks in the northern Balkans and in the Carpathian Basin. For Ecol Manag 156:197–209
Borkowski D, McCleary T, McAllister M, Romero-Severson J (2014) Primers for 52 polymorphic regions in the Quercus rubra chloroplast, 47 of which amplify across 11 tracheophyte clades. Tree Genetics & Genomes:1-9 doi:10.1007/s11295-014-0729-x
Boulanger MT, Lyman RL (2014) Northeastern North American Pleistocene megafauna chronologically overlapped minimally with Paleoindians Quaternary Science Reviews 85:35-46 doi:http://dx.doi.org/10.1016/j.quascirev.2013.11.024
Boys J, Cherry M, Dayanandan S (2005) Microsatellite analysis reveals genetically distinct populations of red pine (Pinus resinosa, Pinaceae). Am J Bot 92:833–841. doi:10.3732/ajb.92.5.833
Breen AL, Murray DF, Olson MS (2012) Genetic consequences of glacial survival: the late Quaternary history of balsam poplar (Populus balsamifera L.) in North America. J Biogeogr 39:918–928. doi:10.1111/j.1365-2699.2011.02657.x
Broders KD, Boland GJ (2011) Reclassification of the butternut canker fungus, Sirococcus clavigignenti-juglandacearum, into the genus Ophiognomonia. Fungal Biol 115:70–79. doi:10.1016/j.funbio.2010.10.007
Bryan CF (1965) Butternut (Juglans cinerea L.). In: Fowells HA (ed) Silvics of forest trees of the United States. vol handbook 271. U.S. Department of Agriculture, Washington, DC, pp 208–210
Clark JS et al (1998) Reid's paradox of rapid plant migration—dispersal theory and interpretation of paleoecological records. Bioscience 48:13–24
Cristol DA, Switzer PV (1999) Avian prey-dropping behavior. II. American crows and walnuts. Behav Ecol 10:220–226. doi:10.1093/beheco/10.3.220
Cruzan MB, Templeton AR (2000) Paleoecology and coalescence: phylogeographic analysis of hypotheses from the fossil record. Trends Ecol Evol 15:491–496. doi:10.1016/S0169-5347(00)01998-4
Delcourt HR (1979) Late quaternary vegetation history of the eastern highland rim and adjacent cumberland plateau of Tennessee. Ecol Monogr 49:255–280. doi:10.2307/1942485
Delcourt PA, Delcourt HR, Brister RC, Lackey LE (1980) Quaternary vegetation history of the Mississippi embayment. Quat Res 13:111–132. doi:10.1016/0033-5894(80)90086-1
Demesure B, Sodzi N, Petit RJ (1995) A set of universal primers for amplification of polymorphic noncoding regions of mitochondrial and chloroplast DNA in plants. Mol Ecol 4:129–131
Dumolin-Lapegue S, Pemonge M, Petit R (1997) An enlarged set of consensus primers for the study of organelle DNA in plants. Mol Ecol 6:393–397
Feng Y, Sun W, Romero-Severson J (2008) Heterogeneity and spatial autocorrelation for chloroplast haplotypes in three old growth populations of northern red oak. Silvae Genet 57:212–220
Finkelstein SA, Gajewski K, Viau AE (2006) Improved resolution of pollen taxonomy allows better biogeographical interpretation of post-glacial forest development: analyses from the North American Pollen. Data J Ecol 94:415–430. doi:10.1111/j.1365-2745.2005.01087.x
Furnier GR, Stolz AM, Mustaphi RM, Ostry ME (1999) Genetic evidence that butternut canker was recently introduced into North America. Can J Botan-Rev Canadienne De Botanique 77:783–785
Fyles JG, Hills LV, Matthews JV Jr, Barendregt R, Baker J, Irving E, Jetté H (1994) Ballast brook and beaufort formations (late Tertiary) on Northern Banks Island, Arctic Canada. Quat Int 22–23:141–171. doi:10.1016/1040-6182(94)90010-8
Gill JL, Williams JW, Jackson ST, Lininger KB, Robinson GS (2009) Pleistocene megafaunal collapse, novel plant communities, and enhanced fire regimes in North America. Science 326:1100–1103. doi:10.1126/science.1179504
Godbout J, Jaramillo-Correa JP, Beaulieu J, Bousquet J (2005) A mitochondrial DNA minisatellite reveals the postglacial history of jack pine (Pinus banksiana), a broad-range North American conifer. Mol Ecol 14:3497–3512. doi:10.1111/j.1365-294X.2005.02674.x
Godbout J, Beaulieu J, Bousquet J (2010) Phylogeographic structure of jack pine (Pinus banksiana; Pinaceae) supports the existence of a coastal glacial refugium in northeastern North America. Am J Bot 97:1903–1912. doi:10.3732/ajb.1000148
Goheen JR, Swihart RK (2003) Food-hoarding behavior of gray squirrels and North American red squirrels in the central hardwoods region: implications for forest regeneration. Can J Zool 81:1636–1639
Gonzales LM, Williams JW, Grimm EC (2009) Expanded response-surfaces: a new method to reconstruct paleoclimates from fossil pollen assemblages that lack modern analogues. Quat Sci Rev 28:3315–3332. doi:10.1016/j.quascirev.2009.09.005
Hamilton MB (1999) Four primer pairs for the amplification of chloroplast intergenic regions with intraspecific variation. Mol Ecol 8:521–523
Hickey LJ, Johnson KR, Dawson MR (1988) The stratigraphy, sedimentology, and fossils of the haughton formation: a post-impact crater-fill, Devon Island, N.W.T., Canada. Meteoritics 23:221–231. doi:10.1111/j.1945-5100.1988.tb01284.x
Hills LV, Klovan JE, Sweet AR (1974) Juglans eocinerea n. sp., beaufort formation (Tertiary), southwestern Banks Island, Arctic Canada. Can J Bot 52:65–90. doi:10.1139/b74-011
Hoban SM, Anderson R, McCleary TS, Schlarbaum SE, Romero-Severson J (2008) Thirteen nuclear microsatellite loci for butternut (Juglans cinerea L.). Mol Ecol Resour 8:643–646
Hoban SM, McCleary TS, Schlarbaum SE, Romero-Severson J (2009) Geographically extensive hybridization between the forest trees American butternut and Japanese walnut. Biol Lett 5:324–327. doi:10.1098/rsbl.2009.0031
Hoban SM et al (2010) Range-wide distribution of genetic diversity in the North American tree Juglans cinerea: a product of range shifts, not ecological marginality or recent population decline. Mol Ecol 19:4876–4891. doi:10.1111/j.1365-294X.2010.04834.x
Hoban S, Schlarbaum S, Brosi S, Romero-Severson J (2012a) A rare case of natural regeneration in butternut, a threatened forest tree, is parent and space limited. Conserv Genet 13:1447–1457. doi:10.1007/s10592-012-0386-2
Hoban SM, McCleary TS, Schlarbaum SE, Anagnostakis SL, Romero-Severson J (2012b) Human-impacted landscapes facilitate hybridization between a native and an introduced tree. Evol Appl 5:720–731
Hoban SM, McCleary TS, Schlarbaum SE, Romero-Severson J (2014) Spatial genetic structure in 21 populations of butternut, a temperate forest tree (Juglans cinerea L.), is correlated to spatial arrangement, habitat, and land-use history. For Ecol Manag 314:50–58. doi:10.1016/j.foreco.2013.11.001
Huang SSF, Hwang SY, Lin TP (2002) Spatial pattern of chloroplast DNA variation of Cyclobalanopsis glauca in Taiwan and east Asia. Mol Ecol 11:2349–2358
Ivan JS, Swihart RK (2000) Selection of mast by granivorous rodents of the central hardwood forest region. J Mammal 81:549–562. doi:10.1644/1545-1542(2000)081<0549:SOMBGR>2.0.CO;2
Jackson ST, Weng C (1999) Late quaternary extinction of a tree species in eastern North America. Proc Natl Acad Sci 96:13847–13852. doi:10.1073/pnas.96.24.13847
Jackson ST, Overpeck JT, Webb- Iii T, Keattch SE, Anderson KH (1997) Mapped plant-macrofossil and pollen records of late quaternary vegetation change in Eastern North America. Quat Sci Rev 16:1–70. doi:10.1016/S0277-3791(96)00047-9
Jackson ST, Webb RS, Anderson KH, Overpeck JT, Webb T III, Williams JW, Hansen BCS (2000) Vegetation and environment in Eastern North America during the last glacial maximum. Quat Sci Rev 19:489–508. doi:10.1016/S0277-3791(99)00093-1
Jaramillo-Correa JP, Beaulieu J, Bousquet J (2004) Variation in mitochondrial DNA reveals multiple distant glacial refugia in black spruce (Picea mariana), a transcontinental North American conifer. Mol Ecol 13:2735–2747. doi:10.1111/j.1365-294X.2004.02258.x
Johnson WC, Thompson Webb III (1989) The role of blue jays (Cyanocitta cristata L.) in the postglacial dispersal of fagaceous trees in Eastern North America. J Biogeogr 16:561–571. doi:10.2307/2845211
Le Corre V, Kremer A (1998) Cumulative effects of founding events during colonisation on genetic diversity and differentiation in an island and stepping-stone model. J Evol Biol 11:195–512
Liu Y, Andersen JJ, Williams JW, Jackson ST (2013) Vegetation history in central Kentucky and Tennessee (USA) during the last glacial and deglacial periods. Quat Res 79:189–198. doi:10.1016/j.yqres.2012.12.005
Loehle C (2007) Predicting pleistocene climate from vegetation in North America. Clim Past 3:109–118. doi:10.5194/cp-3-109-2007
Magni CR, Ducousso A, Caron H, Petit RJ, Kremer A (2005) Chloroplast DNA variation of Quercus rubra L. in North America and comparison with other Fagaceae. Mol Ecol 14:513–524
Marsico TD, Hellmann JJ, Romero-Severson J (2009) Patterns of seed dispersal and pollen flow in Quercus garryana (Fagaceae) following post-glacial climatic changes. J Biogeogr 36:929–941. doi:10.1111/j.1365-2699.2008.02049.x
Martínez-Meyer E, Peterson AT (2006) Conservatism of ecological niche characteristics in North American plant species over the pleistocene-to-recent transition. J Biogeogr 33:1779–1789. doi:10.1111/j.1365-2699.2006.01482_33_10.x
McCleary TS, Robichaud RL, Nuanes S, Anagnostakis SL, Schlarbaum SE, Romero-Severson J (2009) Four cleaved amplified polymorphic sequence (CAPS) markers for the detection of the Juglans ailantifolia chloroplast in putatively native J. cinerea populations. Mol Ecol Resour 9:525–527. doi:10.1111/j.1755-0998.2008.02465.x
McCleary T, McAllister M, Coggeshall M, Romero-Severson J (2013) EST-SSR markers reveal synonymies, homonymies and relationships inconsistent with putative pedigrees in chestnut cultivars. Genet Resour Crop Evol 60:1209–1222. doi:10.1007/s10722-012-9912-9
McLachlan JS, Clark JS, Manos PS (2005) Molecular indicators of tree migration capacity under rapid climate change. Ecology 86:2088–2098
Middleton B (2000) Hydrochory, seed banks, and regeneration dynamics along the landscape boundaries of a forested wetland. Plant Ecol 146:167–181. doi:10.1023/A:1009871404477
Moore JE, McEuen AB, Swihart RK, Contreras TA, Steele MA (2007) Determinants of seed removal distance by scatter-hoarding rodents in deciduous forests. Ecology 88:2529–2540. doi:10.2307/27651399
Omar S, Lemonnier B, Jones N, Ficker C, Smith ML, Neema C, Towers GHN, Goel K, Arnason JT (2000) Antimicrobial activity of extracts of eastern North American hardwood trees and relation to traditional medicine. J Ethnopharmacol 73:161–170
Parks A, Jenkins M, Ostry M, Zhao P, Woeste K (2014) Biotic and abiotic factors affecting the genetic structure and diversity of butternut in the southern Appalachian Mountains, USA. Tree Genet Gen 10:541–554. doi:10.1007/s11295-014-0702-8
Petit RJ, El Mousadik A, Pons O (1998) Identifying populations for conservation on the basis of genetic markers. Conserv Biol 12:844–855
Petit RJ et al (2002) Chloroplast DNA variation in European white oaks: phylogeography and patterns of diversity based on data from over 2600 populations. For Ecol Manag 156:5–26. doi:10.1016/S0378-1127(01)00645-4
Petit RJ et al (2003) Glacial refugia: hotspots but not melting pots of genetic diversity. Science 300:1563–1565
Pons O, Petit RJ (1996) Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144:1237–1245
Posada D, Crandall K (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818
Ronquist F et al (2012) MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. doi:10.1093/sysbio/sys029
Royall PD, Delcourt PA, Delcourt HR (1991) Late quaternary paleoecology and paleoenvironments of the Central Mississippi Alluvial Valley. Geol Soc Am Bull 103:157–170. doi:10.1130/0016-7606(1991)103<0157:lqpapo>2.3.co;2
Schlarbaum SE, Hebard F, Spaine PC, Kamalay JC (1997) Three American tragedies: chestnut blight, butternut canker, and Dutch elm disease. In: Britton KO (ed) Proceedings, exotic pests of eastern forests. vol 1997 April 8-10. Tennessee Exotic Pest Plant Council, Nashville, pp 45–54
Schultz J (2003) Conservation assessment for butternut or white walnut (Juglans cinerea L.). USDA Forest Service, Eastern Region, Milwaukee, WI. 76 p. www.fs.fed.us/r9/wildlife/tes/ca-overview/docs/plant_juglans_cinera-Butternut2003.pdf
Shaw J, Lickey EB, Schilling EE, Small RL (2007) Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. Am J Bot 94:275–288. doi:10.3732/ajb.94.3.275
Soltis DE, Morris AB, McLachlan JS, Manos PS, Soltis PS (2006) Comparative phylogeography of unglaciated eastern North America. Mol Ecol 15:4261–4293
Stapanian MA, Smith CC (1986) How fox squirrels influence the invasion of prairies by nut-bearing trees. J Mammal:326-332
Stewart JR, Lister AM (2001) Cryptic northern refugia and the origins of the modern biota. Trends Ecol Evol 16:608–613. doi:10.1016/S0169-5347(01)02338-2
Stewart JR, Lister AM, Barnes I, Dalén L (2010) Refugia revisited: individualistic responses of species in space and time. Proc R Soc B Biol Sci 277:661–671. doi:10.1098/rspb.2009.1272
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi:10.1093/molbev/msr121
Victory ER, Glaubitz JC, Rhodes OE Jr, Woeste KE (2006) Genetic homogeneity in Juglans nigra (Juglandaceae) at nuclear microsatellites. Am J Bot 93:118–126. doi:10.3732/ajb.93.1.118
Wall SBV (2001) The evolutionary ecology of nut dispersal. Bot Rev 67:74–117. doi:10.2307/4354385
Watts WA (1975) Vegetation record for the last 20,000 years from a small marsh on Lookout Mountain, northwestern Georgia. Geol Soc Am Bull 86:287–291. doi:10.1130/0016-7606(1975)86<287:vrftly>2.0.co;2
Watts WA (1979) Late quaternary vegetation of central Appalachia and the New Jersey coastal plain. Ecol Monogr 49:427–469. doi:10.2307/1942471
Whitlock C, Dawson MR (1990) Pollen and vertebrates of the early Neogene Haughton formation, Devon Island, Arctic Canada. Arctic 43:324–330. doi:10.2307/40510958
Williams JW, Jackson ST (2007) Novel climates, no-analog communities, and ecological surprises. Front Ecol Environ 5:475–482. doi:10.1890/070037
Williams JW, Shuman BN, Webb T (2001) Dissimilarity analyses of late-quaternary vegetation and climate in eastern North America. Ecology 82:3346–3362
Wolfe JA (1980) Tertiary climates and floristic relationships at high latitudes in the northern hemisphere. Palaeogeogr Palaeoclimatol Palaeoecol 30:313–323. doi:10.1016/0031-0182(80)90063-2
Zaya D, Howe H (2009) The anomalous Kentucky coffeetree: megafaunal fruit sinking to extinction? Oecologia 161:221–226. doi:10.1007/s00442-009-1372-3
Zhang WH, Chen ZD, Li JH, Chen HB, Tang YC (2003) Phylogeny of the Dipsacales s.l. based on chloroplast trnL-F and ndhF sequences. Mol Phylogenet Evol 26:176–189
Acknowledgments
We thank Bob Anderson, Paul Berrang, Sunshine Brosi, Bryan Connolly, Brice Leech, Scott Schlarbaum and Barb Boysen for assisting in sample collection. We also thank the private landowners, state governments and the provincial governments of Ontario and New Brunswick for allowing us access to state, provincial and private forests. This project was funded in part by an NSF Research Experience for Undergraduates summer program grant to Kristen Laricchia. Sean Hoban is supported as a Postdoctoral Fellow at the National Institute for Mathematical and Biological Synthesis, funded by NSF Award #DBI-1300426, and The University of Tennessee, Knoxville.
Conflict of interest
The authors affirm that they have no conflicts of interest.
Data archiving statement
Sequences for haplotypes 2, 5, 6 and 10, which show all the polymorphisms we detected except one, are deposited at NCBI. Accessions numbers are listed in Table S1. The polymorphic region of ndhK-ndhC, being <200 bp and thus too short to deposit, is reported in Table S2.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by R. Sederoff
Rights and permissions
About this article
Cite this article
Laricchia, K.M., McCleary, T.S., Hoban, S.M. et al. Chloroplast haplotypes suggest preglacial differentiation and separate postglacial migration paths for the threatened North American forest tree Juglans cinerea L.. Tree Genetics & Genomes 11, 30 (2015). https://doi.org/10.1007/s11295-015-0852-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-015-0852-3