Abstract
Most biosynthetic gene clusters (BGCs) of Actinobacteria are either silent or expressed less than the detectable level. The non-genetic approaches including biological interactions, chemical agents, and physical stresses that can be used to awaken silenced pathways are compared in this paper. These non-genetic induction strategies often need screening approaches, including one strain many compounds (OSMAC), reporter-guided mutant selection, and high throughput elicitor screening (HiTES) have been developed. Different types of genetic manipulations applied in the induction of cryptic BGCs of Actinobacteria can be categorized as genome-wide pleiotropic and targeted approaches like manipulation of global regulatory systems, modulation of regulatory genes, ribosome and engineering of RNA polymerase or phosphopantheteine transferases. Targeted approaches including genome editing by CRISPR, mutation in transcription factors and modification of BGCs promoters, inactivation of the highly expressed biosynthetic pathways, deleting the suppressors or awakening the activators, heterologous expression, or refactoring of gene clusters can be applied for activation of pathways which are predicted to synthesize new bioactive structures in genome mining studies of Acinobacteria. In this review, the challenges and advantages of employing these approaches in induction of Actinobacteria BGCs are discussed. Further, novel natural products needed as drug for pharmaceutical industry or as biofertilizers in agricultural industry can be discovered even from known species of Actinobactera by the innovative approaches of metabolite biosynthesis elicitation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The identification of 589 new compounds in only a five year span, shows that Actinobacteria still keep their rank in potential in production of bioactive compounds (Jose et al. 2021). Until the early 20th century, it was assumed that almost all metabolites had been identified and about 70% of these biologically active metabolites obtained to date belong to the genus Streptomyces (Singh et al. 2021). While the development of high-throughput omics methods provides the possibility of genome mining due to the fact that most of the secondary metabolites are still not discovered. The BGCs repositories such as MIBiG, NaPDoS, and IMG-ABC, provide suitable information to connect biosynthetics genes to their final chemical products, and predict which compounds have cryptic pathway and have not yet been discovered (Ziemert et al. 2012; Palaniappan et al. 2020; Terlouw et al. 2022). The improvement of genome mining tools for BGCs identification revealed that approximately 23,000 identified metabolites account for only about 3% of all synthetic metabolites leaving the major fraction of biosynthetic genes still to be discovered (Scherlach and Hertweck 2021). The discovery of new bioactive compounds from known species in recent years, as well as the estimation of about 20–40 gene clusters in each species based on genome data, confirms the existence of other biosynthetic pathways in known strains that can be induced and lead to the discovery of new compounds (Liu et al. 2019b). Molecular methods based on genetic manipulations and culture-based non-genetic methods have been investigated to express silenced biosynthetic gene clusters. In this review, various methods of activating the silent pathways, increasing the expression of BGCs and the advances in the techniques of activating the silent pathways of secondary metabolites are compared.
Approaches in the activation of silent metabolic pathways
Methods to stimulate cells to produce secondary metabolites can be divided into genetic and non-genetic approaches. Non-genetic methods are often blind with unidentified mechanisms and the evaluating inducers are applied during the fermentation. While in genetic methods, conditions for gene expression are provided by manipulating the bacterial genome or genetic elements including the cloning of clusters into a heterologous host. In the following sections, various possible methods for expression of silent gene clusters have been discussed and compared.
Biological induction
Living cells, biomass or cellular components are used to stimulate microorganisms to produce secondary metabolites. Since the physiology of microorganisms is different in the presence of the symbions or competitor agents, diverse types of cell candidates and cellular components can stimulate each microorganism to produce bioactive metabolites. The mechanisms of secondary metabolites production in environment are not clear in most cases. Therefore, for inducing natural products in laboratory, random organisms, biological substances, and inducers must be screened.
Living cells
The production or increase in production of secondary metabolites in symbiotic, parasitic, competitive or mimetic interactions is a natural response to environmental conditions. By carrying out a part of the biosynthetic pathway, secreting some compounds or changing the physical and chemical factors of the culture medium, microorganisms provide the conditions for the neighbouring strains to biosynthesize new metabolites, which is not possible in mono-culture (Kim et al. 2021) like the compounds presented in Table 1. Reports on elicitation of Actinobacteria using live cells can be divided into three general cultivation categories: (i) Cultivation of Actinobacteria with yeasts or fungi; (ii) Cultivation of Actinobacteria with bacteria containing mycolic acid; (iii) Cultivation of Actinobacteria with other bacteria (Peng et al. 2021). The effect of co-culturing on activation of silent pathways can be investigated in various systems including microfluidic system, petri dish, microtiter plates, solid supports, bioreactor systems, and transwell systems (Hug et al. 2018).
In the experiment conducted to identify stimulating strains in the production of secondary metabolites, Streptomyces lividans was used as a model actinobacterium, due to the production of blue and red pigments actinorhodin and undecylprodigiosin, which are visible indicators of the activated pathway (Onaka et al. 2011; Onaka 2017). All inducing strains belonged to MACB (mycolic acid-containing bacteria) and 33 new bioactive metabolites were identified by cultivating 12 different species of Actinobacteria using the combined-culture method (Hoshino et al. 2019). In addition, inducing effect of mycolic-acid molecules has also been shown in Streptomyces lividans.
In addition, the biological interaction between actinobacterial and fungal strains may result in the induction of natural products pathways. As an illustration, borrelidins J and K are induced in marine-derived Streptomyces rochei MB037 when it is co-cultured with a fungus Rhinocladiella similis (Yu et al. 2019). On the other hand, overexpression of secondary metabolites is also reported from co-culturing of actinobacterial strains with non-actinobacteria; as an example, in cultivation of Streptomyces sp. PTY087I2 isolated from a Panamanian tunicate with several species of human pathogens, including methicillin-susceptible S. aureus, the production of granaticin, granatomycin D, and dihydrogranaticin B were increased (Sung et al. 2017). Nevertheless, the detection of new antibiotics can also be due to horizontal gene transfer which was observed in production of rhodostreptomycins by a Rhodococcus strain in co-culture with Streptomyces (Kurosawa et al. 2010).
Since the activation mechanism of many genes in co-culture is unknown, due to the lack of investigation of specific regulatory induction pathways for most metabolites, there is a lack of sufficient details (Okada and Seyedsayamdost 2017) to precisely control their expression (Table 1).
Cellular components
Some cell wall compounds as well as various types of oligosaccharides such as mannan oligosaccharide (MO), alginate-derived oligomanuronate (OM) and polysaccharides can induce the production of secondary metabolites (Pettit 2011). The investigation on the effect of fungal biomass in inducing natamycin production in S. natalensis HW-2 showed the presence of stimulating compounds in the cell wall.
N-acetyleglucosamine (GlcNAc) is a cell wall-derived molecule that was identified in the study of the link between programmed cell death (PCD) and production of secondary metabolite. In starvation condition which leads to cell lysis, N-acetylglucosamine is released into the environment and, as a signal, induces the production of bioactive metabolites with the probable purpose of cell protection (Barka et al. 2016). Moreover, addition of GlcNAc to culture medium of Actinokineospora sp. EG49 led to the production of new actinosporin metabolite (Tomm et al. 2019).
Chemical induction
Chemical elicitors can be categorized into elements (micronutrient and rare earth elements), natural small molecules (intracellular and secreted metabolites), micro/nanoparticles and synthetic chemical compounds.
Natural small molecules
Natural small molecules include signal molecules (antibiotics and their intermediates, hormone-like molecules, siderophores, bioactive secondary metabolites.) and cell wall components (Zhang et al. 2022) which were discussed in the former section (Celuular components).
Hormone-like signaling molecules
Known hormone-like signal molecules are classified into five types based on their structure: gamma-butyrolactones (GBL), furans, gamma-butenolides, pimaricin-inducer (PI) factor and N-methylphenylalanyl-dehydrobutyrine diketopiperazine (Niu et al. 2016). The first three groups have a 5-membered heterocyclic ring containing four carbons and one oxygen (Table 2). The other two groups have completely different structures and their activity has not been precisely characterized (Niu et al. 2016). Unlike gamma butyrolactones, furans are very stable under alkaline conditions. GBL is produced by many Streptomycetes (Onaka 2017) and due to their function in secondary metabolite production and sporulation, GBL are recognized as a signaling molecule for triggering the biosynthetic pathway and differentiation in Actinobacteria.
Antibiotics and their intermediates
Antibiotics can act as inducers and regulators of the biosynthesis of other metabolites in sub-inhibitory concentrations (SICs) (Zarins-Tutt et al. 2016). Some types of antibiotics target regulatory genes, especially the family of pas-luxR regulatory genes, which belong to the group of quorum sensing (QS) molecules and regulate the amount of metabolite production based on its concentration in the environment. Adding 1.5 µg/ml tetracycline in the culture medium can induce and enhance the biosynthesis of different types of streptophenazines in a Streptomyces strain up to 2.8-fold (Mitova et al. 2008). The up to 6-fold increase in actinomycin D4 biosynthesis in Streptomyces, by stimulation using chloramphenicol is another example of the role of antibiotics as inducers (Zhang et al. 2022). Goadsporin is a modified oligopeptide with 19 amino acids produced by Streptomyces sp. TP-A0584 (Onaka et al. 2001; Yoon and Nodwell 2014) that has been reported to activate the prodigiosin production and sporulation in S. lividans (Onaka et al. 2001) (Table 2). It seems that many secondary metabolites like goadsporin act as signal-like molecules at low concentrations and activate some biosynthetic pathways, while they have antimicrobial properties at high concentrations and preventing cell growth in some specific streptomyces (Onaka et al. 2001).
Presumably, all regulatory molecules which are under the control of environmental changes or changes in cell physiology, transmit signals to cluster regulators, leading to the expression of (cluster situated regulators) CSR (Niu et al. 2016). As a result of the expression of these regulators, the production of secondary metabolites is switched. Signaling molecules such as Streptomyces coelicolor butanolides (SCBs), methylenomycin furans (MMFs), N-butanoyl-L-hemoserine lactone (C4-HSL), A-factor, factor P1 (enhancer of pimaricin) interact with the scb1 cognate receptor. In addition, antibiotics such as actinorhodin and undecylprodigiosin react with pseudoreceptors (JadP2/ScbR2) and activate a series of events leading to morphogenetic changes and the production of secondary metabolites (Salwan and Sharma 2020).
Elements
Changing the amount of micronutrients namely phosphate, nitrogen, sulfur and iron (Martín and Liras 2020), and the presence of metal stress in the culture medium (Tomm et al. 2019; Zong et al. 2021) can stimulate or inhibit the synthesis of secondary metabolites depending on the type of synthesized metabolite, bacterial strain and other environmental stimuli (Wohlleben et al. 2017).
Carbon
Its high concentration often inhibits secondary metabolism through carbon catabolite repression (CCR). The high-density microarray evaluation in S. coelicolor M145 showed expression of 651 genes under glucose suppression condition. Other carbon sources which cause CCR include glycerol, maltose, mannose, xylose, sucrose and citrate (Wohlleben et al. 2017).
Moreover, the sugar uptake system is reduced during secondary metabolism of Streptomyces, which prevents the optimal production of natural products. The manipulation of two sugar transporter systems, TP2 and TP5 using temporal promoters in S. bingchenggensis led to increased production of milbemycin (a group of macrolide biopesticides). In this way, TP2 increased the production of milbemycin A3, and TP5 increased the production of milbemycin A4 up to 29.7% and 32%, respectively (Jin et al. 2020).
Phosphate
The role of phosphate in stimulating or inhibiting the activity of some enzymes involved in secondary metabolism and gene expression has been reported. The response to phosphate deficiency is carried out through the PhoR-PhoP two-component system as ΔphoP mutants of S. coelicolor M145 and shows an increase or decrease in the expression of 551 genes under phosphate limitation (Rodríguez-García et al. 2007; Rodríguez et al. 2013)
The optimal concentration of phosphate for secondary metabolite production varies in bacteria (Wohlleben et al. 2017), while phosphate limitation often leads to stimulation of secondary metabolite production (Hoskisson and Fernández-Martínez 2018). However, exceptions have also been reported (Wohlleben et al. 2017). An example is the activation of PhoR-PhoP system under phosphate limitation that inactivates the transcription of aveR regulatory gene and thus prevents the production of avermectin in S. avermitilis (Martín et al. 2017).
Nitrogen
Ammonium is the preferred source of inorganic nitrogen in Actinobacteria, while nitrate and nitrite (Martín and Liras 2020) are considered as alternative sources (Wohlleben et al. 2017). Ammonium deficiency reduces the growth and stimulates secondary metabolism in many bacteria. Nevertheless, high concentration of nitrate increases the production of secondary metabolite, which is called “stimulating effect of nitrate” (Wohlleben et al. 2017). The main regulator of nitrogen metabolism in Actinobacteria is GlnR (Wohlleben et al. 2017; Hoskisson and Fernández-Martínez 2018), which has regulatory association with PhoP regulatory proteins (Martín and Liras 2020).
Rare earth elements (REEs)
REEs comprise 17 elements such as yttrium, scandium, lanthanum (Ochi and Hosaka 2013; Zong et al. 2021), some of which are essential elements for microbial metabolism (Zong et al. 2021). Microorganisms probably acquired the ability to react to small amounts of these elements as environmental stress during their evolution (Zhang et al. 2022).
The production of streptomycin by S. griseus in the presence of Sc element increased up to 4-fold and the production of actinorhodin in S. coelicolor increased up to 25-fold (Ochi and Hosaka 2013; Zong et al. 2021). It is suggested that molecules such as ppGpp, ribosome and RNA polymerases are the targets of these elements and through these macromolecules they regulate the biosynthesis of secondary metabolites (Zong et al. 2021). It has been shown that lanthanum can activate silent pathways of antibacterial compounds produced by Promicromonospora kermanensis at concentrations up to 50 µM, while at 100 µM it cannot awaken the antimicrobial biosynthetic pathways (Mohammadipanah et al. 2020). Rare elements of ScCl3 and LaCl3 are also reported as transcription amplifiers of cryptic metabolite pathways in Streptomyces griseus. Likewise, several rare elements by their stress stimulating effect induce the transcription of the BGCs in an explicit Sub-MIC concentration range in Actinobacteria (Ochi et al. 2014).
Micro and nanoparticles
Investigation of micro- and nanoparticle has shown their ability to activate pathways and increase the production of secondary metabolites. The usage of talc microparticles in the culture medium of S. lividans led to an increase in the production of bottromycin A2 up to two-fold (Table 3), which was even up to 13- fold in other actinobacterial strains (Kuhl et al. 2021). Liu et al. (2019b) showed that S. coelicolor M145 produces different amounts of actinorhodin in response to the treatment with CuO particles, Al2O3 and carbon nanomaterials. To determine the role of carbon nanomaterial in actinorhodin biosynthesis, carbon nanoparticles in the form of ball-milled biochar, graphene oxide and carbon nanotubes were used. Among them, disordered spherical ball-milled biochar was the most effective for damaging cells and stimulating the expression of gene clusters (Liu et al. 2019a). Therefore, during the biosynthesis of metabolites, the concentration of particles should be precisely controlled (Zong et al. 2021). In addition to concentration of the nanoparticles, particle size influences the production of secondary metabolites (Niu et al. 2016).
Chemical agents
Antibiotic remodeling compounds (ARCs) were identified in a survey aimed at finding effective chemical molecules in secondary metabolism. In a study, more than 30,000 chemical molecules were used against S. coelicolor and 19 compounds induced the production of actinorhodin pigment. Four compounds were structurally related to the triclosan, including ARC 2, 3, 4 and 5 (Ochi and Hosaka 2013; Yoon and Nodwell 2014). These ARCs inhibit the FabI enzyme which is related to the final stage of fatty acid biosynthesis, and as a result, acyl-coenzyme A can support the polyketide biosynthesis (Ochi and Hosaka 2013).
Dimethyl sulfoxide is a toxic chemical that at explicit concentrations, induces the biosynthesis of secondary metabolites by changing metabolic pathways (Zong et al. 2021; Zhang et al. 2022), which is called the DMSO effect (Zong et al. 2021). For instance, 3% DMSO in the culture medium of S. azureus ATCC14921 caused a two-fold increase in the biosynthesis of thiostrepton (Chen et al. 2000; Pettit 2011; Zong et al. 2021). Wang et al. (2018) showed a 30% increase in the accumulation of tacrolimus in S. tsukubensis in the presence of DMSO in combination with LaCl3. Metabolomic studies on the accumulation of tacrolimus showed the role of DMSO in increasing the production of precursors through metabolic pathways such as pentose phosphate (Zong et al. 2021). This effect was also shown in ascomycin biosynthesis in S. hygroscopicus (Zong et al. 2021).
Ethanol at certain concentrations has also shown elicitation or inhibition effect on secondary metabolite biosynthesis, which depends on various conditions such as bacterial culture medium, global regulatory systems (such as AfsR and GlnR) and ROS signal (Zong et al. 2021). The biosynthesis of chloramphenicol and jadomycin in S. venezuelae ATCC 10,712 in the presence of 6% ethanol was increased (Sekurova et al. 2016). The production of validamycin A (an antifungal against sheath blight disease of rice and wheat) by S. hygroscopicus could be increased up to 60% by adding ethanol at a concentration of 10 mM for 10 h (Zhou et al. 2012). H2O2 is another inducing agent that has increased the production of ascomycin by S. hygroscopicus up to 26% at the optimal concentration (Wang et al. 2019b).
Physical induction
Accumulation of microorganisms or physical contact between cells is one of the factors that stimulate cells to produce secondary metabolites. Fluctuations of the physical parameters are considered as environmental stress for the microorganism, which stimulates the production of secondary metabolites (Fig. 1).
Physical contact
The physical contact can stimulate the production of secondary metabolites in some microorganisms. Therefore, artificial physical contacts such as scaffolds can be used with the aim of inducing the silent BGCs. Cotton balls are generally used by imitating the symbiotic relationship between sponges or sea corals and some microorganisms in their habitats. With the growth of bacteria around cotton fibers, formation of biofilms is facilitated, which can stimulate the production of some secondary metabolites. Timmerman et al. (2019) reported a significant difference in the production of violacein in the presence of cotton balls compared to the absence of balls in Pseudoalteromonas luteoviolacea. Considering the correlation between biofilm formation and its effect on the activation of the Quorum Sensing system, many secondary metabolites that are produced at low and undetectable concentration, can be induced using physical entrapment and accumulation of signal molecules (Timmermans et al. 2019).
Environmental stress cues
Sometimes Actinobacterial strains produce specific secondary metabolites in response to adaptation to tough envionmental condition. By mimicking this fact and disrupting the optimal growth conditions by changing the oxygen level, thermal shock, pH shock, changing the ambient light, and hydrostatic pressure (Yoon and Nodwell 2014) can stimulate bacteria to produce secondary metabolites.
Oxygen
Changes in the concentration of dissolved oxygen (DOC) in culture medium can affect the growth and the production of secondary metabolites (Zong et al. 2021). For example, increasing DOC above the optimal concentration in culture medium of S. parvulus caused a decrease in manumycin production or an increase in purple pigment production under oxygen deficiency in Streptomyces CNQ-525 (Zong et al. 2021).
Temperature
Thermal shock is a widely investigated environmental stress, which may cause activation of secondary metabolism by damage to the cell membrane or accumulating unfolded or misfolded proteins (Yoon and Nodwell 2014). The optimum temperature for the growth of microorganisms might be different from the temperature required for secondary metabolite production (Tomm et al. 2019). The biosynthesis of jadomycin in S. venezulae is a classic example of the effect of temperature shift from 27 to 42 °C (Yoon and Nodwell 2014).
Light
As biosynthetic pathways of Actinobacteria inhabit dark areas, UV irradiation can be an environmental stress activating the expression of some silent pathways (Rule and Cheeptham 2013). The antimicrobial activity was induced in 17% of the actinobacterial strains following UV treatment.
pH
In evaluating the effect of pH on the production of secondary metabolites, other than the physicochemical parameters, factors including the time of pH change and the slope of pH changes over time should also be considered (Tomm et al. 2019). The increased production of methylomycin by S. coelicolor under acidic conditions is reported (Hayes et al. 1997), and the inducing effect of alkaline pH is also reported in Promicromonospora kermanensis (Mohammadipanah et al. 2020).
The effect of some chemical and physical inducers on regulatory pathways of secondary metabolites and activation of gene clusters. For instance, in phosphate limitation, P is transferred into the cell through the PstSCAB transferring system and activates the PhoP-PhoR two-component system. Then, PhoP/R positively/negatively affects the gene cluster expression. AfsQ1/Q2 have a role in nitrogen metabolism and the P/N/C ratio is probably effective for stimulating this regulatory system. The ammonium ions are transported into the cells by the amtB transporter, which is clustered with genes glnK and glnD. MtrA/B and GlnR enforce opposite effects on expression of nitrogen metabolism. DasR global regulator exerts negative effect on actII-ORF4. GlcNAc activates actII-ORF4 by suppressing DasR. The use of other carbon sources except glucose in the production of secondary metabolites in carbon deficiency is related to cAMP-CRP
Methods in screening of the induced pathways
OSMAC approach and environmental cues
Environmental stress or signal molecules affect the expression of secondary metabolites by changing the precursors or the activity of biosynthetic enzymes, as well as affecting regulatory genes and microbial metabolism. The large scale screening method of OSMAC (one strain many compounds) or the so-called “brothological” method is a conventional strategy in which several stimuli consisting of physical factors (temperature, pH, shaking), nutrition (carbon, nitrogen, phosphorus sources, salinity), and chemical inducers (mediator molecules, antibiotics) are used to investigate the effect of inducers on secondary metabolite production (Covington et al. 2018). Since the inducing effects of different BGCs are unknown, multiple variables should be screened in this method, and based on metabolomic, proteomic, and transcriptomic data, selection of the favorable conditions of cultivation will be possible (Fig. 2).
Different chemical or biological agents can be added or removed from cultural condition in this method. Covington et al. (2018) used a set of environmental stimuli including two types of antibiotics, lanthanum, scandium and microbial culture mixture containing MACB, to induce the metabolites of cave actinobacterial isolates. Isolation and identification of the produced metabolites showed a new structure called funisamine (linear polyketide) was identified from Streptosporangium with a new aminopolyol polyketide structure. Further investigations showed a significant increase in the production of funisamine in the culture mixture containing Bacillus strains (Covington et al. 2018). Furthermore, some novel tools have been used to optimize OSMAC technique, like micro-fermentation. Using this strategy, evaluation of 528 conditions in 44 marine Microccocaceae strains led to the discovery of a new thiazolyl peptide from a Kocuria sp. with anti-MRSA activity (Palomo et al. 2013). Another new tool for OSMAC is microfluidics-chips in which various parameters can be adjusted to assess the simulatory effect of environmental condition on induction of silent pathways. Using novel complementary tools of analysis and comparison, the scale of induction analysis can be expanded (Romano et al. 2018).
One of the beneficial points in applying OSMAC method is triggering the diversity of chemical structures of compounds as shown in a study on metabolomics of sea-derived Actinobacteria under OSMAC experiments. This method is not only helpful for production compounds from silent BGCs, but also may lead to altering the chemical structures of well-known natural products (Gamaleldin et al. 2020).
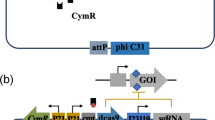
Using OSMAC (One Strain-Many Compounds) method for induction strains with silent pathways. Engineered or wild strains are optimized for growth and production of secondary metabolites under laboratory condition. Applying OSMAC methods on these strains increase the probability of BGCs expression specifically for metagenome-derived BGCs. (1) Extracting BGCs from metagenome samples or unculturable strains. (2) BGCs are inserted in Actinobacteria strains by different genetical tools like CRISPR-CAS 9. (3) Selecting hosts like engineered strains of S. coelicolor, S. lividans, and E. coli with optimum known conditions in the lab. (4) Activaiting or increasing secondary metabolites production. (5) Using NMR, HPLC-HRMS for detection of new secondary metabolites
Reporter-guided mutant selection
The basis of Reporter-Guided Mutant Selection (RGMS) consists of inducing mutation and generation of phenotypic libraries in which expression of the reporter genes determines the expression level of the targeted BGCs in the mutant strains.
Different candidates were used as reporter genes in different studies. In one study, neo gene was integrated with PKS biosynthetic gene sgnS1 as a reporter gene. Mutant strains were selected after three rounds of chemical mutagenesis, and the level of natamycin production by Streptomyces gilvosporeus increased to 340% (Wang et al. 2016).
In an analogous manner, for re-activation of jadomycin in S. venezuelae, kanamycin resistance gene (neo) as a reporter, and color-based detection by catechol oxidase (xylE) were used. Two new anthraquinone aminoglycosides gaudimycin D and E were discovered by activating pga cryptic gene in Streptomyces sp. PGA64 (Guo et al. 2015). Moreover, this technique has been applied for increasing the production level of clavulanic acid in S. clavuligerus and natamycin in S. gilvosporeus (Xiang et al. 2009; Wang et al. 2016).
It was seen that even biosynthetic genes can be used as reporter in similar studies. The biosynthetic gene bpsA of non-ribosomal peptide indigoidine was used as reporter gene based on its detectability. The bpsA gene was introduced to engineered strains for evaluating targeted BGC expression. In continuation of this study, it was proved that TetR-like repressors genes can be applied for evaluating coelimycin production (Sun et al. 2017b).
High-throughput elicitor screening (HiTES)
The High-Throughput Elicitor Screening (HiTES) method includes increasing the expression of BGCs by elicitors and evaluating the omics data using reporter assays. The HiTES were designed with the aim of inducing promoters, expression of positive regulatory genes, or inhibition of repressor genes involved in SMs pathways by chemical inducers. In this method, effect of expanded types of chemical or biological inducers were analyzed in large scale, simultaneously. In a HiTES study, an unexplored NRPS gene cluster in S. albidoflavus was selected, and two types of reporters were considered. Initially, a silenced sur (pSur) promoter was fused to a triple eGFP cassette (pSur-eGFPx3) and placed at a neutral site in the S. albidoflavus chromosome. In the second strain, the insertion of the eGFPx3 specific site was performed directly downstream of the native pSur promoter. The signals of ivermectin (anti-fungal) and etoposide (anti-cancer drug) were indicators of finding inducer molecules for sur gene cluster (Mao et al. 2018). With this approach different libraries of components with potential to induce BGCs can be constructed.
HiTES, a high-throughput natural products discovery system, leading to identification of various types and numbers of natural products in one set of analysis. Detailed analysis of metabolites in the presence and absence of inducers led to the discovery of 14 new metabolites from four compound families. These families include surogamids, acylated surogamids, albucyclones and a group of linear decameric peptides. Albuquinone A is another new metabolite that was identified and did not belong to the sur gene cluster. These findings indicated the effect of ivermectin and etoposide on other gene clusters (Xu et al. 2017a).
Some specific chemical agents seem to have a general activatory or suppressory impact on BGCs expressions in various Actinobacterial genera. Atenolol was introduced as a global inducer compound that increased the expression of cryptic BGCs, leading to the identification of unknown compounds like taylorflavins A and B, hiroshidine, and pyridindolol in S. hiroshimensis. HiTES can be upgraded by reporting systems for increased accuracy and validity of the analysis. The use of Imaging Mass spectrometry (IMS) as a complement of HiTES (HiTES-IMS) can provide a more accurate analysis of metabolomic profile of strains (Xu et al. 2019).
Besides, the analytical, biological hyphenation of the HiTES can be used (bioactivity-HiTES), in which induced metabolites are screened based on biological activity, and finally, new metabolites are isolated and identified (Moon et al. 2019).
Genome-wide pleiotropic methods
Approaches that can awaken several BGCs in a non-targeted manner are considered pleotropic or genome-wide methods. Since some enzymes or regulators initiate various reactions in non-specific substrates, alterations in their gene sequences or function can activate multiple silent pathways (McLean et al. 2019).
Manipulation of global regulatory systems
There are two main groups of regulator genes that control the transcription and expression of BGCs. These regulators include global regulatory genes (GRGs) or pleotropic regulatory genes and cluster situated regulators (CSRs). Global regulatory genes are located upstream or downstream of BGCs and affect the expression of a wide range of gene clusters, while CSRs are within gene clusters and direct increase in expression of BGC. These regulatory proteins affect the expression of a wide range of gene clusters and regulate various reactions related to differentiation, cell growth and secondary metabolite production. For example, DasR regulatory protein, a member of GntR-family, causes the expression of genes involved in the decomposition of vegetative mycelia in the presence of the N-acetylglucosamine. Global regulators can also control the expression of cluster situated regulators (McLean et al. 2019). If regulatory genes are randomly manipulated, new unpredictable pathways are likely to be awakened. By targeting global or pleotropic regulatory genes, various types of mechanisms or cluster might be affected. For example, manipulation of adpA gene has resulted in the activation of germicidin and oviedomycin gene clusters (Baral et al. 2018). AdpA transcription factor (TF) as a global regulator is a practical and useful tool for the expression of BGCs and has the ability to bind to interacting sequences on the genome and regulate biosynthesis of various secondary metabolites. The use of plasmid pGM4181 with adpA under the control of a strong promoter in Streptomyces cyanogenus S136 led to a significant increase in the production of an antifungal metabolite (Yushchuk et al. 2021). In another example, BldD regulatory proteins adjust the biosynthesis of actinorhodin and undecylprodigiosin in addition to the formation of aerial mycelium by binding to BldC protein from MerR family (McLean et al. 2019). AfsR is another global regulatory protein that is activated by the phosphorylation of serine/threonine residues by a membrane kinase in the presence of environmental sensors and causes the expression of the regulatory gene afsS, which consequently leads to the increase in the production of actinorudin and undecylprodigiosin (Bibb 2005). Among global regulators, two-component systems (TCSs) are regulators that include two parts: sensing kinase (mainly histidine) in the inner membrane and cytoplasmic regulator (Xia et al. 2020). PhoP/R and AfsQ1/Q2 are the well-known TCSs in activating/blocking biosynthesis pathways of secondary metabolites (Fig. 1).
The cluster situated regulators (CSRs) are local regulatory genes that are highly influenced by environmental tensions and pleiotropic effects and are activated or repressed in the presence of environmental signals and other global regulators. Streptomyces antibiotic regulatory proteins (SARPs) are the largest group of local regulators, mainly in the genus Streptomyces (Bibb 2005). In the experiments conducted by Wu et al. (2021), the genes encoding SARPs as activators of biosynthetic clusters were cloned into pLM1 plasmid and integrated into S. tsukubensis genome. The positive regulatory effect of SARP led to the activation of the tsu gene cluster and biosynthesis of a new anthracycline, tsukubarubicin (Wu et al. 2021). Another major regulator family, the LuxR-family (LAL) (Bibb 2005), binds to the helix-turn-helix motif in the N-terminal region of the LuxR regulator, then initiates the transcription by binding the C-terminal part of regulator protein to DNA (Salwan and Sharma 2020). A CSR within a gene cluster, in addition to regulating the transcription of nearby genes, is likely to play a role in the expression of distant genes (McLean et al. 2019).
Modulation of regulatory genes
The regulatory elements of BGCs in bacteria gene clusters include various promoters, sigma factors, ribosomal binding-sites, regulatory RNAs (riboswitches, ribozymes, RNA termometers and non-coding RNA) (Lotz and Suess 2018), untranslated region (UTR), and terminator sequences (Myronovskyi and Luzhetskyy 2016). Inducing BGCs by these elements (Fig. 3) is mostly untargeted because most of the time the explicit impact of these elements are unknown, and modulation of these elements has unpredictable results in secondary metabolic pathways.
Promoter engineering Considering the necessity of a strong promoter for gene expression at significant levels, various promoter libraries have been constructed and designed. The most important promoters used for gene expression in actinomycetes are ermE, actΙΙ orf4 and sf14P, some synthetic promoters such as tcp830 and modified derivatives of native promoter such as kasOP (Myronovskyi and Luzhetskyy 2016). Production of Toyocamycin by S. diastatochromogenes 1628 as a fungicide was induced up to 5.2-fold using the promoter spl-21 (Xu et al. 2017b). Some promoters can be engineered by new engineering tools such as CRISPR Cas9.
Sigma factors have different frequencies in Streptomyces genus from 35 in S. albidoflavus to more than 60 in S. avermitilis and S. coelicolor (Myronovskyi and Luzhetskyy 2016). Considering the role of sigma factors in identifying the transcription starting sites (TSSs), deletion or overexpression of this gene can increase the formation of transcription complex and expression of biosynthetic gene clusters. For instance, an approximately 96% increase in avermectin production is observed in the sig8 deletion mutant (Dsig8) compared to the wild S. avermitilis (Sun et al. 2017a). Moreover, manipulation or replacement of (σ) factors can have a significant upregulating effect on BGCs expression. For example, the adaption of sigma (σ) factors from actinobacterial source in E. coli up-regulates the expression of oxyB biosynthetic gene of oxytetracycline in E.coli (Stevens et al. 2013).
5ʹ- untranslated region (UTR) sequence is located in the region between the TSS and the start codon and contains important genetic instructions for regulating transcription under certain circumstances. It also contains a ribosome binding site (RBS) that partly pairs with the 16 S rRNA sequence and causes the ribosome to be correctly positioned on the mRNA and translation is initiated exactly at the start codon (Lee et al. 2019). For the first time, investigation of the 5ʹ-UTR in S. lividans TK24 showed that AT-richness of of this region correlate with higher efficiency in the translation process (Myronovskyi and Luzhetskyy 2019).
Ribosomal binding-site (RBS), like promoters, have variable capabilities in gene expression, and multiple RBSs can be used. It is shown that the presence of A/U-rich sequences with various lengths and mRNA stem-loop structure upstream of RBS is associated with higher efficiency of translation (Alagesan et al. 2018). The investigation of the 5ʹ-UTR in S. lividans TK24 showed that nucleotide changes in this region can affect the quality of gene expression (Myronovskyi and Luzhetskyy 2019). In another research that was conducted to increase the sterol production in Mycobacterium neoaurum ATCC 25,795, different RBSs were investigated and the presence of RBS with medium strength was more efficient in enhancing the production of metabolites (Sun et al. 2020).
Riboswitches are short-stranded RNA sequences that are mainly located in the 5ʹ-UTR region of mRNA. These small sequences change mRNA folding and regulate gene expression at the translational level by binding to a ligand molecule. Ligands can be small molecules such as ions, free radicals, vitamins, which are normally present at low concentrations in the cell. Guanidine ΙІІ is a riboswitch present in most Actinobacteria and has a role in the regulation of the emrE and sugE genes and the SMR efflux pump (Lotz and Suess 2018).
Ribosome engineering
The idea of ribosome engineering was formed when the production of large amounts of actinorhodin was reported in a strain of Streptomyces lividans with a modified S12 ribosomal protein (RpsL) that was related to streptomycin resistance. Therefore, point mutations in ribosome can be used as a practical method to stabilize the ribosome, express gene clusters and increase their productivity (Shima et al. 1996; Abdelmohsen et al. 2015). Resistance mutations to other antibiotics such as tetracycline, erythromycin (Covington et al. 2021), streptomycin and gentamicin in Streptomyces coelicolor led to a 180-fold increase in actinorhodin production compared to the wild strain (Wang et al. 2008). The site of action of antibiotics in the ribosome includes decoding center (DC), peptidyl (P) and exit (E) sites in the 30 S subunit, Peptidyl Transferase Center (PTC) in the 50 S subunit, Nascent Peptide Exit Tunnel (NPET), translation factors of EF-TU, EF-G and IF2 in the 50 S subunit (Lin et al. 2018).
The investigation of ribosome engineering technique in 1068 Actinobacteria strains showed that 43% of Streptomyces and 6% of non-Streptomycetes after spontaneous mutation in rpsL or rpoB acquired the ability to produce secondary metabolites (Ochi and Hosaka 2013).
RNA polymerase engineering
Application of this method is more usual in fungal strains than actinobacterial strains. The bacterial alarmon ppGpp which is formed by ppGpp synthetase in response to stresses (Baral et al. 2018), binds to RNA polymerase (RNAP) and ultimately initiates the synthesis of the secondary metabolites. This suggests that modification of RNAP, by introducing a rifampicin-resistant mutation in rpoß, may mimic the ppGpp-bound form and activate the expression of BGCs (Ochi and Hosaka 2013). Generation of rifampicin-resistant mutations in the rpoB gene related to the β subunit of the polymerase enzyme in Streptomyces somaliensis SCSIO ZH66 led to an increase in the production of fredericamycin metabolite up to 3-fold (Zhang et al. 2015).
Phosphopantheteine transferases
In the initial stage of biosynthesis of polyketides and non-ribosomal peptides, acyl transferase and peptidyl transferase should be activated by a post-translational modification, which can be fostered by the activation of phosphopantetheinyl transferases (PPTases). There are two types of PPTases in prokaryotes. The first type is responsible for Acyl Protein Carrier (ACP) modification and the second type is present in biosynthesis pathways of secondary metabolites, and modifies the Carrier Proteins (CPs). This approach is limited to inducing the PKS and NRPS genes, but has shown high efficiacy in most of the studies.
In heterologous expression of a cyanobacterial natural product in E. coli, five different PPTases were investigated. The results of this study showed that efficiency of PPTase depends on different factors like type of pathway, selection of the proper type of PPTase, CP interactions, induction conditions in the host, and increasing the concentration of PPTase, which has upregulation effect on the expression of BGCs (Liu et al. 2018). Insertion of PPTase genes (svp and sfp) isolated from Streptomyces verticillus and Bacillus subtilis, respectively, into 33 Actinomysetes genomes caused overexpression of various PKS and NRPS BGCs in 23 strains (Zhang et al. 2017a).
In another study, the increased expression of these two PPTase genes in S. alboniger NRRL B-1832 was introduced using the constitutive promoter ermE by conjugation. This approach of induction resulted in the isolation of three cryptic nucleosides puromycin A, B, and C (Yan et al. 2018).
Targeted genome-wide methodologies
Using CRISPR-Cas system in targeted modification
The CRISPR-Cas genomic tool can be used in various genetic-induction approaches such as heterologous expression, BGC refactoring, gene knockout or knock in, and cryptic BGC conjugation. Homology Directed Repair (HDR) is a CRISPR-based method which is directed by plasmid transfer containing elements such as cas nuclease, gRNA, and homologus repair template. Moreover, approaches such as Non-Homolgous End Joining (NHEJ) and Base editor are the other CRISPR-based methods that have been used for genome editing in Streptomyces spp. (Karthik et al. 2022).
The CRISPR-Cas9 can be widely used to induce cryptic BGCs by replacing the inactive promoter of cryptic genes with active promoters and BGC refactoring. In a study, CRISPR-Cas9 strategy was used to activate two unexplored BGCs in Streptomyces roseosporous, one of them was homologous to topolycyclic tetramate macrolactam BGC in S. griseus. Moreover, in another study, the cryptic type I polyketide synthesis was activated by insertion of kasO*. This strong promoter induces the expression of aurR1 which is the activating gene in the production of Auroramycin (Lim et al. 2018). The replacement of constitutive kasO*p with CRISPR-Cas 9 genetic tool triggered the expression of cryptic type II polyketide BGC in S. viridochromogenes, type III polyketide BGC in S. venezuelae, and indigoidine compound BGC in S. albidoflavus (Zhang et al. 2017b). In addition to promoter replacement, CRISPR methods can be used for the addition or deletion of regulatory genes. The pathway specific regulator gene papR3, which had repression effect on pristinamycin I production in Streptomyces pristinaespiralis, has been deleted from wild strain by CRISPR Cas genome editing system that lead to an increased production of pristinamycin I to more than 130 mg/L (Meng et al. 2017).
Currently, CRISPR-Cas9 has a crucial role in genetic induction methods like cloning, heterologous expression, and whole BGC refactoring. Homology-directed repair method has been used in three Streptomyces strains with 70–100% efficacy, and with a deletion range from 20 bp to 30 K bp (Cobb et al. 2015). The efficiency of this method depends on various parameters like sites of interest, targeted strains, and the type of Cas9 that cause some limitations for its adaption in different actinobacterial species (Yeo et al. 2019).
There are challenges in using this approach, like the creation of DNA double-strand breaks (DSBs) that cause genome instability, and toxicity. To solve these challenges, CRISPR-BEST (CRISPR-Base Editing System) has been used as C-to-T, and (CRISPR-aBEST) as A-to-G base editor. This strategy has been used to introduce a stop codon that led to inactivation of two copies of kirN, a gene involved in kirromycin production in Streptomyces collinus Tü365. This strategy has also been used in other Streptomyces spp. (Tong et al. 2019a, b).
The other important limitation of CRISPR-Cas9 for genome editing in actinomycetes genomes is the toxicity of Cas9 protein. It has been proved that overexpression of Cas-9 hindered the exoconjugation of plasmid pCRISPomyces-2. Therefore, replacement of weaker constitutive promoter for cas9 gene in novel designed plasmid pCM4.4 leads to reduction in Cas9 expression and increases probability of success in genome engineering. In this study, ACT BGC that encode type II polyketide actinorhodin was employed inefficacy evaluation of genome editing by this approach (Ye et al. 2020).
Transcription factors and promoter exchange/modifications
In most promoter engineering studies, native weak promoters of inactive BGCs have been replaced with naturally active promoters, or synthetic promoters which are effective in induction of BGCs. Most natural promoters used in promoter engineering have been isolated from productive species. Expression of cryptic gene bpsA, which was silent under normal laboratory condition, was increased after inserting the erythromycin resistance gene promoter (ermE*p) in front of this gene (Olano et al. 2014).
TFDs (Transcription Factor Decoys) have been applied for inducing the expression of silent NRPS and PKS BGCs. TDFs are engineered sequences which mimic the specific sequences that bind to regulatory factors and act as competitors for regulatory DNAs. This de-repression strategy can be used as large-scale method without any genetic manipulation. This method has been applied in induction of eight silent BGCs in different Streptomyces spp. that led to the discovery of a new oxazole compound (Wang et al. 2019a).
The multi-omics data collected from species under the biological, chemical and physical inducing agents need to be interpreted to identify transcriptional network and expression pattern of BGCs in Actinobacteria. In a related study, four regulatory elements and one sigma factor responsible for the expression of BGCs were discovered based on multi-omics data from ribosome profiling, Term-Seq, RNA-seq, and dRNA-seq in Streptomyces griseus NBRC 13,350 (Hwang et al. 2022).
Inactivation of the highly expressed biosynthetic pathways
This strategy is based on changing the flow of metabolic of secondary metabolites in Actinobacteria, and blocking common compounds to facilitate the production of rare and unique compounds that are cryptic. Many of the pathways that produce secondary metabolites overlap in regulatory proteins, transporter enzymes, precursor molecules, and other effective factors in biosynthetic pathways. Thus, blocking competing pathways with targeted BGCs may induce expression of silent BGCs.
By using pCRISPR-Cas9 and pCRISPomyces-2 genomic tool, streptothricin and streptomycin compounds were knocked out as the most prevalent encoding BGCs from 11 actinobacterial strains. This strategy resulted in the isolation of unknown compounds like amicetin, 5-chloro-3-formylindole, thiolactomycin, and phenanthroviridin (Culp et al. 2019).
Furthermore, production of anti-diabetic secondary metabolite acarbose can be increased by decreasing the flux to valienol and 1-epi-valienol shunt metabolites in Actinoplanes sp. SE50/110 (Zhao et al. 2020).
Deletion of the suppressors or awakening the activators
Regulatory genes are responsible for modifying the time and level of core biosynthetic gene expressions. In regular natural condition of actinobacterial strains, these genes are under control of the secondary metabolite concentration, environmental stimuli, and interaction with other species. In most cases, ploeotropic and global regulators activate/deactivate the pathway-specific regulators, and pathway-specific regulators which directly control the expression of BGCs. The metabolic profile comparison in wild-type and mutant strains, analyzing the BGCs expression under different cultural condtions, and effect of chemical, physical, and biological elicitors and multi-omics data of manipulations like overexpression or knocking out targeted genes aid in mapping the regulatory network and identifying mechanism of their effect on BGCs expression (Fig. 3).
The common strategy is based on manipulation of regulatory genes by inducing the expression of genes encoding activator proteins and deleting or blocking genes encoding repressor proteins. A hybrid of PKS and NRPS gene clusters producing three Totopotensamides, can be activated by overexpression of totR1 (LuxR-type protein) or knockout of totR5 and totR3 which are negative regulators of SM in Streptomyces pactum SCSIO 02999 (Chen et al. 2017). Although finding these genes and their roles are challenging, advances in machine learning twill improve the detection of these genes in BGCs.
The main methods for regulatory genes discovery. (1) A Overexpression of activator genes, or deletion of suppressor genes, and evaluation of the on the production. B Using various types of cultures with different chemical, biological, and physical inducers. C Inducing mutation by different agents, and assessment of their effect on BGCs expression. (2) Analyzing effect of regulatory genes on BGC expression by omic approach (genomics, transcriptomics, proteomics, and metabolomics techniques). (3) Creation of regulatory network models using systems biology and bioinformatical tools from multi-omics data
In a recent study, the effect of two regulator genes in formocamycins production by Streptomyces formicae KY5 has been reported. It was observed that the deletion of the suppressor gene forJ from MarR-family regulators reduced the expression of formocamycins BGC. On the other hand, another discovered regulator gene forGF showed an upregulation effect on production of formocamycins. However, the simultaneous overexpression of forGF, and forJ deletion resulted in an increased formocamycins production of up to to 10-fold (Devine et al. 2021).
Manipulation of regulatory genes can also be effective for inducing BGCs in heterologous expression and optimized engineered hosts for secondary metabolites production. In a recent study, cryptic manumycin-type BGCs of Saccharothrix espanaensis were induced in heterologous expression in E. coli using two regulatory genes, espR1 and espR2, under the control of ermE* constitutive promoter (Gorniaková et al. 2021).
Finding cis elements to control transcriptional factors led to controlling the expression of targeted pathways based on the type (activator/repressor) of factors. The training sets from several omics data of different actinobacterial genera allow the in silico prediction of transcription sites using tools like PREDetector. With high-throughput proteomics methods like DAP-seq (DNA affinity purification sequencing) identification of transcription factors based on DNA-protein interaction, and creating libraries from them is feasible (Hiard et al. 2007; Rigali et al. 2018).
Another method called semi-targeted approach has been used to activate mayamycin A in Streptomyces sp. TÜ17 which was conducted by introducing cluster situated regulators (CSRs) and Streptomyces antibiotic regulatory proteins (SARPs) on a plasmid. These plasmids triggered the production of chartreusin-like compound and warkmycin in Streptomyces sp. TÜ102 and Streptomyces sp. TÜ10, respectively (Mingyar et al. 2021) (Table. 4).
Heterologous expression in surrogate hosts
Characterization of silent BGCs construction by genome mining tools provides required information for their cloning into a host with optimized condition for BGC expression. Transfering the whole genome or a cluster to another host in this way has resulted in the creation of strains with higher productivity (such as cephalosporin production), the biosynthesis of new compounds (such as indolizomycin), or even the activation of silenced gene clusters (Shitut et al. 2020).
Different BGCs from metagenomes and whole genomes have been cloned in fosmid, cosmid, and E. coli libraries such as the large size PKS BGC of quinolidomicin A1 with more than 20 Kb genome size have been cloned in BAC library (Hashimoto et al. 2018).
Different heterologous expression techniques have been used for induction of BGCs. The instability of the heterologous genome and the limitation of the recombination time due to the rapid reconstruction of the cell wall after the fusion of cells are among the challenges of recombinant BGCs (Shitut et al. 2020). In addition to the recombination of chromosomes, the fusion of L-forms causes polyploidy, and the presence of multiple gene copy that can have beneficial effects on the enzymes, antibiotic production and biological control.
Heterologous expression studies demonstrated that the most common hosts like E. coli and S. cerevisiae, despite their simple genomic manipulation or cultivation, are not appropriate hosts for the production of natural products, because they cannot support the production of natural products with complicated chemical structures. In recent years, S. coelicolor M1146 has been considered as an alternative host for BGC expressions. The BGC of RNA polymerase inhibitor Pseudouridimycin (PUM) was identified in Streptomyces sp. DSM 26,212 by knock-out method and successfully expressed in S. coelicolor in heterologous expression (Böhringer et al. 2021). In another example, three novel lasso peptide BGCs were discovered from Streptomyces leeuwenhoekii C34 by genome mining method, and unlike the lp1 (predicted to belong to citrulassin family) and the lp2 BGC was successfully expressed in S. coelicolor. The results of the study show that the active promoter ermE* can activate the BGC expression in the host, and there is no regulatory mechanism for leepeptin biosynthesis in native strain (Gomez-Escribano et al. 2019; Böhringer et al. 2021).
The engineered strain Streptomyces griseofuscus DSM 40,191 has also been introduced as a candidate host for heterologous expression which has been optimized for heterologous expression using CRISPR-Cas 9, CRISPR-cBEST and GusA vectors (Gren et al. 2021). The novel method of large DNA fragment cloning (NaBLC) has been used to insert BGCs of anaerobic strains into Streptococcus mutans UA159 as a heterologous host. In this strategy, the targeted genome was inserted into the competent host by homologous recombination and then screened using counter selection marker (Hao et al. 2019).
Sequencing, transcriptomic, and genomic analysis of S. albidoflavus have demonstrated that this strain can also be an appropriate strain for heterologous expression of actinobacterial BGCs (Zaburannyi et al. 2014). The construction of spinozad artificial gene cluster by refactoring the gene cluster and placing it in the heterologous host S. albidoflavus increased the biosynthesis of spinozad by 328-fold (Li et al. 2023). Although S. coelicolor is one of the most studied and applicable actinobacterial hosts, as this strain has potential for the production of PKS and NRPS biosynthetic pathways precursors (Gomez-Escribano and Bibb 2011, 2014), the selection of proper host with desired properties must be based on the targeted BGC.
Refactoring of gene clusters
BGC refactoring is the rearrangement of important transcription elements such as promoters, terminators, ribosomal binding sites, regulatory factors, and transcriptional elements and other expression-related factors which have been used to design specific hosts for expression of the targeted BGCs (Alam et al. 2021). To use the BGC refactoring strategy, BGCs should have two characteristics: first, the sequenced and annotated genomes should be tractable, and second the targeted BGCs should be under the control of a single operon (Ogura et al. 2018).
BGC refactoring (Fig. 4) has been considered as a scaleable method for increasing the production of well-known compounds, or designing optimized condition for expression of silent BGCs which lack expression condition. The expression of four glycosins cryptic BGCs from Actinobacteria has been triggered by heterologous expression in E. coli BL21 after two Golden Gate assembly steps (Shantanam et al. 2018).
Streptophenazines were produced by cryptic hybrid (PKS/NRPS) BGC spz from Streptomyces sp. CNB-091. In a study, various types of promoters like strong synthetic promoters’ p21 and sp44 were used to increase expression of spz, and refactored BGCs were assembled in Streptomyces coelicolor M1146. Results of RT-PCR showed that sp44 had the most triggering effect on spz expression. Interestingly, metabolomic comparison from LC-MS results revealed strains with refactored BGC have produced more than 35 different streptophenazines with various chemical structures (Bauman et al. 2019).
The heterologous expression of ato gene cluster isolated from Amycolatopsis tolypomycina NRRL B-24,205 in yeast led to production of two anticancer compounds, atolypene A and atolypene B. In this study, a combined method from CRISPR Cas9 was used for cleavaging off the native promoter, and transformation assisted recombination (TAR) was applied to reassemble ato BGC with synthetic promoters. This method, named miCRISTAR (multiplex Cas9-TAR), which enables exchanging various promoters was possible across ato BGC (Gustavson et al. 2018).
Process of BGC refactoring: A Genomes of actinobacterial strains acquired from whole genome sequencing or metagenomics analysis. B Genome mining of secondary metabolites pathways using tools like antiSMASH. C Recognition of cryptic BGCs and selection of non-orphan cryptic BGCs. D Cloning of cryptic BGCs by novel methods like Gibson, TAR, or Golden Gate assembly methods. E Targeted manipulation of BGC in different genetic elemensts like strong promoters (native or synthetic), and upregulation, insertion, or deletion of suppressor genes. F Optimization of expression by heterologous host engineering methods. G Structure elucidation or detection of the induced cryptic metabolite
Pathway induction by epigenomic modifications
The epigenetic modulating agents of the actinobacterial genome include chemical and conformational modifications. Known chemical modifications of the bacterial epigenome include histone deacetylases, DNA methyltransferases and modifying nucleoid-associated proteins. Conformational dynamic of the bacterial genome varies the exposure of the clusters to the transcription which itself is influenced by the chemical modification of the nucleoid associated protein and deoxyribonucleic acids. Among these four types of epigenomic approaches discussed in below subsections, only inhibitors of histone deacetylaseare frequently used for pathway elicitation in actinobacteria.
Manipulating the nucleoid-associated proteins in actinobacteria
There are reported nucleoid-associated proteins in Actinobacteria that their activation or deletion be used for universal BGC induction in these group of bacteria. The Transcription of secondary metabolites in Streptomyces is related to a bifunctional nucleoid-related protein (DdbA) with a DNA-binding histone H1-like domain and an interacting domain with RNA polymerase which itself is regulated by ppGpp (Aldridge et al. 2013).
Another recently reported nucleoid-associated protein, Lsr2, inhibits the expression of gene clusters by binding to the AT-rich sequence (generally species-specific genes which are presumably newly obtained by horizontal gene transfer). Thus, deletion of the lsr2 gene in Actinobacteria can lead to the synthesis of new metabolites (Gehrke et al. 2019). Lsr2 is assumed to suppress the cluster by its polymerization on the BGCs. In contrast, Lsr2 repression is neutralized by CmlR by promoting the RNA polymerase and removal of Lsr2 polymers from genome. Engineering of these two counteracting nucleoid related proteins led to a 130 fold increase in chloramphenicol production by S. venezuelae (Zhang et al. 2021).
Modulating the DNA methylation of actinobacteria
Modifying the activity of methylation using S-adenosylmethionine (SAM)-dependent methyltransferases (MTs) or the level of SAM can be suggested as a non-targeted or semi-targeted approach for awakening the cryptic pathways as it can contribute to the secondary metabolite biosynthesis in four ways. First, through activation of signaling pathways like AfsK serine/threonine kinase in actinorhodin biosynthesis (Jin et al. 2011); by natural addition of the methyl group to the substrate; by artificial inclusion of the methyl group for improving the bioactivity or bioavailability of the biosynthesizing compound and lastly epigenetic modification of the bacterial genome by increasing the methylation substrate of DNA methyltransferases.
Modifying the chromosomal conformation
The three dimension (3D) structure of chromosomal organization in S. coelicolor, shows remarkable change by shift from primary to secondary metabolism and some BGCs in special local conformations can be expressed. Therefore, an inserted single reporter gene or whole BGC can have a higher transcription after relocation into the hotspots, which may indicate a new approach for induction of secondary metabolites (Deng et al., 2023). Future advances in machine learning may aid in mimicking the conditions inducing the favored 3D of the chromosome in the position of BGCs without the need to relocate the cluster to the positions with favored spatial structure through chromosome remodeling.
Inhibiting the histone deacetylation of genome in actinobacteria
The limited access of RNA polymerase to BGCs due to the supercoiling can be ameliorated by inhibiting the polyamine deacetylase of Actinobacteria. This approach is the most frequently employed epigenetic-based approach for awakening the cryptic pathways in Actinobacteria. For instance, compounds like sodium butyrate and valproic acid have shown inducing effect on secondary metabolites in S. coelicolor (Zheng et al. 2022)d kermanensis (Mohammadipanah et al. 2020) assumed by the inhibition of histone deacetylation, respectively.
Concluding remarks
Discrepancies in the gene clusters (BGCs) identified through genome mining, compared to the known metabolic profiles of secondary metabolites in Actinobacteria species, indicate the presence of compounds that remain unexpressed in natural or laboratory conditions. Addressing the challenge of many gene clusters either not transcribed or expressed at undetectable levels, two primary objectives are pursued in the induction and activation of actinobacterial secondary metabolite biosynthetic pathways. These objectives encompass boosting the production of known secondary metabolites or eliciting the expression of cryptic/silent genes to facilitate the discovery of novel compounds.
The use of chemical substances, such as hormone-like molecules, secondary metabolites, elements, and micro/nanoparticles, can be considered relatively cost-effective method for stimulating the production of secondary metabolites. Among these chemical inducers, talc microparticles has reported with the most significant impact, resulting in a 13-fold increase in secondary metabolite production (Kuhl et al. 2021). Nevertheless, their practical application on a large scale is hindered by the toxicity of these particles at high concentrations to microorganisms and the environment.
Both the RGMS and HiTES methods offer extensive screening capabilities for various chemical and other inducers simultaneously. Additionally, advanced complementary tools, like the use of Imaging Mass Spectrometry (IMS) with HiTES, have enhanced the accuracy of data analysis and the discovery of novel inducers. However, they face two primary challenges. Firstly, their effectiveness is highly reliant on the regulatory and transcription factors associated with BGCs, and their impact varies across different BGCs. Secondly, their influence on metabolic pathways remains ambiguous due to gaps in our knowledge of secondary metabolite metabolic pathways. Transposon genes can be utilized to awake the silent pathways of Actinobacteria which need complementary analytical approaches such as IMS (Imaging Mass Spectrometry) and SOM (Self-Organizing Map) in evaluating metabolomics profile of mutants in Actinobacterial strains. The most beneficial advantages of this method are identification, analyzing and regulation of several BGC pathways instead of using single reporter system for a pathway (Yoshimura et al. 2020).
The other screening approach of OSMAC, is effective in activating hidden BGCs in different strains, but is often time-consuming. In this strategy, the co-culture approach simulates microbial communication through signal molecules or cellular components derived from either living or deceased biomass. The primary challenge within this strategy revolves around the purification of compounds, especially in co-culture studies where multiple compounds are simultaneously generated and accumulate. Additionally, the multitude of variables involved makes altering these variables for different strains a complex task, which can increase the number of experimental setups while reducing the focus on specific BGCs and strains in an OSMAC study. Adopting similar conditions in the OSMAC method for strains sharing genetic, physiological, and chemotactic characteristics can enhance the likelihood of BGC expression. Combining heterologous expression with OSMAC methods can also lead to induction of silent BGCs.
Employing the expression strategy of PPTases can be regarded as a non-targeted approach to stimulate the NRPS and PKS or their hybrid. Nevertheless, there are certain challenges associated with utilizing these enzymes for the upregulation of secondary including the mutations in PPTase genes due to the instability of actinomycete genomes and the inefficacy of some PPTases introduced via horizontal gene transfer. Additionally, it’s worth noting that this technique, like HiTES and RGMS, is not all-encompassing in its ability to comprehensively increase or induce the expression of various types of BGCs.
The genetic manipulation of Actinobacteria presents challenges due to factors such as a high GC% content, slow growth, and specific physiological traits. Moreover, most genetic engineering methods necessitate tailored adjustments for different strains to achieve satisfactory expression of BGCs. For instance, heterologous expression has encountered difficulties in numerous Actinobacterial species. Obstacles like the cumbersome cloning of large DNA fragments, the inability to coordinate the expression of cloned genes, limitations in the availability of precursors in the surrogate host, and the potential toxicity of final products or intermediates to the surrogate host constrain the utility of heterologous expression for induction of cryptic BGC. Additionally, not all promoters exhibit uniform effectiveness in activating BGC expression. While some robust synthetic or natural promoters can effectively induce BGCs, the key issue lies in the absence of a discernible relationship between promoters and other genetic or epigenetic factors. The development of optimized Actinobacteria hosts for pathway induction necessitates careful host selection, the use of potent promoters, the overexpression of positive regulators, and the removal of negative regulators. So far, E. coli, S. lividans, and S. coelicolor have extensive use as hosts for heterologous expression of silent BGCs due to attributes like manipulability and the ease of maintaining growth conditions in a laboratory setting despite their other disadvantages. The analogue of approaches have been used in BGCs induction in fungal species, or other eukaryotic species like chromatin remodeling, and mutation of transcription factors can be assessed in Actinobacteria. Using targeted induction for pharmaceutical or medical applications is more reasonable, as the less complex metabolite extract is more important compared to compounds that are intended for agricultural applications and can be commercialized as semi-pure products. Moreover, methods like OSMAC and co-culture methods are compatible for induction of pathways producing compounds with agricultural applications as these strategies mimic the ecological conditions which intervene in the communication and networks of strains.
Furthermore, all traditional methods such as co-culturing, the utilization of physical, chemical, and biological inducers, and the OSMAC techniques are applicale for strains that thrive in laboratory settings while the majority of Biosynthetic Gene Clusters (BGCs) are reported from uncultivated Actinobacteria from metagenomic samples. Therefore, the primary focus should be directed towards addressing the challenges associated with genetic manipulation of non-expressing pathways, including the substantial genome size of BGCs for heterologous expression, the high-GC content in actinobacterial strains, the development of engineering tools, and the accumulation of systems biology data.
Despite the emerging potential of CRISPR Cas-mediated modulation in BGC induction, limitations arise when attempting to introduce various CRISPR Cas complement elements such as recombinases, Cas effector proteins, and guide RNAs concurrently. Additionally, BGCs refactoring and heterologous expression face challenges, including the cytotoxic effects of Cas9, limited genomic accessibility in the transformation of recombinant DNA in certain actinobacterial strains, and decreased efficiency in multiplex genome editing (Musiol-Kroll et al. 2019).
Moreover, applying BGC refactoring and heterologous expression to induce BGC expression can result in increased chemical diversification within compounds, even for known BGCs.
In summary, each induction approach possesses limitations, and there is no all-encompassing method capable of inducing all cryptic pathways as universal BGC inducers. Most non-genetic induction approaches lack specificity and often yield random results. In contrast, targeted induction methods typically apply a rational framework to design optimal engineered hosts or BGCs for the increased production of known or cryptic BGCs. Despite the simplicity of non-genetic methods, except for the purification phase, they tend to be more cost-effective for scaling up the production of natural products, especially for industrial and agricultural applications. However, it’s evident that non-genetic methods have limited success, lack a discernible pattern, and do not consistently induce BGCs.
Consequently, a combination of biological, chemical, and physical factors, as exemplified in methods like OSMAC, can mimic the natural stimulants of the inheret regulatory cascade of the BGCs. On the other hand, for medical applications that require high-purity products due to the challenges in compound separation, genetic methods or designing engineered hosts may be necessary. Nevertheless, developing new host models for heterologous expression of BGCs sourced from metagenomic samples is crucial. Although these methods are not universally successful for all BGCs, modifying epigenetic factors and employing screening methods like OSMAC increases the likelihood of BGC expression.
Future perspectives
Due to the necessity of discovering new bioactive compounds for pharmaceutical and agricultural sectors, new approaches need to be devised based on genome data to induce metabolics pathways. However, still the untargeted approaches such as OSMAC and HiTES are effective in expressing silent pathways especially in various strains simultaneously. Furthermore, the probable inducing conditions can be predicted by the aid of machine learning methods from omic data obtained from OSMAC studies. Considering the role of environmental microorganisms as a stimulus in the production of bioactive compounds, pathogens and pest secretory metabolites is suggested to be evaluated in co-culture technique and chemical inducers methods as inducers in the biosynthesis of antimicrobials, pesticides and biofertilizers, respectively. Moreover, increasing the taxonomic diversity of co-cultivated strains can increase the chemical diversity of the induced SMs.
One of the least investigated parameters in induction of cryptic pathways in the physical texture of the sorounding microenvironment of the actinobacterial cells. Nanofiber with various chemistry, porosity and plasticity can be generated using electrospinning and be evaluated as inducing agents of BGCs in Actinobacteria. Moreover, investigating the inducing effect of dark and light conditions and their sequence or intervals on the inducton of silent pathways is suggessted.
On the flip side, improving the cloning techniques or introducing novel methods like CRISPR Cas9 will increase the efficiency of BGC engineering and pathway induction. In addition, developing the optimally engineered actinobacterial hosts will decrease the challenges of cloning and overexpression of BGCs. However, the limitation of cloning large pieces of DNA, the inability to coordinately express cloned genes, the limitation in the supply of precursors in the surrogate host and the possibility of the final product or intermediates being toxic to the surrogate host need to be resolved.
Although using active PPTases in optimally engineered species for increasing polyketides and non-ribosomal peptides production or their hybrid sounds promising, PPTases with comprehensive efficiency on broad types of PKS and NRPS are still needed. Overall, for development of universal method, there is a requirement for discovering enzymes, regulatory genes, and TFs or PPTases which are common and effective in various Actinobacterial strains.
Meanwhile, terminators and UTR sequences are regulatory sequences which, despite their important role in various stages of gene expression, have not been well studied in Actinobacterial strains. The multi-omics data that is collected after elicition in OSMAC and HiTES methods is suitable to find correlation between elicitors and regulatory genes. This data may be applicable to generate networks from regulatory genes and transcription factors by systems biology models with higher accuracy.
The genome mining analysis of Actinobacteria shows that there are some unique BGCs in Actinobacteria with no assigned product because of lacking the core biosytnesis genes or deficiency in regulatory genes or both. In addition, the regulatory systems of BGCs expression demonstrate that most of the BGCs are affected by pathway-specific regulator genes. Therefore, unexpressed BGCs may be the result of partial gene transfer of BGCs in horizontal gene transfer, and absence of important biosynthetic genes, or regulatory genes of targeted BGC. Applying the genome mining tools like antiSMASH to map the structure of targeted BGC is needed as the prerequisit step of inducing cyptic BGCs to explore thebiosynthesis genes, regulatory genes, transporter genes, and other complementary genetic elements which are essential for BGCs expression (Skinnider et al. 2017; Blin et al. 2021).
Notwithstanding novel genome editing, gene transferring, and cloning techniques like CRISPR Cas9, TAR cloning, Golden Gate gene assembly, there are still a limitations in genomes manipulation of Actinobacteria. One of the most important challenges is transferring BGCs and large-sized genetic elements. On the other hand, the effect of specific phages and prophages in horizontal gene transferring that lead to actinobacterial BGCs variation have been proved. Using these prophages as genome vectors may be helpful for manipulation of Actinobacteria, but different factors like programmability and being resistant in laboratory condition are necessary (Seshadri et al. 2022).
One of the issues that has paramount importance in heterologous expression of BGCs is selecting an appropriate host. All non actinomycete hosts for BGCs expression had some limitations and the number of BGCs that are expressed in E. coli are not significant, or they are only produced in low titers. Nevertheless, producing actinobacterial natural products in yeast or other eukaryotic hosts have some challenges such as codon optimization. In addition, studies show that well-known Streptomcyes strains like S. lividans, S. avermitilis, S. albidoflavus, and S. coelicolor are still not efficient for heterologous expression of large BGCs (Hwang et al. 2021). The advancements in synthetic biology methods enable the design of genetic elements like TFs, regulatory genes, promoters, ribosome binding sites, and even biosynthetic genes for control of BGC expressions. By advance editing, assembling, and genome engineering methods, developing optimized and specific engineered hosts for BGC expression will be feasible in future.
As most of the detected BGCs are identified from environmental DNA (36 times the number of BGCs in MIBiG dataset) (Ma et al. 2023), before applying inducing methods, first, these BGCs should be coloned in an engineered, surrogate, and culturable host. One of the reasons behind the failure in induction of the silent pathways or heterologous expression of environmental BGC is that their transcriptional factor is unclustered with their BGC and often cannot be found or assigned in BGC prediction algorithms. In it shown that comparative analysis of mutual rank and Spearman-derived coexpression networks can aid in identification of non-resident transcriptional factors located outside of the BGCs in fungi (Kwon et al., 2021). Thus, future analysis of gene coexpression for characterizing the transcriptional networks are needed for the actinobacterial species with cryptic pathways (Kwon et al. 2021). The effect of cultural condition, growth elements, and their corresponding regulatory genes is concludable from the effect of inducers that are applied in non-genetic methods. The interactomic data in parallel with transcriptomic and metabolomic are also required to elucidate the molecular regulatory mechanisms of the pathway awakening by the aid of cultural techniques.
The recent report on the promotion of differentiation in S. venezuelae following phage infection (Luthe et al. 2023), support the assumption that the induction of secondary metabolites upon phage infection is presumably a conventional response in Streptomyces. Function of the induced secondary metabolites can be attributed to the antagonistic effect against other bacteriophages or acting as an alarmon of being infected to the surrounding cell (Kronheim et al. 2023). Thus, actinophage can be employed as global or specific elicitors of secondary metabolism in Actinobacteria. Another untargeted approach that we can propose for the triggering the BGC in Actinobacteria is using the outer membrane vesicle (OMV) of other microorganisms which contain the signal molecules with the nature of protein or nucleotide. Another targeted approach that still has not been applied for pathway elicitation in Actinobacteria is employment of the Non-coding RNA (ncRNA) for which can be used for inhibiting the trival or known compounds or activating the transcription of the gene cluster by chromosome remodeling. However, failure in induction such experiments can be due the incompleteness of the cluster or the necessity of a tailoring or regulatory protein which is located outside of the gene clusters of that compound. Thus, one of the demanding applications in the BGC analysis is the tools that can predict the functionality of the BGC and differentiate the pathways that are silent from the BGC which are orphan and incomplete.
Data availability
There is no generated experimental data report in this paper and all discussed data here are from the publocations referred in this paper.
References
Abdelmohsen UR, Grkovic T, Balasubramanian S, Kamel MS, Quinn RJ, Hentschel U (2015) Elicitation of secondary metabolism in actinomycetes. Biotechnol Adv 33:798–811
Adnani N, Chevrette MG, Adibhatla SN, Zhang F, Yu Q, Braun DR, Nelson J, Simpkins SW, McDonald BR, Myers CL, Piotrowski JS, Thompson CJ, Currie CR, Li L, Rajski SR, Bugni TS (2017) Coculture of Marine Invertebrate-Associated Bacteria and Interdisciplinary technologies Enable Biosynthesis and Discovery of a New Antibiotic, Keyicin. ACS Chem Biol 12(12):3093–3102. https://doi.org/10.1021/acschembio.7b00688
Alagesan S, Hanko EKR, Malys N, Ehsaan M, Winzer K, Minton NP (2018) Functional genetic elements for controlling gene expression in Cupriavidus necator H16. Appl Environ Microbiol 84(19):e00878-18. https://doi.org/10.1128/AEM.00878-18
Alam K, Hao J, Zhang Y, Li A (2021) Synthetic biology-inspired strategies and tools for engineering of microbial natural product biosynthetic pathways. Biotechnol Adv 49:107759. https://doi.org/10.1016/j.biotechadv.2021.107759
Aldridge M, Facey P, Francis L, Bayliss S, Del Sol R, Dyson P (2013) A novel bifunctional histone protein in Streptomyces: a candidate for structural coupling between DNA conformation and transcription during development and stress? Nucleic Acids Res 41(9):4813–4824. https://doi.org/10.1093/nar/gkt180
Baral B, Akhgari A, Metsä-Ketelä M (2018) Activation of microbial secondary metabolic pathways: avenues and challenges. Synth Syst Biotechnol 3(3):163–178. https://doi.org/10.1016/j.synbio.2018.09.001
Barka EA, Vatsa P, Sanchez L, Gaveau-Vaillant N, Jacquard C, Klenk H-P, Clément C, Ouhdouch Y, van Wezel GP (2016) Taxonomy, physiology, and Natural products of Actinobacteria. Microbiol Mol Biol Rev 80(1):1–43. https://doi.org/10.1128/mmbr.00019-15
Bauman KD, Li J, Murata K, Mantovani SM, Dahesh S, Nizet V, Luhavaya H, Moore BS (2019) Refactoring the cryptic streptophenazine Biosynthetic Gene Cluster unites Phenazine, Polyketide, and nonribosomal peptide Biochemistry. Cell Chem Biol 26(5):724–736e7. https://doi.org/10.1016/j.chembiol.2019.02.004
Bibb MJ (2005) Regulation of secondary metabolism in Streptomycetes. Curr Opin Microbiol 8(2):208–215. https://doi.org/10.1016/j.mib.2005.02.016
Blin K, Shaw S, Kloosterman AM, Charlop-Powers Z, Van Wezel GP, Medema MH, Weber T (2021) AntiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res 49(W1):W29–W35. https://doi.org/10.1093/nar/gkab335
Böhringer N, Patras MA, Schäberle TF (2021) Heterologous expression of pseudouridimycin and description of the corresponding minimal biosynthetic gene cluster. Molecules 26(2):1–10. https://doi.org/10.3390/molecules26020510
Chen G, Wang GYS, Li X, Waters B, Davies J (2000) Enhanced production of microbial metabolites in the presence of dimethyl sulfoxide. J Antibiot (Tokyo) 53(10):1145–1153. https://doi.org/10.7164/antibiotics.53.1145
Chen R, Zhang Q, Tan B, Zheng L, Li H, Zhu Y (2017) Genome mining and activation of a silent PKS/NRPS gene cluster direct the production of totopotensamides. Org Lett 19(20):5697–5700
Cobb RE, Wang Y, Zhao H (2015) High-Efficiency Multiplex Genome Editing of Streptomyces Species Using an Engineered CRISPR/Cas System. ACS Synth Biol 4(6):723–728. https://doi.org/10.1021/sb500351f
Corre C, Song L, O’Rourke S, Chater KF, Challis GL (2008) 2-Alkyl-4-hydroxymethylfuran-3-carboxylic acids, antibiotic production inducers discovered by Streptomyces coelicolor genome mining. Proc Natl Acad Sci U S A 105(45):17510–17515. https://doi.org/10.1073/pnas.0805530105
Covington BC, Spraggins JM, Ynigez-Gutierrez AE, Hylton ZB, Bachmann BO, Parales RE (2018) Response of secondary metabolism of hypogean actinobacterial genera to chemical and biological stimuli. Environmental microbiology. Crossm. aem.asm.org 1. Appl Environ Microbiol Downloaded 84:1125–1143. https://doi.org/10.1128/AEM
Covington BC, Xu F, Seyedsayamdost MR (2021) A natural product chemist’s guide to Unlocking Silent Biosynthetic Gene clusters. Annu Rev Biochem 2021 June 20:90: 763–788. https://doi.org/10.1146/annurev-biochem-081420-102432
Cui H, Ni X, Shao W, Su J, Su J, Ren J, Xia H (2015) Functional manipulations of the tetramycin positive regulatory gene ttmRIV to enhance the production of tetramycin A and nystatin A1 in Streptomyces Ahygroscopicus. J Ind Microbiol Biotechnol 42(9):1273–1282. https://doi.org/10.1007/s10295-015-1660-3
Culp EJ, Yim G, Waglechner N, Wang W, Pawlowski AC, Wright GD (2019) Hidden antibiotics in actinomycetes can be identified by inactivation of gene clusters for common antibiotics. Nat Biotechnol 37(10):1149–1154. https://doi.org/10.1038/s41587-019-0241-9
Derewacz DK, Covington BC, McLean JA, Bachmann BO (2015) Mapping Microbial Response metabolomes for Induced Natural Product Discovery. ACS Chem Biol 10(9):1998–2006. https://doi.org/10.1021/acschembio.5b00001
Devine R, McDonald HP, Qin Z, Arnold CJ, Noble K, Chandra G, Wilkinson B, Hutchings MI (2021) Re-wiring the regulation of the formicamycin biosynthetic gene cluster to enable the development of promising antibacterial compounds. Cell Chem Biol 28(4):515–523e5. https://doi.org/10.1016/j.chembiol.2020.12.011
Gamaleldin NM, Bakeer W, Sayed AM, Shamikh YI, El-Gendy AO, Hassan HM, Horn H, Abdelmohsen UR, Hozzein WN (2020) Exploration of chemical diversity and antitrypanosomal activity of some red sea-derived actinomycetes using the OSMAC approach supported by LC-MS-based metabolomics and molecular modelling. Antibiotics 9(9):1–16. https://doi.org/10.3390/antibiotics9090629
Gao C, Hindra, Mulder D, Yin C, Elliot MA (2012) Crp is a global regulator of antibiotic production in Streptomyces. MBio. https://doi.org/10.1128/mBio.00407-12
Gehrke EJ, Zhang X, Pimentel-Elardo SM, Johnson AR, Rees CA, Jones SE, Hindra, Gehrke SS, Turvey S, Boursalie S, Hill JE, Carlson EE, Nodwell JR, Elliot MA (2019) Silencing cryptic specialized metabolism in Streptomyces by the nucleoid-associated protein Lsr2. Elife 8:1–28. https://doi.org/10.7554/eLife.47691.001
Gomez-Escribano JP, Bibb MJ (2011) Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb Biotechnol 4(2):207–215. https://doi.org/10.1111/j.1751-7915.2010.00219.x
Gomez-Escribano JP, Bibb MJ (2014) Heterologous expression of natural product biosynthetic gene clusters in Streptomyces coelicolor: from genome mining to manipulation of biosynthetic pathways. J Ind Microbiol Biotechnol 41(2):425–431. https://doi.org/10.1007/s10295-013-1348-5
Gomez-Escribano JP, Castro JF, Razmilic V, Jarmusch SA, Saalbach G, Ebel R, Jaspars M, Andrews B, Asenjo JA, Bibb MJ (2019) Heterologous expression of a cryptic gene cluster from Streptomyces Leeuwenhoekii C34T yields a novel Lasso peptide, Leepeptin. Appl Environ Microbiol 85(23):e01752–e01719. https://doi.org/10.1128/AEM.01752-19
Gorniaková D, Petříček M, Kahoun D, Grabic R, Zelenka T, Chroňáková A, Petříčková K (2021) Activation of a cryptic manumycin-type biosynthetic gene cluster of saccharothrix espanaensis dsm44229 by series of genetic manipulations. Microorganisms 9(3):1–15. https://doi.org/10.3390/microorganisms9030559
Gou L, Han T, Wang X, Ge J, Liu W, Hu F, Wang Z (2017) A novel TetR family transcriptional regulator, CalR3, negatively controls calcimycin biosynthesis in Streptomyces chartreusis NRRL 3882. Front Microbiol 8(NOV):1–10. https://doi.org/10.3389/fmicb.2017.02371
Gren T, Whitford CM, Mohite OS, Jørgensen TS, Kontou EE, Nielsen JB, Lee SY, Weber T (2021) Characterization and engineering of Streptomyces griseofuscus DSM 40191 as a potential host for heterologous expression of biosynthetic gene clusters. Sci Rep 11(1):1–14. https://doi.org/10.1038/s41598-021-97571-2
Guo F, Xiang S, Li L, Wang B, Rajasärkkä J, Gröndahl-Yli-Hannuksela K, Ai G, Metsä-Ketelä M, Yang K (2015) Targeted activation of silent natural product biosynthesis pathways by reporter-guided mutant selection. Metab Eng 28:134–142. https://doi.org/10.1016/j.ymben.2014.12.006
Guo J, Zhang X, Lu X, Liu W, Chen Z, Li J, Deng L, Wen Y (2018) SAV4189, a MarR-family regulator in Streptomyces avermitilis, activates avermectin biosynthesis. Front Microbiol 9(JUN):1–15. https://doi.org/10.3389/fmicb.2018.01358
Gustavson DE, Miyake A, Kim S-H, Lu W, Ahmadi MK, Montiel D, Ternei MA, Brady SF (2018) Atolypenes, tricyclic bacterial sesterterpenes discovered using a multiplexed in vitro Cas9-TAR gene cluster refactoring approach. ACS Synth Biol 8(1):109–118. https://doi.org/10.1021/acssynbio.8b00361.Atolypenes
Hao T, Xie Z, Wang M, Liu L, Zhang Y, Wang W, Zhang Z, Zhao X, Li P, Guo Z, Gao S, Lou C, Zhang G, Merritt J, Horsman GP, Chen Y (2019) An anaerobic bacterium host system for heterologous expression of natural product biosynthetic gene clusters. Nat Commun 10(1):1–13. https://doi.org/10.1038/s41467-019-11673-0
Hashimoto T, Hashimoto J, Kozone I, Amagai K, Kawahara T, Takahashi S, Ikeda H, Shin-Ya K (2018) Biosynthesis of Quinolidomicin, the largest known Macrolide of Terrestrial Origin: identification and heterologous expression of a Biosynthetic Gene Cluster over 200 kb. Org Lett 20(24):7996–7999. https://doi.org/10.1021/acs.orglett.8b03570
Hayes A, Hobbs G, Smith CP, Oliver SG, Butler PR (1997) Environmental signals triggering methylenomycin production by Streptomyces coelicolor A3(2). J Bacteriol 179(17):5511–5515. https://doi.org/10.1128/jb.179.17.5511-5515.1997
Hiard S, Marée R, Colson S, Hoskisson PA, Titgemeyer F, van Wezel GP, Joris B, Wehenkel L, Sébastien R (2007) PREDetector: a new tool to identify regulatory elements in bacterial genomes. Biochem Biophys Res Commun 357(4):861–864. https://doi.org/10.1016/j.bbrc.2007.03.180
Higo A, Hara H, Horinouchi S, Ohnishi Y (2012) Genome-wide distribution of AdpA, a global regulator for secondary metabolism and morphological differentiation in Streptomyces, revealed the extent and complexity of the AdpA regulatory network. DNA Res 19(3):259–273. https://doi.org/10.1093/dnares/dss010
Hoshino S, Okada M, Wakimoto T, Zhang H, Hayashi F, Onaka H, Abe I (2015a) Niizalactams A-C, Multicyclic Macrolactams isolated from Combined Culture of Streptomyces with Mycolic Acid-Containing Bacterium. J Nat Prod 78(12):3011–3017. https://doi.org/10.1021/acs.jnatprod.5b00804
Hoshino S, Wakimoto T, Onaka H, Abe I (2015b) Chojalactones A-C, cytotoxic butanolides isolated from streptomyces sp. cultivated with mycolic acid containing bacterium. Org Lett 17(6):1501–1504. https://doi.org/10.1021/acs.orglett.5b00385
Hoshino S, Zhang L, Awakawa T, Wakimoto T, Onaka H, Abe I (2015c) Arcyriaflavin E, a new cytotoxic indolocarbazole alkaloid isolated by combined-culture of mycolic acid-containing bacteria and Streptomyces cinnamoneus NBRC 13823. J Antibiot (Tokyo) 68(5):342–344. https://doi.org/10.1038/ja.2014.147
Hoshino S, Okada M, Awakawa T, Asamizu S, Onaka H, Abe I (2017) Mycolic acid containing bacterium stimulates Tandem Cyclization of Polyene Macrolactam in a Lake Sediment Derived Rare Actinomycete. Org Lett 19(18):4992–4995. https://doi.org/10.1021/acs.orglett.7b02508
Hoshino S, Ozeki M, Awakawa T, Morita H, Onaka H, Abe I (2018a) Catenulobactins a and B, heterocyclic peptides from culturing Catenuloplanes sp. with a mycolic acid-containing bacterium. J Nat Prod 81(9):2106–2110. https://doi.org/10.1021/acs.jnatprod.8b00261
Hoshino S, Ozeki M, Wong CP, Zhang H, Hayashi F, Awakawa T, Morita H, Onaka H, Abe I (2018b) Mirilactams C-E, Novel Polycyclic macrolactams isolated from combined-culture of Actinosynnema mirum NBRC 14064 and Mycolic Acid-Containing Bacterium Highlighted Paper selected by editor-in-Chief. Chem Pharm Bull 66(6):660–667
Hoshino S, Wong CP, Ozeki M, Zhang H, Hayashi F, Awakawa T, Asamizu S, Onaka H, Abe I (2018c) Umezawamides, new bioactive polycyclic tetramate macrolactams isolated from a combined-culture of Umezawaea sp. and mycolic acid-containing bacterium. J Antibiot (Tokyo) 71(7):653–657. https://doi.org/10.1038/s41429-018-0040-4
Hoshino S, Onaka H, Abe I (2019) Activation of silent biosynthetic pathways and discovery of novel secondary metabolites in actinomycetes by co-culture with mycolic acid-containing bacteria. J Ind Microbiol Biotechnol 46:363–374
Hoskisson PA, Fernández-Martínez LT (2018) Regulation of specialised metabolites in Actinobacteria – expanding the paradigms. Environ Microbiol Rep 10:231–238
Huang Y, Yang D, Pan G, Tang GL, Shen B (2016) Characterization of LnmO as a pathway-specific Crp/Fnr-type positive regulator for leinamycin biosynthesis in Streptomyces atroolivaceus and its application for titer improvement. Appl Microbiol Biotechnol 100(24):10555–10562. https://doi.org/10.1007/s00253-016-7864-2
Huang X, Ma T, Tian J, Shen L, Zuo H, Hu C, Liao G, Palareti G, Legnani C, Cosmi B, Antonucci E, Erba N, Poli D, Testa S, Tosetto A (2017) wblA, a pleiotropic regulatory gene modulating morphogenesis and daptomycin production in Streptomyces roseosporus. J Appl Microbiol 123(3):669–677. https://doi.org/10.1111/ijlh.12426
Hug JJ, Bader CD, Remškar M, Cirnski K, Müller R (2018) Concepts and methods to access novel antibiotics from actinomycetes. Antibiot Jun 7(2):44. https://doi.org/10.3390/antibiotics7020044
Hwang S, Lee Y, Kim JH, Kim G, Kim H, Kim W, Cho S, Palsson BO, Cho BK (2021) Streptomyces as microbial chassis for heterologous protein expression. Front Bioeng Biotechnol. https://doi.org/10.3389/fbioe.2021.804295
Hwang S, Lee N, Choe D, Lee Y, Kim W, Kim JH, Kim G, Kim H, Ahn NH, Lee BH, Palsson BO, Cho BK (2022) System-Level Analysis of Transcriptional and Translational Regulatory Elements in Streptomyces griseus. Front Bioeng Biotechnol 10(February):1–18. https://doi.org/10.3389/fbioe.2022.844200
Jin YY, Cheng J, Yang SH, Meng L, Palaniyandi SA, Zhao XQ, Suhs JW (2011) S-Adenosyl-l-methionine activates actinorhodin biosynthesis by increasing autophosphorylation of the ser/thr protein kinase afsk in Streptomyces coelicolor A3(2). Biosci Biotechnol Biochem 75(5):910–913. https://doi.org/10.1271/bbb.100873
Jin P, Li S, Zhang Y, Chu L, He H, Dong Z, Xiang W (2020) Mining and fine-tuning sugar uptake system for titer improvement of milbemycins in Streptomyces Bingchenggensis. Synth Syst Biotechnol 5(3):214–221. https://doi.org/10.1016/j.synbio.2020.07.001
Jose PA, Maharshi A, Jha B (2021) Actinobacteria in natural products research: Progress and prospects. Microbiol Res 246:126708. https://doi.org/10.1016/j.micres.2021.126708
Kang Y, Wang Y, Hou B, Wang R, Ye J, Zhu X, Wu H, Zhang H (2019) Adpalin, a pleiotropic transcriptional regulator, is involved in the cascade regulation of lincomycin biosynthesis in Streptomyces lincolnensis. Front Microbiol 10(OCT):1–13. https://doi.org/10.3389/fmicb.2019.02428
Karthik L, Arivarasan VK, Vignesh MR, Anitha P (2022) CRISPR ERA: Current Applications and Future Perspectives on Actinobacteria. Actinobacteria. https://doi.org/10.1007/978-981-16-5835-8_10
Kawai K, Wang G, Okamoto S, Ochi K (2007) The rare earth, scandium, causes antibiotic overproduction in Streptomyces spp. FEMS Microbiol Lett 274(2):311–315. https://doi.org/10.1111/j.1574-6968.2007.00846.x
Kim JH, Lee N, Hwang S, Kim W, Lee Y, Cho S, Palsson BO, Cho BK (2021) Discovery of novel secondary metabolites encoded in Actinomycete genomes through coculture. J Ind Microbiol Biotechnol Jun. https://doi.org/10.1093/jimb/kuaa001
Kronheim S, Solomon E, Ho L, Glossop M, Davidson AR, Maxwell KL (2023) Complete genomes and comparative analyses of Streptomyces phages that influence secondary metabolism and sporulation. Sci Rep 13(1):1–11. https://doi.org/10.1038/s41598-023-36938-z
Kuhl M, Rückert C, Gläser L, Beganovic S, Luzhetskyy A, Kalinowski J, Wittmann C (2021) Microparticles enhance the formation of seven major classes of natural products in native and metabolically engineered Actinobacteria through accelerated morphological development. Biotechnol Bioeng 118(8):3076–3093. https://doi.org/10.1002/bit.27818
Kurosawa K, MacEachran DP, Sinskey AJ (2010) Antibiotic biosynthesis following horizontal gene transfer: New milestone for novel natural product discovery? Expert Opin Drug Discov 5(9):819–825. https://doi.org/10.1517/17460441.2010.505599
Kwon MJ, Steiniger C, Cairns TC, Wisecaver JH, Lind AL, Pohl C, Regner C, Rokas A, Meyer V (2021) Beyond the biosynthetic gene cluster paradigm: genome-wide coexpression networks connect clustered and unclustered transcription factors to secondary metabolic pathways. Microbiol Spectr. https://doi.org/10.1128/spectrum.00898-21
Lee Y, Lee N, Jeong Y, Hwang S, Kim W, Cho S, Palsson BO, Cho BK (2019) The transcription Unit Architecture of Streptomyces lividans TK24. Front Microbiol 10(September):1–13. https://doi.org/10.3389/fmicb.2019.02074
Li C, He H, Wang J, Liu H, Wang H, Zhu Y, Wang X, Zhang Y, Xiang W (2019) Characterization of a LAL-type regulator NemR in nemadectin biosynthesis and its application for increasing nemadectin production in Streptomyces Cyaneogriseus. Sci China Life Sci 62(3):394–405. https://doi.org/10.1007/s11427-018-9442-9
Li X, Guo R, Luan J, Fu J, Zhang Y, Wang H (2023) Improving spinosad production by tuning expressions of the forosamine methyltransferase and the forosaminyl transferase to reduce undesired less active byproducts in the heterologous host Streptomyces albus J1074. Microb Cell Fact 22(1):1–7. https://doi.org/10.1186/s12934-023-02023-3
Lim YH, Wong FT, Yeo WL, Ching KC, Lim YW, Heng E, Chen S, Tsai DJ, Lauderdale TL, Shia KS, Ho YS, Hoon S, Ang EL, Zhang MM, Zhao H (2018) Auroramycin: a potent antibiotic from Streptomyces roseosporus by CRISPR-Cas9 activation. ChemBioChem 19(16):1716–1719. https://doi.org/10.1002/cbic.201800266
Lin J, Zhou D, Steitz TA, Polikanov YS, Gagnon MG (2018) Ribosome-targeting antibiotics: modes of Action, mechanisms of Resistance, and implications for Drug Design. Annu Rev Biochem 87:451–478. https://doi.org/10.1146/annurev-biochem-062917-011942
Lin CY, Pang AP, Zhang Y, Qiao J, Zhao GR (2020) Comparative transcriptomic analysis reveals the significant pleiotropic regulatory effects of LmbU on lincomycin biosynthesis. Microb Cell Fact 19(1):1–16. https://doi.org/10.1186/s12934-020-01298-0
Liu T, Mazmouz R, Neilan BA (2018) An in Vitro and in vivo study of Broad-Range Phosphopantetheinyl transferases for Heterologous expression of Cyanobacterial Natural products. ACS Synth Biol 7(4):1143–1151. https://doi.org/10.1021/acssynbio.8b00091
Liu X, Tang J, Wang L, Giesy JP (2019a) Al2O3 nanoparticles promote secretion of antibiotics in Streptomyces coelicolor by regulating gene expression through the nano effect. Chemosphere 226:687–695. https://doi.org/10.1016/j.chemosphere.2019.03.156
Liu X, Tang J, Wang L, Liu Q, Liu R (2019b) A comparative analysis of ball-milled biochar, graphene oxide, and multi-walled carbon nanotubes with respect to toxicity induction in Streptomyces. J Environ Manage 243(January):308–317. https://doi.org/10.1016/j.jenvman.2019.05.034
Liu X, Tang J, Wang L, Liu R (2019c) Mechanism of CuO nano-particles on stimulating production of actinorhodin in Streptomyces coelicolor by transcriptional analysis. Sci Rep 9(1):1–11. https://doi.org/10.1038/s41598-019-46833-1
Lotz TS, Suess B (2018) Small-Molecule-Binding Riboswitches. Microbiol Spectr 6(4):1–12. https://doi.org/10.1128/microbiolspec.rwr-0025-2018
Luthe T, Kever L, Hänsch S, Hardy A, Tschowri N, Weidtkamp-Peters S, Frunzke J (2023) Streptomyces development is involved in the efficient containment of viral Infections. microLife 4(January):1–13. https://doi.org/10.1093/femsml/uqad002
Ma D, Wang C, Chen H, Wen J (2018) Manipulating the expression of SARP family regulator BulZ and its target gene product to increase tacrolimus production. Appl Microbiol Biotechnol 102(11):4887–4900. https://doi.org/10.1007/s00253-018-8979-4
Ma B, Lu C, Wang Y, Yu J, Zhao K, Xue R, Ren H, Lv X, Pan R, Zhang J, Zhu Y, Xu J (2023) A genomic catalogue of soil microbiomes boosts mining of biodiversity and genetic resources. Nat Commun. https://doi.org/10.1038/s41467-023-43000-z
Mao D, Okada BK, Wu Y, Xu F, Seyedsayamdost MR (2018) Recent advances in activating silent biosynthetic gene clusters in bacteria. Curr Opin Microbiol Oct 45:156–163. https://doi.org/10.1016/j.mib.2018.05.001
Martín JF, Liras P (2020) The Balance Metabolism Safety Net: integration of stress signals by interacting transcriptional factors in Streptomyces and related Actinobacteria. Front Microbiol 10:3120. https://doi.org/10.3389/fmicb.2019.03120
Martín JF, Rodríguez-García A, Liras P (2017) The master regulator PhoP coordinates phosphate and nitrogen metabolism, respiration, cell differentiation and antibiotic biosynthesis: comparison in Streptomyces coelicolor and Streptomyces avermitilis. J Antibiot (Tokyo) 70(5):534–541. https://doi.org/10.1038/ja.2017.19
McKenzie NL, Nodwell JR (2007) Phosphorylated AbsA2 negatively regulates antibiotic production in Streptomyces coelicolor through interactions with pathway-specific regulatory gene promoters. J Bacteriol 189(14):5284–5292. https://doi.org/10.1128/JB.00305-07
McLean TC, Wilkinson B, Hutchings MI, Devine R (2019) Dissolution of the disparate: co-ordinate regulation in antibiotic biosynthesis. Antibiot Jun 8(2):83. https://doi.org/10.3390/antibiotics8020083
Meng J, Feng R, Zheng G, Ge M, Mast Y, Wohlleben W, Gao J, Jiang W, Lu Y (2017) Improvement of pristinamycin I (PI) production in Streptomyces pristinaespiralis by metabolic engineering approaches. Synth Syst Biotechnol 2(2):130–136. https://doi.org/10.1016/j.synbio.2017.06.001
Mingyar E, Mühling L, Kulik A, Winkler A, Wibberg D, Kalinowski J, Blin K, Weber T, Wohlleben W, Stegmann E (2021) A regulator based semi-targeted approach to activate silent biosynthetic gene clusters. Int J Mol Sci. https://doi.org/10.3390/ijms22147567
Mitova MI, Lang G, Wiese J, Imhoff JF (2008) Subinhibitory concentrations of antibiotics induce phenazine production in a marine Streptomyces sp. J Nat Prod 71(5):824–827. https://doi.org/10.1021/np800032a
Mohammadipanah F, Kermani F, Salimi F (2020) Awakening the secondary metabolite pathways of Promicromonospora Kermanensis using Physicochemical and Biological Elicitors. Appl Biochem Biotechnol 192(4):1224–1237. https://doi.org/10.1007/s12010-020-03361-3
Moon K, Xu F, Zhang C, Seyedsayamdost MR (2019) Bioactivity-HiTES unveils cryptic antibiotics encoded in ctinomycete bacteria. ACS Chem Biol 14(4):767–774
Musiol-Kroll EM, Tocchetti A, Sosio M, Stegmann E (2019) Challenges and advances in genetic manipulation of filamentous actinomycetes-the remarkable producers of specialized metabolites. Nat Prod Rep 36(9):1351–1369. https://doi.org/10.1039/c9np00029a
Myronovskyi M, Luzhetskyy A (2016) Natural product reports product discovery †. Nat Prod Rep 00:1–14. https://doi.org/10.1039/C6NP00002A
Myronovskyi M, Luzhetskyy A (2019) Heterologous production of small molecules in the optimized: Streptomyces hosts. Nat Prod Rep 36(9):1281–1294. https://doi.org/10.1039/c9np00023b
Niu G, Chater KF, Tian Y, Zhang J, Tan H (2016) Specialised metabolites regulating antibiotic biosynthesis in Streptomyces spp. FEMS Microbiol Rev 40(4):554–573. https://doi.org/10.1093/femsre/fuw012
Ochi K, Hosaka T (2013) New strategies for drug discovery: activation of silent or weakly expressed microbial gene clusters. Appl Microbiol Biotechnol 97:87–98
Ochi K, Tanaka Y, Tojo S (2014) Activating the expression of bacterial cryptic genes by rpoB mutations in RNA polymerase or by rare earth elements. J Ind Microbiol Biotechnol 41(2):403–414. https://doi.org/10.1007/s10295-013-1349-4
Ogura Y, Parsons WH, Kamat SS, Cravatt BF, Mao D, Okada BK, Wu Y, Xu F, Seyedsayamdost MR (2018) Recent advances in activating silent biosynthetic gene clusters in bacteria. Curr Opin Microbiol 45(10):156. https://doi.org/10.1016/j.mib.2018.05.001.Recent
Okada BK, Seyedsayamdost MR (2017) Antibiotic dialogues: induction of silent biosynthetic gene clusters by exogenous small molecules. FEMS Microbiol Rev 41(1):19–33. https://doi.org/10.1093/femsre/fuw035
Olano C, García I, González A, Rodriguez M, Rozas D, Rubio J, Sánchez-Hidalgo M, Braña AF, Méndez C, Salas JA (2014) Activation and identification of five clusters for secondary metabolites in Streptomyces albus J1074. Microb Biotechnol 7(3):242–256. https://doi.org/10.1111/1751-7915.12116
Onaka H (2017) Novel antibiotic screening methods to awaken silent or cryptic secondary metabolic pathways in Actinomycetes. J Antibiot (Tokyo) 70(8):865–870. https://doi.org/10.1038/ja.2017.51
Onaka H, Tabata H, Igarashi Y, Saroa Y, Furumai T (2001) Goadsporin, a chemical substance which promotes secondary metabolism and Morphogenesis in Streptomycetes I. Purification and Characterization. Antibiot 54(12):1036–1044. https://doi.org/10.7164/antibiotics.54.1036
Onaka H, Mori Y, Igarashi Y, Furumai T (2011) Mycolic acid-containing bacteria induce natural-product biosynthesis in Streptomyces species. Appl Environ Microbiol 77(2):400–406. https://doi.org/10.1128/AEM.01337-10
Palaniappan K, Chen IMA, Chu K, Ratner A, Seshadri R, Kyrpides NC, Ivanova NN, Mouncey NJ (2020) IMG-ABC v.5.0: an update to the IMG/Atlas of biosynthetic gene clusters knowledgebase. Nucleic Acids Res 48(D1):D422–D430. https://doi.org/10.1093/nar/gkz932
Palomo S, González I, De La Cruz M, Martín J, Tormo JR, Anderson M, Hill RT, Vicente F, Reyes F, Genilloud O (2013) Sponge-derived Kocuria and Micrococcus spp. as sources of the new thiazolyl peptide antibiotic kocurin. Mar Drugs 11(4):1071–1086. https://doi.org/10.3390/md11041071
Park HB, Park JS, Lee S, Il, Shin B, Oh DC, Kwon HC (2017) Gordonic Acid, a polyketide glycoside derived from bacterial coculture of Streptomyces and Gordonia Species. J Nat Prod 80(9):2542–2546. https://doi.org/10.1021/acs.jnatprod.7b00293
Peng XY, Wu JT, Shao CL, Li ZY, Chen M, Wang CY (2021) Co-culture: stimulate the metabolic potential and explore the molecular diversity of natural products from microorganisms. Mar Life Sci Technol 3:363–374
Pettit RK (2011) Small-molecule elicitation of microbial secondary metabolites. Microb Biotechnol 4:471–478. https://doi.org/10.1111/j.1751-7915.2010.00196.x
Rigali S, Titgemeyer F, Barends S, Mulder S, Thomae AW, Hopwood DA, van Wezel GP (2008) Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep 9(7):670–675. https://doi.org/10.1038/embor.2008.83
Rigali S, Anderssen S, Naômé A, van Wezel GP (2018) Cracking the regulatory code of biosynthetic gene clusters as a strategy for natural product discovery. Biochem Pharmacol 153:24–34. https://doi.org/10.1016/j.bcp.2018.01.007
Rodríguez H, Rico S, Díaz M, Santamaría RI (2013) Two-component systems in Streptomyces: key regulators of antibiotic complex pathways. Microbial Cell Factories 12:127
Rodríguez-García A, Barreiro C, Santos-Beneit F, Sola-Landa A, Martín JF (2007) Genome-wide transcriptomic and proteomic analysis of the primary response to phosphate limitation in Streptomyces coelicolor M145 and in a ∆phoP mutant. Proteomics 7(14):2410–2429. https://doi.org/10.1002/pmic.200600883
Romano S, Jackson SA, Patry S, Dobson ADW (2018) Extending the one strain many compounds (OSMAC) principle to marine microorganisms. Mar Drugs 16(7):1–29. https://doi.org/10.3390/md16070244
Rule D, Cheeptham N (2013) The effects of UV light on the antimicrobial activities of cave actinomycetes. Int J Speleol 42(2):147–153. https://doi.org/10.5038/1827-806X.42.2.7
Salwan R, Sharma V (2020) Molecular and biotechnological aspects of secondary metabolites in Actinobacteria. Microbiol Res 231:126374. https://doi.org/10.1016/j.micres.2019.126374
Scherlach K, Hertweck C (2021) Mining and unearthing hidden biosynthetic potential. Nat Commun 12:3864. https://doi.org/10.1038/s41467-021-24133-5
Sekurova ON, Zhang J, Kristiansen KA, Zotchev SB (2016) Activation of chloramphenicol biosynthesis in Streptomyces venezuelae ATCC 10712 by ethanol shock: insights from the promoter fusion studies. Microb Cell Fact 15(1):1–10. https://doi.org/10.1186/s12934-016-0484-9
Seshadri R, Roux S, Huber KJ, Wu D, Yu S, Udwary D, Call L, Nayfach S, Hahnke RL, Pukall R, White JR, Varghese NJ, Webb C, Palaniappan K, Reimer LC, Sardà J, Bertsch J, Mukherjee S, Reddy TBK, Hajek PP, Huntemann M, Chen I-MA, Spunde A, Clum A, Shapiro N, Wu Z-Y, Zhao Z, Zhou Y, Evtushenko L, Thijs S, Stevens V, Eloe-Fadrosh EA, Mouncey NJ, Yoshikuni Y, Whitman WB, Klenk H-P, Woyke T, Göker M, Kyrpides NC, Ivanova NN (2022) Expanding the genomic encyclopedia of Actinobacteria with 824 isolate reference genomes. Cell Genomics:100213. https://doi.org/10.1016/j.xgen.2022.100213
Shantanam S, Ren MUELLER, Biswas H, Ho S, Van Der Donk S, Zhao WA H (2018) Rapid discovery of glycocins through pathway refactoring in Escherichia coli. ACS Chem Biol 13(10):2966–2972. https://doi.org/10.1021/acschembio.8b00599.Rapid
Shima J, Hesketh A, Okamoto S, Kawamoto S, Ochi K (1996) Induction of actinorhodin production by rpsL (encoding ribosomal protein S12) mutations that confer streptomycin resistance in Streptomyces lividans and Streptomyces coelicolor A3(2). J Bacteriol 178(24):7276–7284. https://doi.org/10.1128/jb.178.24.7276-7284.1996
Shitut S, Bergman GÖ, Kros A, Rozen DE, Claessen D (2020) Use of permanent wall-deficient cells as a system for the discovery of new-to-nature metabolites. Microorganisms 8(12):1–8. https://doi.org/10.3390/microorganisms8121897
Shu D, Chen L, Wang W, Yu Z, Ren C, Zhang W, Yang S, Lu Y, Jiang W (2009) afsQ1-Q2-sigQ is a pleiotropic but conditionally required signal transduction system for both secondary metabolism and morphological development in Streptomyces coelicolor. Appl Microbiol Biotechnol 81(6):1149–1160. https://doi.org/10.1007/s00253-008-1738-1
Singh TA, Passari AK, Jajoo A, Bhasin S, Gupta VK, Hashem A, Alqarawi AA, Abd-Allah EF (2021) Tapping into actinobacterial genomes for natural product discovery. Front Microbiol 12:655620. https://doi.org/10.3389/fmicb.2021.655620
Skinnider MA, Merwin NJ, Johnston CW, Magarvey NA (2017) PRISM 3: expanded prediction of natural product chemical structures from microbial genomes. Nucleic Acids Res 45(W1):W49–W54. https://doi.org/10.1093/nar/gkx320
Smanski MJ, Peterson RM, Rajski SR, Shen B (2009) Engineered Streptomyces platensis strains that overproduce antibiotics platensimycin and platencin. Antimicrob Agents Chemother 53(4):1299–1304. https://doi.org/10.1128/AAC.01358-08
Stevens DC, Conway KR, Pearce N, Villegas-Peñaranda LR, Garza AG, Boddy CN (2013) Alternative sigma factor over-expression enables heterologous expression of a Type II polyketide biosynthetic pathway in Escherichia coli. PLoS ONE. https://doi.org/10.1371/journal.pone.0064858
Sugiyama R, Nishimura S, Ozaki T, Asamizu S, Onaka H, Kakeya H (2015) 5-Alkyl-1,2,3,4-tetrahydroquinolines, new membrane-interacting lipophilic metabolites produced by combined culture of Streptomyces nigrescens and Tsukamurella pulmonis. Org Lett 17(8):1918–1921. https://doi.org/10.1021/acs.orglett.5b00607
Sugiyama R, Nishimura S, Ozaki T, Asamizu S, Onaka H, Kakeya H (2016) Discovery and Total Synthesis of Streptoaminals: Antimicrobial [5,5]-Spirohemiaminals from the Combined-Culture of Streptomyces nigrescens and Tsukamurella pulmonis. Angew Chemie 55(35):10278–10282
Sun D, Wang Q, Chen Z, Li J, Wen Y (2017a) An alternative σ factor, σ8, Controls avermectin production and multiple stress responses in Streptomyces avermitilis. Front Microbiol 8(APR):1–16. https://doi.org/10.3389/fmicb.2017.00736
Sun YQ, Busche T, Rückert C, Paulus C, Rebets Y, Novakova R, Kalinowski J, Luzhetskyy A, Kormanec J, Sekurova ON, Zotchev SB (2017b) Development of a Biosensor Concept to detect the production of cluster-specific secondary metabolites. ACS Synth Biol 6(6):1026–1033. https://doi.org/10.1021/acssynbio.6b00353
Sun H, Yang J, Song H (2020) Engineering Mycobacteria artificial promoters and ribosomal binding sites for enhanced sterol production. Biochem Eng J 162:107739. https://doi.org/10.1016/j.bej.2020.107739
Sung AA, Gromek SM, Balunas MJ (2017) Upregulation and identification of antibiotic activity of a marine-derived Streptomyces sp. via co-cultures with human pathogens. Mar Drugs. https://doi.org/10.3390/md15080250
Takano E (2006) γ-Butyrolactones: Streptomyces signalling molecules regulating antibiotic production and differentiation. Curr Opin Microbiol 9(3):287–294. https://doi.org/10.1016/j.mib.2006.04.003
Tanaka Y, Hosaka T, Ochi K (2010) Rare earth elements activate the secondary metabolite-biosynthetic gene clusters in Streptomyces coelicolor A3(2). J Antibiot (Tokyo) 63(8):477–481. https://doi.org/10.1038/ja.2010.53
Terlouw BR, Blin K, Navarro-Muñoz JC, Avalon NE, Chevrette MG, Egbert S, Lee S, Meijer D, Recchia MJJ, Reitz ZL, van Santen JA, Selem-Mojica N, Tørring T, Zaroubi L, Alanjary M, Aleti G, Aguilar C, Al-Salihi SAA, Augustijn HE, Avelar-Rivas JA, Avitia-Domínguez LA, Barona-Gómez F, Bernaldo-Agüero J, Bielinski VA, Biermann F, Booth TJ, Carrion Bravo VJ, Castelo-Branco R, Chagas FO, Cruz-Morales P, Du C, Duncan KR, Gavriilidou A, Gayrard D, Gutiérrez-García K, Haslinger K, Helfrich EJN, van der Hooft JJJ, Jati AP, Kalkreuter E, Kalyvas N, Kang K, Bin, Kautsar S, Kim W, Kunjapur AM, Li Y, Lin G, Loureiro C, Louwen JJR, Louwen NLL, Lund G, Parra J, Philmus B, Pourmohsenin B, Pronk LJU, Rego A, Rex DAB, Robinson S, Rosas-Becerra LR, Roxborough ET, Schorn MA, Scobie DJ, Singh KS, Sokolova N, Tang X, Udwary D, Vigneshwari A, Vind K, Vromans SPJM, Waschulin V, Williams SE, Winter JM, Witte TE, Xie H, Yang D, Yu J, Zdouc M, Zhong Z, Collemare J, Linington RG, Weber T, Medema MH (2022) MIBiG 3.0: a community-driven effort to annotate experimentally validated biosynthetic gene clusters. Nucleic Acids Res. https://doi.org/10.1093/nar/gkac1049
Timmermans ML, Picott KJ, Ucciferri L, Ross AC (2019) Culturing marine bacteria from the genus Pseudoalteromonas on a cotton scaffold alters secondary metabolite production. Microbiologyopen 8(5):1–10. https://doi.org/10.1002/mbo3.724
Tomm HA, Ucciferri L, Ross AC (2019) Advances in microbial culturing conditions to activate silent biosynthetic gene clusters for novel metabolite production. J Ind Microbiol Biotechnol 46(9–10):1381–1400. https://doi.org/10.1007/s10295-019-02198-y
Tong Y, Robertsen HL, Blin K, Klitgaard AK, Weber T, Lee SY (2019aa) CRISPR-BEST: a highly efficient DSB-free base editor for filamentous Actinomycetes. bioRxiv. https://doi.org/10.1101/582403
Tong Y, Whitford CM, Robertsen HL, Blin K, Jørgensen TS, Klitgaard AK, Gren T, Jiang X, Weber T, Lee SY (2019b) Highly efficient DSB-free base editing for Streptomycetes with CRISPR-BEST. Proc Natl Acad Sci U S A 116(41):20366–20375. https://doi.org/10.1073/pnas.1913493116
Tyurin AP, Alferova VA, Korshun VA (2018) Chemical elicitors of antibiotic biosynthesis in Actinomycetes. Microorganisms 6(2):52. https://doi.org/10.3390/microorganisms6020052
Wang G, Hosaka T, Ochi K (2008) Dramatic activation of antibiotic production in Streptomyces coelicolor by cumulative drug resistance mutations. Appl Environ Microbiol 74(9):2834–2840. https://doi.org/10.1128/AEM.02800-07
Wang Y, Tao Z, Zheng H, Zhang F, Long Q, Deng Z, Tao M (2016) Iteratively improving natamycin production in Streptomyces gilvosporeus by a large operon-reporter based strategy. Metab Eng 38(July):418–426. https://doi.org/10.1016/j.ymben.2016.10.005
Wang C, Huang D, Liang S (2018) Identification and metabolomic analysis of chemical elicitors for tacrolimus accumulation in Streptomyces tsukubaensis. Appl Microbiol Biotechnol 102(17):7541–7553. https://doi.org/10.1007/s00253-018-9177-0
Wang B, Guo F, Dong SH, Zhao H (2019a) Activation of silent biosynthetic gene clusters using transcription factor decoys. Nat Chem Biol 15(2):111–114. https://doi.org/10.1038/s41589-018-0187-0
Wang C, Wang J, Yuan J, Jiang L, Jiang X, Yang B, Zhao G, Liu B, Huang D (2019b) Generation of Streptomyces hygroscopicus cell factories with enhanced ascomycin production by combined elicitation and pathway-engineering strategies. Biotechnol Bioeng 116(12):3382–3395. https://doi.org/10.1002/bit.27158
Wei K, Wu Y, Li L, Jiang W, Hu J, Lu Y, Chen S (2018) MilR2, a novel TetR family regulator involved in 5-oxomilbemycin A3/A4 biosynthesis in Streptomyces hygroscopicus. Appl Microbiol Biotechnol 102(20):8841–8853. https://doi.org/10.1007/s00253-018-9280-2
Wohlleben W, Bera A, Mast Y, Stegmann E (2017) Regulation of secondary metabolites of Actinobacteria. Biology and Biotechnology of Actinobacteria. Springer, Heidelberg, pp 181–232
Wu H, Liu W, Shi L, Si K, Liu T, Dong D, Zhang T, Zhao J, Liu D, Tian Z, Yue Y, Zhang H, Xuelian B, Liang Y (2017) Comparative genomic and Regulatory analyses of Natamycin production of Streptomyces lydicus A02. Sci Rep 7(1):1–12. https://doi.org/10.1038/s41598-017-09532-3
Wu Q, Bin, Chen XA, Lv ZY, Zhang XY, Liu Y, Li YQ (2021) Activation and discovery of tsukubarubicin from Streptomyces tsukubaensis through overexpressing SARPs. Appl Microbiol Biotechnol 105(11):4731–4741. https://doi.org/10.1007/s00253-021-11344-5
Xia H, Li X, Li Z, Zhan X, Mao X, Li Y (2020) The Application of Regulatory Cascades in Streptomyces: Yield Enhancement and Metabolite Mining. Front Microbiol 11:1–14. https://doi.org/10.3389/fmicb.2020.00406
Xiang SH, Li J, Yin H, Zheng JT, Yang X, Wang H, Bin, Luo JL, Bai H, Yang KQ (2009) Application of a double-reporter-guided mutant selection method to improve clavulanic acid production in Streptomyces clavuligerus. Metab Eng 11(4–5):310–318. https://doi.org/10.1016/j.ymben.2009.06.003
Xu F, Nazari B, Moon K, Bushin LB, Seyedsayamdost MR (2017a) Discovery of a cryptic antifungal compound from Streptomyces albus J1074 using high-throughput Elicitor Screens. J Am Chem Soc 139(27):9203–9212. https://doi.org/10.1021/jacs.7b02716
Xu X, Wang J, Bechthold A, Ma Z, Yu X (2017b) Selection of an efficient promoter and its application in toyocamycin production improvement in Streptomyces diastatochromogenes 1628. World J Microbiol Biotechnol 33(2):0. https://doi.org/10.1007/s11274-016-2194-1
Xu F, Wu Y, Zhang C, Davis KM, Moon K, Bushin LB, Seyedsayamdost MR (2019) A genetics-free method for high-throughput discovery of cryptic microbial metabolites. Nat Chem Biol 15(2):161–168. https://doi.org/10.1038/s41589-018-0193-2
Yan X, Zhang B, Tian W, Dai Q, Zheng X, Hu K, Liu X, Deng Z, Qu X (2018) Puromycin A, B and C, cryptic nucleosides identified from Streptomyces alboniger NRRL B-1832 by PPtase-based activation. Synth Syst Biotechnol 3(1):76–80. https://doi.org/10.1016/j.synbio.2018.02.001
Yan YS, Yang YQ, Zhou LS, Zhang L, Xia HY (2022) MilR3, a unique SARP family pleiotropic regulator in Streptomyces Bingchenggensis. Arch Microbiol 204(10):1–16. https://doi.org/10.1007/s00203-022-03240-x
Ye S, Enghiad B, Zhao H, Takano E (2020) Fine-tuning the regulation of Cas9 expression levels for efficient CRISPR-Cas9 mediated recombination in Streptomyces. J Ind Microbiol Biotechnol 47(4–5):413–423. https://doi.org/10.1007/s10295-020-02277-5
Yeo WL, Heng E, Tan LL, Lim YW, Lim YH, Hoon S, Zhao Zhang M M, Wong FT (2019) Characterization of Cas proteins for CRISPR-Cas editing in streptomycetes. Biotechnol Bioeng 116(9):2330–2338. https://doi.org/10.1002/bit.27021
Yoo YJ, Hwang Jyeon, Shin Hluyung, Cui H, Lee J, Yoon YJ (2015) Characterization of negative regulatory genes for the biosynthesis of rapamycin in Streptomyces rapamycinicus and its application for improved production. J Ind Microbiol Biotechnol 42(1):125–135. https://doi.org/10.1007/s10295-014-1546-9
Yoon V, Nodwell JR (2014) Activating secondary metabolism with stress and chemicals. J Ind Microbiol Biotechnol 41:415–424. https://doi.org/10.1007/s10295-013-1387-y
Yoshimura A, Covington BC, Gallant É, Zhang C, Li A, Seyedsayamdost MR, Brenowitz AGRB (2020) Unlocking cryptic metabolites with mass spectrometry-guided transposon mutant selection. ACS Chem Biol 15(10):2766–2774. https://doi.org/10.1021/acschembio.0c00558.Unlocking
Yu M, Li Y, Banakar SP, Liu L, Shao C, Li Z, Wang C (2019) New metabolites from the co-culture of marine-derived Actinomycete Streptomyces rochei MB037 and fungus rhinocladiella similis 35. Front Microbiol 10(MAY):1–11. https://doi.org/10.3389/fmicb.2019.00915
Yushchuk O, Ostash I, Mösker E, Vlasiuk I, Deneka M, Rückert C, Busche T, Fedorenko V, Kalinowski J, Süssmuth RD, Ostash B (2021) Eliciting the silent lucensomycin biosynthetic pathway in Streptomyces Cyanogenus S136 via manipulation of the global regulatory gene adpA. Sci Rep 11(1):1–14. https://doi.org/10.1038/s41598-021-82934-6
Zaburannyi N, Rabyk M, Ostash B, Fedorenko V, Luzhetskyy A (2014) Insights into naturally minimised Streptomyces albus J1074 genome. BMC Genom. https://doi.org/10.1186/1471-2164-15-97
Zarins-Tutt JS, Barberi TT, Gao H, Mearns-Spragg A, Zhang L, Newman DJ, Goss RJM (2016) Prospecting for new bacterial metabolites: a glossary of approaches for inducing, activating and upregulating the biosynthesis of bacterial cryptic or silent natural products. Nat Prod Rep 33(1):54–72. https://doi.org/10.1039/c5np00111k
Zhang YY, Zou ZZ, Niu GQ, Tan HR (2013) jadR* and jadR2 act synergistically to repress jadomycin biosynthesis. Sci China Life Sci 56(7):584–590. https://doi.org/10.1007/s11427-013-4508-y
Zhang Y, Huang H, Xu S, Wang B, Ju J, Tan H, Li W (2015) Activation and enhancement of Fredericamycin A production in deepsea-derived Streptomyces somaliensis SCSIO ZH66 by using ribosome engineering and response surface methodology. Microb Cell Fact 14(1):1–11. https://doi.org/10.1186/s12934-015-0244-2
Zhang B, Tian W, Wang S, Yan X, Jia X, Pierens GK, Chen W, Ma H, Deng Z, Qu X (2017a) Activation of Natural products Biosynthetic pathways via a protein modification level regulation. ACS Chem Biol 12(7):1732–1736. https://doi.org/10.1021/acschembio.7b00225
Zhang MM, Wong FT, Wang Y, Luo S, Lim YH, Heng E, Yeo WL, Cobb RE, Enghiad B, Ang EL, Zhao H (2017b) CRISPR-Cas9 strategy for activation of silent Streptomyces biosynthetic gene clusters. Nat Chem Biol 13(6):607–609. https://doi.org/10.1038/nchembio.2341
Zhang X, Andres SN, Elliot MA (2021) Interplay between nucleoid-associated proteins and transcription factors in controlling specialized metabolism in streptomyces. MBio. https://doi.org/10.1128/mBio.01077-21
Zhang J, Hassan HA, Abdelmohsen UR, Zahran EM (2022) A glossary for chemical approaches towards unlocking the trove of metabolic treasures in Actinomycetes. Molecules 27(1):142. https://doi.org/10.3390/molecules27010142
Zhao Q, Luo Y, Zhang X, Kang Q, Zhang D, Zhang L, Bai L, Deng Z (2020) A severe leakage of intermediates to shunt products in acarbose biosynthesis. Nat Commun 11(1):1–15. https://doi.org/10.1038/s41467-020-15234-8
Zheng J, Li Y, Liu N, Zhang J, Liu S, Tan H (2022) Multi-omics Data reveal the Effect of Sodium Butyrate on Gene expression and protein modification in Streptomyces. Genomics Proteom Bioinf. https://doi.org/10.1016/j.gpb.2022.09.002
Zhou WW, Ma B, Tang YJ, Zhong JJ, Zheng X (2012) Enhancement of validamycin A production by addition of ethanol in fermentation of Streptomyces hygroscopicus 5008. Bioresour Technol 114:616–621. https://doi.org/10.1016/j.biortech.2012.03.124
Ziemert N, Podell S, Penn K, Badger JH, Allen E, Jensen PR (2012) The natural product domain seeker NaPDoS: a phylogeny based bioinformatic tool to classify secondary metabolite gene diversity. PLoS ONE 7(3):1–9. https://doi.org/10.1371/journal.pone.0034064
Zong G, Fu J, Zhang P, Zhang W, Xu Y, Cao G, Zhang R (2021) Use of elicitors to enhance or activate the antibiotic production in Streptomyces. Crit. Rev. Biotechnol 42(8):1260–1283. https://doi.org/10.1080/07388551.2021.1987856
Funding
The work was not funded by any funding agency.
Author information
Authors and Affiliations
Contributions
SK and NF data: collection and writing the manuscript draft & FM: Conceptualization and manuscript edition.
Corresponding author
Ethics declarations
Conflict of interest
Authors state no commercial declarations on the content of the paper.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Karimian, S., Farahmandzad, N. & Mohammadipanah, F. Manipulation and epigenetic control of silent biosynthetic pathways in actinobacteria. World J Microbiol Biotechnol 40, 65 (2024). https://doi.org/10.1007/s11274-023-03861-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-023-03861-4