Abstract
The entomopathogenic nematode Heterorhabditis bacteriophora (Nematoda: Rhabditidae) is used in biological insect control. Their dauer juveniles (DJs) are free-living and developmentally arrested, invading host insects. They carry cells of their bacterial symbiont Photorhabdus spp. in the intestine. Once inside the insect´s hemolymph the DJs perceive a food signal, triggering them to exit the DJ stage and regurgitate the Photorhabdus cells into the insect’s haemocoel, which kill the host and later provide essential nutrients for nematode reproduction. The exit from the DJ stage is called “recovery”. For commercial pest control, nematodes are industrially produced in monoxenic liquid cultures. Artificial media are incubated with Photorhabdus before DJs are added. In absence of the insect’s food signal, DJs depend on unknown bacterial food signals to trigger exit of the DJ stage. A synchronized and high DJ recovery determines the success of the industrial in vitro production and can significantly vary between nematode strains, inbred lines and mutants. In this study, fourteen bacterial strains from H. bacteriophora were isolated and identified as P. laumondii, P. kayaii and P. thracensis. Although the influence of bacterial supernatants on the DJ recovery of three inbred lines and two mutants differed significantly, the bacterial impact on recovery has a subordinate role whereas nematode factors have a superior influence. Recovery of inbred lines decreased with age of the DJs. One mutant (M31) had very high recovery in bacterial supernatant and spontaneous recovery in Ringer solution. Another mutant (M88) was recovery defective.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The entomopathogenic nematode (EPN) Heterorhabditis bacteriophora (Nematoda: Rhabditidae) is commercially mass produced in liquid culture industrial scale bioreactors (Ehlers 2001). These safe biocontrol agents (Ehlers 2003) are widely used in pest management to substitute synthetic insecticides, mainly of soil-dwelling insect larvae (Grewal et al. 2005; Lacey et al. 2015). The nematode is symbiotically associated with bacteria of the genus Photorhabdus (Gammaproteobacteria: Morganellaceae) (Adeolu et al. 2016). The third nematode stage, the dauer juvenile (DJ) of EPN is free-living, non-feeding and packed with fat reserves, providing it with energy for a long-term survival in the soil. It is developmentally arrested, carrying cells of its bacterial symbiont in the gut. The DJs search for suitable hosts and once inside the hemolymph of an insect, the DJs perceive a yet unknown food signal, which induces the exit of the dauer stage and the development to self-fertilizing hermaphrodites. This process is called DJ recovery. During recovery the DJs regurgitate cells of their symbiont into the hemocoel of its host. Both partners overcome the insect’s immune response, after 1–2 days the bacteria proliferate and kill the host by septicaemia. The bacterial cells act as nutritional source for the nematodes (Han and Ehlers 2000), which produce offspring. Once the cadaver is consumed, starvation induces the formation of dauers, which leave the host on search for new victims.
The nematode-bacterium interaction is obligatory. The bacteria rely on the nematode for the transmission between insect hosts, and the nematode requires the bacterial symbiont to kill the insect host and to develop and reproduce. Bacteria-free nematodes are non-pathogenic (Han and Ehlers 2000).
Different to the in vivo life cycle, the commercial mass production of EPNs starts with a Photorhabdus pre-culture for minimum 24 h. Afterwards DJs are inoculated (Ehlers 2001). Under these conditions, the DJ recovery is triggered by a food signal emitted by their bacteria (Strauch and Ehlers 1998). However, the recovery in bacterial cultures is lower than inside an insect. Whereas almost 100% of DJs recover within a day inside an insect, the recovery in in vitro culture is highly variable and often scattered over a period of 2–5 days resulting in a non-synchronous population development causing lower yields and occurrence of a second generation of males and females, which are unable to mate in liquid culture, thus not producing offspring (Dolan et al. 2002; Han et al. 2000; Strauch and Ehlers 1998). During industrial production the liquid culture can undergo two scenarios. Either the recovery of inoculated DJs is high and synchronized, leading to high DJ yields at the end of the process, with very few non-desired stages such as males, unfertilized females and small juveniles (optimal process) or recovery is low, resulting in many second-generation adults and low DJ yields (suboptimal process). Thus, the rapid, synchronized and high DJ recovery is the key to a successful production of a high DJ yield of good quality (Johnigk et al. 2004).
For many years DJ recovery was less of a problem as the commercial strain recovered well due to selection for high recovery over many years of continuous subculturing. Since H. bacteriophora was subjected to intensive breeding activities targeting the improvement of beneficial traits, like virulence, longevity and stress-resistance (e.g. Godina et al. 2023; Mukuka et al. 2010; Sumaya et al. 2017, 2018), superior hybrids are now considered for commercialisation. However, their variable DJ recovery presents a major obstacle during industrial production. Therefore Wang et al. (2023) phenotyped the recovery of mutant inbred lines and wild types (WT) of H. bacteriophora by exposing DJs to symbiont culture supernatants of strain P. laumondii (DE2). A large variability of DJ recovery was observed (2% up to 90%) and nematode DNA polymorphisms with potential association to the DJ recovery trait were identified. The results emphasized the relative low DJ recovery in WTs in comparison to the commercial line. Thus, research is crucial to further elucidate factors influencing DJ recovery.
Concerning specificity of H. bacteriophora to different Photorhabdus spp., the nematode appears to be relatively flexible. Until today, ten Photorhabdus species and subspecies have been isolated from H. bacteriophora: P. cinerea (Kazimierczak et al. 2017), P. kayaii, P. thracensis (Hazir et al. 2004), P. laumondii subsp. laumondii and clarkei and P. luminescens subsp. luminescens (Saux et al. 1999), P. caribbeanensis, P. khaini subsp. khaini, P. stackebrandtii (Tailliez et al. 2010) and kleinii (Machado et al. 2018). Whether this flexibility concerning the bacterial association can bring advantages to the nematode-bacteria complex, has already been demonstrated for nematode virulence. For instance, Machado et al. (2020) improved virulence of H. bacteriophora to western corn rootworm (WCR) by increasing benzoxazinoid resistance of Photorhabdus through bacterial symbiont engineering. Concerning bacterial natural products, secondary metabolites have been linked to traits like insect pathogenicity, protection of the insect cadaver and support of nematode development (Shi and Bode 2018). Several conservative biosynthetic gene clusters for natural products have been identified from Photorhabdus spp. (Shi et al. 2022). Concerning DJ recovery in H. bacteriophora, two compounds of less than 20 kDa (non-identified) were reported to influence this recovery (Aumann and Ehlers 2001). Joyce et al. (2008) and Wang et al. (2022) reported that stilbenes produced by Photorhabdus trigger H. bacteriophora DJ recovery, however, the compounds were not sufficient in absence of bacteria cells or supernatant. Thus, the bacterial food signal appears to be of complex composition and individual elements may not fully trigger DJ recovery. To stabilize the industrial production of novel H. bacteriophora strains and lines, it is crucial to determine whether certain WT Photorhabdus strains possess a better potential to support DJ recovery.
Up to date there is no report concerning phenotypic variability related to different Photorhabdus strains causing DJ recovery. In this study, we (i) isolated fourteen Photorhabdus bacterial strains from previously phenotyped nematode WT strains, (ii) we identified the strains to species level based on 16 S rRNA and four housekeeping genes, (iii) we produced sterile bacterial supernatants and tested five previously phenotyped H. bacteriophora lines/strains included in the work of Wang et al. (2023) for DJ recovery upon exposure to supernatants, (iv) we tested whether DJ age (storage in 4 °C) influences the DJ recovery and (v) tested whether DJ recovery can be influenced by potential supernatant volatiles.
Materials and methods
Cultivation of Heterorhabditis bacteriaphora lines
Five H. bacteriaphora lines were used in this study, including two commercial lines (EN01 and HB4), the inbred line IL3 selected from EN01 (Sumaya et al. 2018) and two mutant inbred lines M31 & M88. The mutant lines derived from donor line IL3 and have been phenotyped for their DJ recovery (Wang et al. 2023). DJs of each line were grown in monoxenic liquid cultures established by egg isolation and surface-sterilization (Lunau et al. 1993), and subsequent incubation in Wouts agar medium (16 g Bacto nutrient broth, 12 g Bacto agar, 5 g corn-oil or sunflower-oil, 1 L distilled water). Wouts agar plates were transferred after ~ 14 days into 24 h liquid cultures of P. laumondii (strain DE2) into LM medium (15 g yeast extract, 20 g soy powder, 4 g NaCl, 0.35 g KCl, 0.15 g CaCl2, 0.1 g MgSO4, 6 g lecithin diluted in same volume of rapeseed oil, 40 g rapeseed oil, 1 L distilled water, pH 7 ± 0.1) and cultured on a rotatory shaker (180 rpm, rotation diam. 4 cm) (Ehlers et al. 1998; Hirao and Ehlers 2010). Subcultures were carried out by the same procedure, inoculating DJs at a final density of 4000 DJs mL−1 using cultures of previous generations. To check for monoxenic conditions and excluding contaminated cultures, samples were streaked on NBTA agar (10 g Bacto tryptic soy broth, 0.025 g bromothymolblue sodium salt, 14 g Bacto agar, 1 L distilled water, 4 mL of 1% sterile filtered 2,3,5-triphenyltetrazoliumchloride) (Akhurst 1980). DJs were harvested by pouring cultures on cotton traps and cleaned by vacuum sieved. Ringer’s solution (9 g NaCl, 0.42 g KCl, 0.37 g CaCl2 × 2 H2O, 0.2 g NaHCO3, 1 L distilled water) was used to store DJs at 4 °C.
Isolation of bacteria from WT strains
Symbiotic Photorhabdus bacteria were isolated from different H. bacteriophora strains (Table 1). DJs from each line/strain were used to infect last instar wax moth larvae (Galleria mellonella) at a dose of 50 DJs per insect. After 24 h incubation at 25 °C, the insect larvae were surface sterilized with 70% ethanol for 20 s, and a drop of insect haemolymph was taken by piercing the hind leg of the insect. The protruding drop of hemolymph was streaked on NBTA agar plates, which were then incubated at 25 °C for 48 h. Then single bacterial colonies were selected and re-streaked on NBTA several times until pure colonies were obtained (Ehlers et al. 1991). Subsequently, single colonies were picked from NBTA and grown in LM medium for 24 to 48 h at 25 °C on a rotatory shaker (180 rpm, rotation diam. 4 cm). Thereafter, Photorhabdus culture stocks were preserved in glycerol 15% (v/v) at − 80 °C. Aliquots of the bacterial cultures were used for DNA isolation and subsequent species determination. The Photorhabdus strains in this study are available at e-nema GmbH upon request and have been submitted to Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures GmbH (DSMZ).
Bacterial supernatant production
Bacterial supernatants were produced from each of the isolated bacteria, as well as from P. laumondii strain DE2 as reference. Bacterial cultures were carried out in LM medium (50 mL media in a 250 mL volume flask) for 24 h. Subsequently the cultures were centrifuged at 10,000 rpm at 4 °C for 10 min. Resulting supernatants were sterilized by passing the liquid through 5 μm and then 0.2 μm pore size filters and stored at − 80 °C until used (Wang et al. 2023). The presence of contaminants was tested by inoculation of samples on NBTA plates.
Bacterial species identification
Extraction of DNA was done with the PureLink™ Microbiome DNA purification kit (Thermo Fisher Scientific, MA, USA) following the manufacturer’s instructions. DNA quantity and quality was assessed with a Nanodrop™ Lite Spectrophotometer (Thermo Fisher Scientific, MA, USA) and diluted to 5 ng µL−1 for PCR amplification. The 16 S rRNA and four housekeeping genes, viz. glutamyl tRNA synthetase (gltX), recombinase protein encoding gene (recA), beta subunit of DNA polymerase III holoenzyme encoding gene (dnaN), and DNA gyrase subunit encoding gene (gyrB) were selected for bacterial species identification by sequencing of PCR products. Primers used for the gene amplifications were published by (Sergeant et al. 2006; Tailliez et al. 2010) (Table 2). PCR reactions were carried out in 1× reaction buffer, 400 µM mM dNTPs, 7.5 pmol of each primer (forward and reverse), 25 mM MgCl2, one unit of Taq DNA polymerase and 10 ng of template DNA (final volume 20 µL). PCR conditions were as follow: 94 °C for 2 min, 40 cycles of 94 °C for 20 s followed by 57 °C for 1.5 min and 72 °C for 1 min, and a final extension 72 °C for 3 min in a Gene Touch™ Thermocycler (Bioer, Hangzhou, China). The PCR products were separated by electrophoresis in a 1% TAE-agarose gel stained with GelRed™ staining (Biotuim, USA), gel-purified (QIAquick gel purification Kit, Qiagen) and sequenced (StarSeq GmbH, Mainz, Germany). The sequences were BLASTed against NCBI data (https://www.ncbi.nlm.nih.gov/) to search for the highest homology with other Photorhabdus species accessions. In parallel, the sequences of the four housekeeping genes were aligned, trimmed and concatenated per separate into a single combined sequence (for each of the fourteen bacterial isolates). A large set of sequences from the same genes was retrieved from NCBI and was used as reference concatemers (Supplementary Table S1). All concatenated sequences were analysed by pairwise alignment using MUSCLE (Edgar 2004), followed by post alignment trimming with G-Blocks as implemented in SeaView 5.0 (Gouy et al. 2020). The TPM2uf + I + G & TIM1 + I + G model was selected as the best-fit model of DNA evolution for 16 S & housekeeping gene setups respectively using jModelTest2 (Darriba et al. 2012) according to the Akaike Information Criterion (AIC). Subsequently, phylogenetic trees were obtained using MrBayes 3.2.7a (Ronquist and Huelsenbeck 2003) at platform CIPRES Science Gateway (https://www.phylo.org/) (Miller et al. 2010). The 50% majority rule consensus phylogenetic trees were visualized using FigTree v. 1.4.4 (Rambaut 2018) and Inkscape (Bah 2011).
DJ recovery assay
Recovery of DJs was assessed based on morphological changes of the head region in cell wells under an inverted microscope (Zeiss, Germany) (Strauch and Ehlers 1998). DJs (100–200) of the nematode lines/strains EN01, HB4, IL3, M31 and M88 were inoculated in randomized arrangement into 24-cell-well plates, each cell filled with 200 µL bacterial supernatant containing 0.2% streptomycin sulphate. Controls were carried out in Ringer’s solution. Recovery was evaluated three days after incubation at 25 °C by counting recovered and unrecovered DJs in each cell well. DJ recovery assays were carried out in four independent experiments, each with four replicates per each nematode line/strain and supernatant.
Influence of ageing on DJ recovery
Nematode DJs of the five strains/lines were stored in Ringer’s solution at 4 ℃ for up to 60 days. The DJ recovery was evaluated periodically, and data was adjusted to a regression model. Experiments were repeated three times (in different batches of DJs), each with five evaluation time points and four replications for each time point.
Influence of volatiles from supernatants on DJ recovery
To test whether DJ recovery can be also induced by volatile substances released by the bacterial supernatant of P. laumondii (strain DE2), three different conditions were tested: (i) DJs in bacterial supernatant (SN), (ii) DJs in Ringer’s solution in neighbouring cell wells with bacterial supernatant (RN), (iii) DJs in Ringer’s not neighbouring supernatant cells (RS). The experimental setup is depicted in Fig. 1. DJs were stored less than a week before inoculation. From each of the variants (SN, RN, RS) four replicates were evaluated and four independent experiments were conducted.
Experimental setup for testing the influence of volatiles from bacterial supernatants of Photorhabdus laumondii (strain DE2) on Heterorhabditis bacteriophora DJ recovery. DJs were incubated in bacterial supernatant (SN), Ringer’s solution neighbouring wells with supernatant (RN) and in Ringer’s solution separate from wells with supernatant (RS)
Statistical analysis
The Shapiro–Wilk test was used to check for normal distribution and equal-variance of the data before analyses by parametric tests (two- and one-way ANOVA followed by Tukey’s HSD test). Non-normally distributed or unequal-variance data were log-transformed or analysed by non-parametric tests (Kruskal–Wallis or Mann–Whitney U). The correlation between phenotypes was assessed by the Pearson correlation coefficient (normally distributed data) or Spearman’s rank correlation coefficient (non-normal-distributed data). The DJ recovery was calculated as percentage of recovered DJs in each cell well. The recovery over storage time was modelled by n-parameter logistic regressions according to Commo and Bot (2016). All data were collated and pre-treated in Microsoft Excel. Data normality & variances test, significance analysis and linear models were performed using GraphPad Prism 12.0 software (San Diego, California, USA). Correlations were assessed with R v4.2.3 in Rstudio v2023.3.0 (Boston, Massachusetts, USA) development environment. The R packages nplr v0.1.7, ggpubr v0.6.0 and ggplot2 v3.3.6 were used to implement the analysis.
Results
Species determination for native bacterial isolates
We isolated and identified the symbiotic bacteria from fourteen H. bacteriophora WT strains by analysis of sequences of the 16 S rRNA (1070 bp), gltX (1200 bp), recA (400 bp), dnaN (1065 bp) and gyrB (950 bp) genes. Concerning BLAST results, two isolates (DE6 and HU2) showed the highest homology to P. kayaii (percent identity (PI) > 96%), eleven isolates (AU1, DE2, DE8, HB1, HY1, IT4, MM14, MM8, PT2, SCR1, XX2) to P. laumondii, and one isolate (PT1) to P. thracensis NCBI accessions. An overview of the sequence identities of the BLAST hits is deposited in supplementary Table S2. Additionally, we carried out sequence pairwise comparisons of our strains and a set of identified Photorhabdus accessions for the given housekeeping genes. The local alignment results were congruent with our BLAST results. The eleven isolates showing homology to P. laumondii grouped in the same clade with accessions of this species. The expected grouping was also confirmed for the isolates with high BLAST homology to P. kayaii and P. thracensis. Phylogenetic trees showing the major groupings are provided in Fig. 2 (16 S rRNA) and Fig. 3 (gltX, dnaN, recA and gyrB). Each nematode strain carried a single Photorhabdus sp.
Phylogenetic relationship of Photorhabdus spp. strains built on 16 S rRNA sequences. The sequences of nearly 1000 bp length were considered for the analysis. Posterior probability values indicate branch support based on Bayesian inference (BI) analysis. Bar: 0.2 nucleotide substitutions per sequence position. Morganella morganii strain SC01 and Escherichia coli strain K12 were used as outgroup. Fourteen new sequences generated in this study are highlighted in red. (Color figure online)
Phylogenetic relationship of Photorhabdus spp. strains built from concatenated nucleotide sequences of four protein-coding genes (gltX, dnaN, recA and gyrB). The concatenated sequences of ~ 3000 bp length were considered for the analysis. Posterior probability values indicate branch support based on Bayesian inference (BI) analysis. Bar: 0.1 nucleotide substitutions per sequence position. Morganella morganii strain SC01 and Escherichia coli strain K12 were used as outgroup. Fourteen new sequences generated in this study are highlighted in red. (Color figure online)
Influence of different bacterial supernatants on DJ recovery
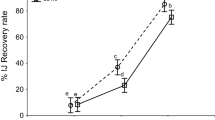
Five H. bacteriophora strains/lines were exposed to supernatants of fourteen identified Photorhabdus sp. isolates to determine their potential to support DJ recovery. Significant differences were observed between the recovery of the nematode materials (Two-way ANOVA; F = 535.1; df = 4, 210; P < 0.01). The highest mean DJ recovery was recorded for the hybrid line EN01 with 86.6 ± 7.7% ranging between 69 and 97.7% and for the mutant M31 82.1 ± 6.7% ranging between 69.8 and 92.4%. Mutant line M88 had the lowest mean recovery of 5.1 ± 2.7% ranging between 2.6 and 12.2% (Fig. 4) Within each H. bacteriophora material, significant differences were also determined for the recovery in different bacterial strain supernatants (Two-way ANOVA; FEN01 = 46.4, FHB4 = 106.2, FIL3 = 61.2, FM31 = 25.9, FM88 = 28.6; df = 13, 168; P < 0.01). The P. laumondii strain HY1 induced the highest DJ recovery in average throughout all nematode lines: EN01 (97.7 ± 2.5%), HB4 (75.3 ± 16.9%), IL3 (60.8 ± 17%), M31 (92.4 ± 5.4%) & M88 (12.2 ± 8.1%) (Fig. 4). P. laumondii strain S-CR1 supernatant was often the lowest in support of DJ recovery, e.g. in EN01 with 69 ± 19.1%, HB4 with 30.2 ± 8.9% and M88 with 2.6 ± 1.7%. Recovery in P. laumondii strain supernatants were generally higher than in supernatants of P. kayaii and P. thracensis. For the nematode lines IL3 and M31, the lowest recovery was observed upon exposure to P. kayaii strain HU2 supernatant (17.1 ± 17.5 and 69.8 ± 16, respectively). The heritability of the trait DJ recovery in bacterial supernatant was h2 < 0.6 despite the differences induced by the different bacterial supernatants. This indicates that the nematode-derived genetic background has a larger influence on DJ recovery, than factors originating from the symbiotic bacterium. However, the bacterial factor(s) can amplify or supress the nematode intrinsic DJ recovery. An overview of the DJ recovery of different nematode materials in fourteen bacterial supernatants, as well as the correlation between the nematode phenotypes and variation between four independent experimental repeats is presented in Fig. 4.
Heterorhabditis bacteriophora DJ recovery (%) of hybrid lines EN01 (A), HB4 (B) & IL3 (C) and mutant lines M31 (D) & M88 (E) in fourteen different Photorhabdus spp. supernatants, and the correlation (F) between DJ recovery level of the nematode lines. The heatmap (G) indicates consistency of the data between repetitions of experiments and allows a side-by-side comparison for all nematode strains/lines. The x-axis scales (A–E) and the colored bar (G) depict DJ recovery (%). The DJ recovery was assessed after 72 h incubation at 25 °C. The assessment was repeated four times, each with four replicates. Different letters behind the bars denote significant differences between nematode lines (p < 0.05). Error bars depict the SD. Stars denote significant differences between treatments (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001). The heritability (h2) is shown for DJ recovery (A–E) on top of the figure
A two-way ANOVA was performed to analyse the effect of H. bacteriophora lines and Photorhabdus strains on DJ recovery. As the ethyl-methane-sulfonate (EMS) mutant lines M31 and M88 represent two extremes in the recovery in bacterial supernatants, another analysis was performed excluding the two mutant lines. Both analyses provide the same results. Significant differences were recorded between nematode strains, as well as between bacteria strains (P < 0.01), but a statistically significant interaction between the nematode and bacteria (P > 0.05) was not recorded. The main source of variation originates from H. bacteriophora whether the mutants were included or not (Table 3).
Influence of aging on DJ recovery
To find out whether the age (storage time) of the DJ inoculum has effects on their recovery, DJs of five H. bacteriophora materials were exposed to supernatant of P. laumondii strain DE2. The dynamics of recovery over the storage time followed different patterns. For the three lines EN01, HB4 and IL3, the highest percent recovery was reported at the early observation points (week 1 and 2), whereas the proportion of recovered DJs decreased with increasing storage time. For instance, DJ recovery of EN01 was > 85% within week 1 and 2, while it decreased down to 18.7 ± 12.9% after two months (week 8). Differences in DJ recovery over DJ storage time were significant (Two-way ANOVA; df = 4, 45; P < 0.01) for each of the lines EN01 (F = 618.5), HB4 (F = 218.9) and IL3 (F = 470.1) (Table 4). The DJ recovery of the mutant line M31 was stable over the observation time (79.6 ± 7.5 to 85.9 ± 9.1%), while DJ recovery of line M88 was low over the complete experiment. While four of the nematode lines hardly recovery in Ringer’s solution, the DJ recovery of line M31 increased over the DJs storage time and this difference in DJ recovery resulted significant (Two-way ANOVA; F = 419.2; df = 4, 45; P < 0.01). Both mutants have an abnormal reaction on bacterial supernatant. Whereas M31 seems to be recovery-constitutive, exiting the dauer stage even in Ringer’s solution, mutant line M88 seems to be recovery-defective with recovery < 5% in supernatant.
Influence of volatiles from supernatants on DJ recovery
No evidence was found for an influence of volatiles from supernatants on DJ recovery, except for mutant M31. Whereas bacterial supernatant caused all nematode strains to recovery (except for recovery-defective mutant line M88), DJs in Ringer’s solution neighbouring cell wells with supernatant did not recovery, and the differences resulted significant (Two-way ANOVA; FEN01 = 3385, FHB4 = 2002, FIL3 = 1587, FM31 = 715.3, FM88 = 95.84; df = 2, 36; P < 0.01). Comparing the recovery of DJs in two treatments without direct exposure to bacterial supernatant, no differences were determined between cell wells with or without adjacent wells containing supernatant, except for the recovery-constitutive mutant line M31 which was significantly higher in wells with neighbouring supernatant (26.8 ± 16.7%) compared to wells without adjacent wells containing supernatant (18.4 ± 10.7%) (Tukey’s HSD test, P < 0.01) (Fig. 5). The present results confirm that the food signal is non-volatile.
Influence of volatiles emitted from neighbouring Photorhabdus laumondii (strain DE2) supernatants on DJ recovery of Heterorhabditis bacteriophora lines. The setup of treatments SN, RN and RS were provided in Fig. 1. DJ recovery was accessed after 72 h incubation in 25 °C. The experiments were repeated four times, each time with four technical replications. Stars over the bars denote significant differences between two treatments (∗∗p < 0.01). Error bars depict the SD
Discussion
Prior to testing the impact of bacterial symbionts on DJ recovery, the strains isolated from the different H. bacteriophora nematodes were identified. Initially the 16 S rRNA gene sequence was considered reliable for species identification and phylogenetic analysis, however in some cases species discrimination was impaired. The multi-locus sequence analysis has therefore been proposed to enhance the resolution of sequence analysis for species identification (Gevers et al. 2005; Tailliez et al. 2010). The nucleotide sequences of the fourteen bacterial isolates resulted highly homologous (> 96%) with both, 16 S rRNA and concatenated housekeeping gene sequences (gltX, recA, dnaN & gyrB) of P. laumondii, P. kayaii and P. thracensis. The resulting phylogenetic tree topology (Figs. 2 and 3) agrees with previous reports (Kazimierczak et al. 2017; Machado et al. 2018; Orozco et al. 2013; Sajnaga and Kazimierczak 2020). Of the ten Photorhabdus described species and subspecies isolated from the nematode H. bacteriophora we found three species.
In this study, H. bacteriophora strains/lines reacted differentially to diverse Photorhabdus supernatants. P. laumondii strains from HY1 & MM14 nematodes showed the highest DJ recovery along all lines contrasting with P. thracensis strain PT1, P. laumondii strain S-CR1 and P. kayaii strain HU2. Kazimierczak et al. (2017) reported that Heterorhabditis nematodes can swap their symbionts at intra- and interspecies level within the genus Photorhabdus. Variations along these nematode-bacterial complexes show that nematode performance can be also affected (Chapuis et al. 2009; Kazimierczak et al. 2017). Instead of swapping the bacteria, Machado et al. (2020) increased Photorhabdus benzoxazinoid resistance through experimental evolution, reintroduced engineered Photorhabdus into the native H. bacteriophora and finally enhanced nematode–symbiont virulence to western corn rootworm. We observed that even very related nematode strains can harbour symbionts conferring different phenotypes regarding DJ recovery. For instance, P. laumondii strains MM8 & MM14 were isolated from H. bacteriophora undergoing several passages through Melolontha melolontha (cockchafer white grub) for 8 and 14 generations, respectively (Berner et al. 2001). We found that DJ recovery upon exposure to bacterial supernatant of MM14 was always higher than in MM8 (P < 0.05, Fig. 4), regardless of the nematode line. Thus, whether nematode strains can adapt over generations to the natural product environment provided by the symbiont and modify their DJ recovery and reproductive programs is therefore a question that arises for future attempts for improvement. Considering that until today we find 10 phylogenetically separated taxa of Photorhabdus associated with H. bacteriophora, the question arises about the major selection pressure causing the evolution of so many different bacterial symbionts. The support of DJ recovery is certainly not linked to specification in Photorhabdus spp. and subspecies, since the bacterial food signal is not involved in DJ recovery inside the insect upon arrival of the nematode in the haemolymph, stage in which the bacteria are not yet released (Ciche and Ensign 2003; Ebrahimi et al. 2011). For industrial mass production it is advisable to select for those symbionts that best trigger DJ recovery and promote high DJ yields.
Knowledge on the nature of the bacterial food signal might help to improve process conditions in biotechnology mass production. Several proteins and biosynthetic compounds have been reported to play an active role in regulating bacteria–nematode–insect interactions (Duchaud et al. 2003; Joyce et al. 2008; Shi and Bode 2018; Shi et al. 2022). Besides conserved core genes that are essential for basic biological fuctions, unique biosynthetic gene clusters within species also may play an important role for the association with the nematode. Six of the Photorhabdus strains analysed in the present study (DE6, HU2, DE2, IT4, HB1 & PT1) were also included in the large analysis reported by Shi et al. (2022). According to the authors, the strains differently synthetized natural products like saccharides and terpenes. Thus, the large contrasts in DJ recovery observed between P. laumondii isolates DE2, IT4, and HB1 may be related to strain-specific features in secondary metabolite clusters. However, whether any of these natural products contribute to DJ recovery needs further investigation.
For the same H. bacteriophora strain/line, the support of DJ recovery by different Photorhabdus strains is variable. But these differences are less pronounced than the differences between the three nematode lines EN01, HB4 and IL3. The analysis for variation clearly indicates that the instability is on the side of the nematode material. So once a supportive bacterial symbiont has been selected, further improvements must be made on the nematode side. This variability is also obvious when the results between the experiments one to four are compared. In EN01 and HB4 the results vary within experiments 1 and 2 and of the first two experiments compared to the last two. The future challenge and focus will be on the improvement of the nematode material. One approach, which already resulted in an improvement in DJ in vitro recovery, is manifested in strain EN01, which has been subcultured over several years leading to enhancement of recovery. The DJ phenotype of this strain may have adapted to the industrial production environment. Another solution is to cross strains with improved traits with EN01 to afterwards select lines which carry the DJ recovery traits of EN01. Single Nucleotide Polymorphisms (SNPs) associated to high DJ recovery have already been identified (Wang et al. 2023) and can support such breeding programmes. Possibly, both approaches will contribute to higher DJ recovery in the future together with the identification of a bacterial symbiont strain with major support of the DJ recovery. We can conclude that different H. bacteriophora DJ batches possess a predetermined DJ recovery level, and this level can be increased using a supportive bacterial symbiont in in vitro mass production. Thus, the switch of Heterorhabditis–Photorhabdus partners can be a good complement to optimize the DJ recovery in H. bacteriophora, provide strains with optimal pre-disposition for recovery and reproduction can be selected.
The DJ recovery of lines EN01, HB4 and IL3 was found to decrease along storage time. In C. elegans, worm aging is reported to implicate muscle deterioration, metabolic disorder, accumulation of molecular and cellular damage (Golden and Melov 2007). In EPN, the lipid reserves of Heterorhabditis DJs are depleting during storage, and their mobility also decreases (Fitters and Griffin 2004; Menti et al. 2000). This limitation can be overcome by preventing too long storage periods of the nematode inoculum. Given the increasing demand for heterorhabditid nematodes in biological control also during winter periods (Lacey and Georgis 2012), the problem of inoculum storage might be less important in the future, or early precultures should produce inoculum with more predictable DJ recovery percent before the season starts.
The use of the mutant M31 is certainly not a solution to the problems with unpredictable and low DJ recovery as this line recovers even without bacterial triggers, which would cause problems during storage of the DJ material prior to and during transportation to the users. However, both mutant lines can be used for future studies investigating the genetic background for DJ recovery in H. bacteriophora.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request. The Photorhabdus sequences in this study are available at the National Center for Biotechnology Information (NCBI). The 16 S rDNA sequences can be found at NCBI under accessions OR350573 to OR350586. For the gene sequences of gltX, recA, dnaN, gyrB homologs, accessions numbers from OR364832 to OR364873 and OR393436 to OR393449 have been assigned.
References
Adeolu M, Alnajar S, Naushad S, Gupta RS (2016) Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: proposal for Enterobacterales Ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam nov. Int J Syst Evol Microbiol 66:5575–5599. https://doi.org/10.1099/ijsem.0.001485
Akhurst RJ (1980) Morphological and functional dimorphism in Xenorhabdus spp., bacteria symbiotically associated with the insect pathogenic nematodes Neoaplectana and Heterorhabditis. J Gen Microbiol 121:303–309. https://doi.org/10.1099/00221287-121-2-303
Aumann J, Ehlers R-U (2001) Physico-chemical properties and mode of action of a signal from the symbiotic bacterium Photorhabdus luminescens inducing dauer juvenile recovery in the entomopathogenic nematode Heterorhabditis Bacteriophora. Nematology 3:849–853. https://doi.org/10.1163/156854101753625344
Bah T (2011) Inkscape: guide to a vector drawing program, 5th edn. Prentice Hall Press, Hoboken, NJ
Berner M, Ehlers R-U, Schnetter W (2001) Genetic variability and discrimination of isolates, inbred lines and hybrids of Heterorhabditis bacteriophora via RAPD-PCR. iN: 34th Annual meeting of the society for invertebrate pathology, Noordwijkerhout, 25–30 Auguest 2001
Chapuis É, Emelianoff V, Paulmier V, Brun NL, Pagès S, Sicard M, Ferdy J-B (2009) Manifold aspects of specificity in a nematode–bacterium mutualism. J Evol Biol 22:2104–2117. https://doi.org/10.1111/j.1420-9101.2009.01829.x
Ciche TA, Ensign JC (2003) For the insect pathogen Photorhabdus luminescens, which end of a nematode is out? Appl Environ Microbiol 69:1890–1897. https://doi.org/10.1128/AEM.69.4.1890-1897.2003
Commo F, Bot BM (2016) R package nplr: n-parameter logistic regressions. https://cran.r-project.org/web/packages/nplr/vignettes/nplr.pdf
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9:772. https://doi.org/10.1038/nmeth.2109
Dolan KM, Jones JT, Burnell AM (2002) Detection of changes occurring during recovery from the dauer stage in Heterorhabditis bacteriophora. Parasitology 125:71–81. https://doi.org/10.1017/S0031182002001762
Duchaud E, Rusniok C, Frangeul L, Buchrieser C, Givaudan A, Taourit S, Bocs S, Boursaux-Eude C, Chandler M, Charles J-F, Dassa E, Derose R, Derzelle S, Freyssinet G, Gaudriault S, Médigue C, Lanois A, Powell K, Siguier P, Vincent R, Wingate V, Zouine M, Glaser P, Boemare N, Danchin A, Kunst F (2003) The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat Biotechnol 21:1307–1313. https://doi.org/10.1038/nbt886
Ebrahimi L, Niknam G, Dunphy GB (2011) Hemocyte responses of the Colorado potato beetle, Leptinotarsa decemlineata, and the greater wax moth, Galleria mellonella, to the entomopathogenic nematodes, Steinernema feltiae and heterorhabditis bacteriophora. J Insect Sci 11:75. https://doi.org/10.1673/031.011.7501
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. https://doi.org/10.1093/nar/gkh340
Ehlers R-U (2001) Mass production of entomopathogenic nematodes for plant protection. Appl Microbiol Biotechnol 56:623–633. https://doi.org/10.1007/s002530100711
Ehlers R-U (2003) Biocontrol nematodes. In: Hokkanen HMT, Hajek AE (eds) Environmental impacts of microbial insecticides: need and methods for risk assessment. Progress in biological control. Kluwer Scientific Publishers, Dordrecht, Netherlands. https://doi.org/10.1007/978-94-017-1441-9_10
Ehlers R-U, Deseö KV, Stackebrandt E (1991) Identification of Steinernema spp. (Nematoda) and their symbiotic bacteria Xenorhabdus spp. from Italian and German soils. Nematologica 37:360–364. https://doi.org/10.1163/187529291X00358
Ehlers R-U, Lunau S, Krasomil-Osterfeld K, Oaterfeld KH (1998) Liquid culture of the entomopathogenic nematode-bacterium-complex Heterorhabditis megidis/Photorhabdus luminescens. Biocontrol 43:77–86. https://doi.org/10.1023/A:1009965922794
Fitters PFL, Griffin CT (2004) Spontaneous and induced activity of Heterorhabditis megidis infective juveniles during storage. Nematology 6:911–917. https://doi.org/10.1163/1568541044038597
Gevers D, Cohan FM, Lawrence JG, Spratt BG, Coenye T, Feil EJ, Stackebrandt E, de Vandamme P, Thompson FL, Swings J (2005) Re-evaluating prokaryotic species. Nat Rev Microbiol 3:733–739. https://doi.org/10.1038/nrmicro1236
Godina G, Vandenbossche B, Centurión A, Dörfler V, Barg M, Ehlers R-U, Molina C (2023) New genotyping rescues old phenotypes: beneficial traits in Heterorhabditis Bacteriophora wild type material and association to single nucleotide polymorphisms. Nematology 25:761–773. https://doi.org/10.1163/15685411-bja10255
Golden TR, Melov S (2007) Gene expression changes associated with aging in C. elegans. In: Community TCeR (ed) WormBook. http://www.wormbook.org. Accessed 12 Feb 2007
Gouy M, Tannier E, Comte N, Parsons DP (2020) Seaview version 5: a multiplatform software for multiple sequence alignment, molecular phylogenetic analyses, and tree reconciliation. In: Katoh K (ed) Multiple sequence alignment. Methods in Molecular Biology, vol 2231. Humana, New York, pp 241–260. https://doi.org/10.1007/978-1-0716-1036-7_15
Grewal PS, Ehlers R-U, Shapiro-Ilan DI (2005) Nematodes as biocontrol agents, 1st edn. CAB International, Wallingford, Oxfordshire
Han R, Ehlers R-U (2000) Pathogenicity, development, and reproduction of Heterorhabditis bacteriophora and Steinernema carpocapsae under aenic. J Invertebr Pathol 75:55–58. https://doi.org/10.1006/jipa.1999.4900
Han R, Cao L, He X, Li Q, Liu X, Huang H, Peng Y, He M (2000) Recovery response of Heterirhabditis bacteriophora and Steinernema carpocapsae to different non-symbiotic microorganisms. Insect Sci 7:271–277. https://doi.org/10.1111/j.1744-7917.2000.tb00419.x
Hazir S, Stackebrandt E, Lang E, Schumann P, Ehlers R-U, Keskin N (2004) Two new subspecies of Photorhabdus luminescens, isolated from Heterorhabditis Bacteriophora (Nematoda: Heterorhabditidae): Photorhabdus luminescens subsp. kayaii subsp. nov. and Photorhabdus luminescens subsp. thracensis subsp. nov. Syst Appl Microbiol 27:36–42. https://doi.org/10.1078/0723-2020-00255
Hirao A, Ehlers R-U (2010) Influence of inoculum density on population dynamics and dauer juvenile yields in liquid culture of biocontrol nematodes Steinernema carpocapsae and S. feltiae (Nematoda: Rhabditida). Appl Microbiol Biotechnol 85:507–515. https://doi.org/10.1007/s00253-009-2095-4
Johnigk S-A, Ecke F, poehling M, Ehlers R-U (2004) Liquid culture mass production of biocontrol nematodes, Heterorhabditis bacteriophora (Nematoda: Rhabditida): improved timing of dauer juvenile inoculation. Appl Microbiol Biotechnol 64:651–658. https://doi.org/10.1007/s00253-003-1519-9
Joyce SA, Brachmann AO, Glazer I, Lango L, Schwär G, Clarke DJ, Bode HB (2008) Bacterial biosynthesis of a multipotent stilbene. Angew Chem 47:1942–1945. https://doi.org/10.1002/anie.200705148
Kazimierczak W, Skrzypek H, Sajnaga E, Skowronek M, Waśko A, Kreft A (2017) Strains of Photorhabdus spp. associated with polish Heterorhabditis isolates: their molecular and phenotypic characterization and symbiont exchange. Arch Microbiol 199:979–989. https://doi.org/10.1007/s00203-017-1368-z
Lacey LA, Georgis R (2012) Entomopathogenic nematodes for control of insect pests above and below ground with comments on commercial production. J Nematol 44:218–225
Lacey LA, Grzywacz D, Shapiro-Ilan DI, Frutos R, Brownbridge M, Goettel MS (2015) Insect pathogens as biological control agents: back to the future. J Invertebr Pathol 132:1–41. https://doi.org/10.1016/j.jip.2015.07.009
Lunau S, Stoessel S, Schmidt-Peisker AJ, Ehlers R-U (1993) Establishment of monoxenic inocula for scaling up in vitro cultures of the entomopathogenic nematodes Steinernema spp. and Heterorhabditis spp. Nematologica 39:385–399. https://doi.org/10.1163/187529293X00330
Machado RAR, Wüthrich D, Kuhnert P, Arce CCM, Thönen L, Ruiz C, Zhang X, Robert CAM, Karimi J, Kamali S, Ma J, Bruggmann R, Erb M (2018) Whole-genome-based revisit of Photorhabdus phylogeny: proposal for the elevation of most Photorhabdus subspecies to the species level and description of one novel species Photorhabdus bodei sp. nov., and one novel subspecies Photorhabdus laumondii subsp. clarkei subsp. nov. Int J Syst Evol Microbiol 68:2664–2681. https://doi.org/10.1099/ijsem.0.002820
Machado RAR, Thönen L, Arce CCM, Theepan V, Prada F, Wüthrich D, Robert CAM, Vogiatzaki E, Shi Y-M, Schaeren OP, Notter M, Bruggmann R, Hapfelmeier S, Bode HB, Erb M (2020) Engineering bacterial symbionts of nematodes improves their biocontrol potential to counter the western corn rootworm. Nat Biotechnol 38:600–608. https://doi.org/10.1038/s41587-020-0419-1
Menti H, Wright D, Perry R (2000) Infectivity of populations of the entomopathogenic nematodes Steinernema feltiae and heterorhabditis megidis in relation to temperature, age and lipid content. Nematology 2:515–521. https://doi.org/10.1163/156854100509439
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES science gateway for inference of large phylogenetic trees. 2010 Gatew Comput Environ Workshop (GCE). https://doi.org/10.1109/GCE.2010.5676129. New Orleans, Accessed 14 Nov 2010
Mukuka J, Strauch O, Hoppe C, Ehlers R-U (2010) Improvement of heat and desiccation tolerance in Heterorhabditis bacteriophora through cross-breeding of tolerant strains and successive genetic selection. Biocontrol 55:511–521. https://doi.org/10.1007/s10526-010-9271-4
Orozco RA, Hill T, Stock SP (2013) Characterization and phylogenetic relationships of Photorhabdus luminescens subsp. sonorensis (γ-Proteobacteria: Enterobacteriaceae), the bacterial symbiont of the entomopathogenic nematode Heterorhabditis Sonorensis (Nematoda: Heterorhabditidae). Curr Microbiol 66:30–39. https://doi.org/10.1007/s00284-012-0220-6
Rambaut A (2018) FigTree. http://tree.bio.ed.ac.uk/software/figtree/
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. https://doi.org/10.1093/bioinformatics/btg180
Sajnaga E, Kazimierczak W (2020) Evolution and taxonomy of nematode-associated entomopathogenic bacteria of the genera Xenorhabdus and Photorhabdus: an overview. Symbiosis 80:1–13. https://doi.org/10.1007/s13199-019-00660-0
Saux MF-L, Viallard V, Brunel B, Normand P, Boemare NE (1999) Polyphasic classification of the genus Photorhabdus and proposal of new taxa: P. luminescens subsp. luminescens subsp. nov., P. luminescens subsp. akhurstii subsp. nov., P. luminescens subsp. laumondii subsp. nov., P. temperata sp. nov., P. Temperata subsp. Temperata subsp. nov. and P. asymbiotica sp. nov. Int J Syst Evol Microbiol 49:1645–1656. https://doi.org/10.1099/00207713-49-4-1645
Sergeant M, Baxter L, Jarrett P, Shaw E, Ousley M, Winstanley C, Morgan JAW (2006) Identification, typing, and insecticidal activity of Xenorhabdus isolates from entomopathogenic nematodes in United Kingdom soil and characterization of the xpt toxin loci. Appl Environ Microbiol 72:5895–5907. https://doi.org/10.1128/AEM.00217-06
Shi Y-M, Bode HB (2018) Chemical language and warfare of bacterial natural products in bacteria–nematode–insect interactions. Nat Prod Rep 35:309–335. https://doi.org/10.1039/C7NP00054E
Shi Y-M, Hirschmann M, Shi Y-N, Ahmed S, Abebew D, Tobias NJ, Grün P, Crames JJ, Pöschel L, Kuttenlochner W, Richter C, Herrmann J, Müller R, Thanwisai A, Pidot SJ, Stinear TP, Groll M, Kim Y, Bode HB (2022) Global analysis of biosynthetic gene clusters reveals conserved and unique natural products in entomopathogenic nematode-symbiotic bacteria. Nat Chem. https://doi.org/10.1101/2022.01.21.477171
Strauch O, Ehlers R-U (1998) Food signal production of Photorhabdus luminescens inducing the recovery of entomopathogenic nematodes Heterorhabditis spp. in liquid culture. Appl Microbiol Biotechnol 50:369–374. https://doi.org/10.1007/s002530051306
Sumaya NH, Aryal S, Vandenbossche B, Barg M, Doerfler V, Strauch O, Molina C, Ehlers R-U (2017) Phenotyping dauer juvenile oxidative stress tolerance, longevity and persistence within wild type and inbred lines of the entomopathogenic nematode Heterorhabditis bacteriophora. Nematology 19:971–986. https://doi.org/10.1163/15685411-00003100
Sumaya NH, Gohil R, Okolo C, Addis T, Doerfler V, Ehlers R-U, Molina C (2018) Applying inbreeding, hybridization and mutagenesis to improve oxidative stress tolerance and longevity of the entomopathogenic nematode Heterorhabditis bacteriophora. J Invertebr Pathol 151:50–58. https://doi.org/10.1016/j.jip.2017.11.001
Tailliez P, Laroui C, Ginibre N, Paule A, Pagès S, Boemare N (2010) Phylogeny of Photorhabdus and Xenorhabdus based on universally conserved protein-coding sequences and implications for the taxonomy of these two genera. Proposal of new taxa: X. vietnamensis sp. nov., P. luminescens subsp. caribbeanensis subsp. nov., P. luminescens subsp. hainanensis subsp. nov., P. temperata subsp. khanii subsp. nov., P. temperata subsp. tasmaniensis subsp. nov., and the reclassification of P. luminescens subsp. thracensis as P. temperata subsp. thracensis comb. nov. Int J Syst Evol Microbiol 60:1921–1937. https://doi.org/10.1099/ijs.0.014308-0
Wang J, Cao L, Huang Z, Gu X, Cui Y, Li J, Li Y, Xu C, Han R (2022) Influence of the ascarosides on the recovery, yield and dispersal of entomopathogenic nematodes. J Invertebr Pathol 188:107717. https://doi.org/10.1016/j.jip.2022.107717
Wang Z, Ogaya C, Dörfler V, Barg M, Ehlers R-U, Molina C (2023) Pheno- and genotyping in vitro dauer juvenile recovery in the nematode Heterorhabditis bacteriophora. Appl Microbiol Biotechnol. https://doi.org/10.1007/s00253-023-12775-y
Acknowledgements
Zhen Wang appreciates the support of the China Scholarship Council (CSC) grant No. 201906850084. Manoj Dhakal gratefully acknowledges the scholarship by the Flemish Interuniversities Council (VLIR-UOS) to attend the International MSc Course in Agro- and Environmental Nematology at the University Ghent, Belgium.
Funding
Zhen Wang was supported by the China Scholarship Council (CSC) Grant No. 201906850084. Manoj Dhakal was supported by the scholarship from Flemish Interuniversities Council (VLIR-UOS).
Author information
Authors and Affiliations
Contributions
RUE, CM and BV conceived the experiments and the project. CM, BV and ZW designed the experiments. All authors performed the experiments. ZW and MD analysed the data. ZW wrote the first draft of the manuscript, CM and RUE read and commented on previous versions of the manuscript. All authors approved the manuscript.
Corresponding author
Ethics declarations
Competing interest
The authors declare no competing interests.
Ethical approval
This study does not contain any studies with human participants or vertebrate animals performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, Z., Dhakal, M., Vandenbossche, B. et al. Enhancing mass production of Heterorhabditis bacteriophora: influence of different bacterial symbionts (Photorhabdus spp.) and inoculum age on dauer juvenile recovery. World J Microbiol Biotechnol 40, 13 (2024). https://doi.org/10.1007/s11274-023-03803-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-023-03803-0