Abstract
Burkholderia sp. SP4, isolated from agricultural soils, has a high capability of degrading di-2-ethylhexyl-phthalate (DEHP). It degrades up to 99% of DEHP (300 mg l−1) in minimal salt (MS) media within 48 h without adding additionally auxiliary carbon source. The optimal conditions for SP4 to degrade DEHP are determined to be at 35 °C and pH 6.0. Supplementation of glucose (3.0 g l−1), sodium dodecyl sulfate (SDS) (0.2%), peptone (0.5 g l−1), or non-ionic surfactant Brij 35 (0.2%, 0.5% or 1%) in MS-DEHP media increases the DEHP degradation activity. Furthermore, kinetic analyses for DEHP degradation by SP4 reveals that it is a first-order reaction, and the half-life analyses also demonstrates that SP4 has a better degradative activity compared to other previously identified microbes. By means of HPLC-ESI-QTOF-MS, the metabolic intermediates of DEHP are identified for SP4, which include mono-2-ethylhexylphthalate (MEHP), mono-butyl phthalate (MBP), phthalic acid (PA), salicylic acid (SA), and 4-oxo-hexanoic acid. The presence of SA indicates that SP4 can consume DEHP using a dual biodegradation pathway diverged from the isomeric products of benzoate. Taken together, our study identifies a resilient DEHP-degradable bacterium and characterizes a novel degradation pathway for DEHP biodegradation. We plan to build on this finding in the context of removing DEHP from various environments.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Phthalate esters (PAEs) are synthetic materials extensively used as additives or plasticizers in the production of plastics (Staples et al. 1997; Kashyap and Agarwal 2018). PAEs are aromatic, colorless liquids with low volatility and solubility in water (Clara et al. 2010; Tran et al. 2021). Since PAEs are not covalently attached to the plastic milieu, they can leach out of the products into the surrounding environment (Adeniyi et al. 2011; Net et al. 2015). With high molecular weights and stabilities, these compounds are constantly detected in the environment, such as in samples taken from landfill leachates (Zheng et al. 2007), sediments (Adeniyi et al. 2011), soils (Tran et al. 2015b), natural water, rivers (Tran et al. 2015a), and the atmosphere (Orecchio et al. 2013). Previous studies have indicated that some PAEs are reproductive and progressive toxicants harmful to animals and plants (Rhee et al. 2002; Liao et al. 2009) and considered as endocrine-disrupting chemicals (EDCs) that are detrimental to human health (Meeker et al. 2009; Quintana-Belmares et al. 2018; Wang et al. 2019). For these reasons, concerns about PAEs production and their occurrence in the environment have amounted to risk assessments being conducted (Tran et al. 2022).
Di-(2-ethylhexyl) phthalate (DEHP) is one of the most highly produced and widely used PAEs in the world (Huang et al. 2008), and is found abundantly in the environment (Clara et al. 2010). To have no observed effect, the concentration of DEHP must be 77 μg l−1 in surface water (Naito et al. 2006) and 470 μg kg−1 dry weight in sediment (Yang et al. 2018). Moreover, the environmental risk limit for DEHP is 1000 μg kg−1 fresh weight in soil (VanWezel et al. 2000). A survey of the distribution of DEHP in 14 samples of surface water and six samples of sediment in Taiwan revealed that the average half-lives of DEHP in sediment were about 14.8 days in aerobic circumstances and 34.7 days in anaerobic circumstances, supporting the idea that degradation of phthalates in the aerobic condition is more effective than those in anaerobic conditions in soils, sediments and sludge (Staples et al. 1997; Wang et al. 2000; Yuan et al. 2002; Wang 2004).
PAEs in natural environments can be removed by hydrolysis, photolysis, and microbial degradation (Staples et al. 2000), but with concern for chemical configuration, hydrolysis and photolysis cannot effectively and stably remove PAEs (Chen et al. 2009). Thus, removal of PAEs through metabolism by microorganisms was suggested (Staples et al. 2000; Ren et al. 2018). Many different DEHP-degrading bacteria have been isolated, including Arthrobacter sp. C21 (Wen et al., 2014), Pseudoxanthomonas sp. (Meng et al., 2015), Rhizobium sp. LMB-1 (Tang et al., 2016), Acinetobacter sp. SN13 (Xu et al., 2017), Gordonia alkanivorans (Nahurira et al., 2017), LF consortium (Li et al., 2018), Rhodococcus ruber YC-YT1 (Yang et al., 2018), Rhodococcus pyridinivorans XB (Zhao et al., 2018), Burkholderia pyrrocinia B1213 (Li et al., 2019), Gordonia sp. 5F (Huang et al., 2019), Enterobacter spp. YC-IL1 (Lamraoui et al., 2020), Ochrobactrum anthropi L1-W (Nshimiyimana et al., 2020), and CM9 consortium (Bai et al. 2020). To understand how microbial degradations of DEHP are accomplished, metabolic intermediates from the degradation of DEHP are identified. As notably, the DEHP metabolic route in microorganisms involves first hydrolyzing DEHP into mono-(2-ethylhexyl) phthalate (MEHP), which is then transformed into phthalic acid (PA) (Magdouli et al. 2013). However, two recent studies showed that MEHP can be primarily transformed into mono-butyl phthalate (MBP), instead of being directly converted into PA in LF consortium (Li et al. 2018) and Burkholderia pyrrocinia B1213 (Li et al. 2019). Next, PA can be converted via two pathways: (1) under aerobic conditions, it is converted to 4, 5-dihydroxyphthalate in Gram-negative bacteria or to 3, 4-dihydroxyphthalate by dioxygenase in Gram-positive bacteria, or (2) under anaerobic conditions, to benzoate (Magdouli et al. 2013). Both routes generate protocatechuate (PCA) using any of the aforementioned intermediary compounds, which is subsequently converted into β-ketoadipate (Naumova et al. 1986; Magdouli et al. 2013). β-ketoadipate is then cleaved and transformed into 4-oxo-hexanoic acid, which enters the tricarboxylic acid (TCA) cycle to generate CO2 and H2O (Li et al. 2019). However, in the second pathway, with benzoate as an intermediate, ortho-hydroxybenzoate [2-hydroxybenzoate, salicylic acid (SA)] can be derived, since the hydroxylation of benzoate in microorganisms favors the ortho- and para- positions (Omori and Yamada 1973). In one bacterium (Chen et al. 2007) and two fungi (Rocheleau et al. 2019; Lubbers et al. 2021), SA is converted into β-ketoadipate, a step-wise manner through intermediates such as catechol and cis–cis muconic acid. Notably, each microbe may adopt different pathways. Thus, the identification of degradation metabolites and the annotation of the degradative pathway are often performed when a new microorganism that can biodegrade DEHP is discovered (Lamraoui et al. 2020).
In search for more microorganisms that can degrade DEHP in the locally polluted environments in Taiwan, we have isolated an indigenous bacterium Burkholderia sp. SP4 that degrades DEHP. In our study, we analyzed the environmental factors and kinetics underlying the degradation activities of the bacterium and also examined the degradation mechanism of SP4, which is likely to be a dual-degradation route.
Materials and methods
Chemical reagents
Di(2-ethylhexyl) phthalate (DEHP), mono(2-ethylhexyl) phthalate (MEHP), mono-butyl-phthalate (MBP), phthalic acid (PA, phthalate), benzoic acid (benzoate), protocatechuate (PCA), salicylic acid (SA, salicylate), and 4-oxohexanoic acid, were manufactured from Sigma-Aldrich (St. Louis, MO) and were purchased from Uni-Onward Co., Ltd. (Taipei, Taiwan).
Bacteria isolation and culture conditions
Soil samples were collected from Taoyuan Tea Research & Extension Station in Taoyuan City of Taiwan in the summer of 2013. The soil type from this district is red-brown soils with a topsoil of light clay loam. A total of five soil samples were acquired from the top layer of soil at different sites in the station. The criteria for picking sample collection sites were based on extensive human activities that can lead to potential plasticizer contaminations. The soil samples were then inoculated for further enrichment into the mineral salt (MS) media [1 g (NH4)2SO4, 1 g NH2CONH2, 1 g KH2PO4, 0.1 g NaCl, 0.1 g MgSO4.7H2O, 40 mg CaCl2, 40 mg FeSO4.7H2O, 10 mg CuSO4.7H2O, 5 mg MnSO4.H2O, 10 mg ZnSO4.7H2O, 0.5 mg CoCl2.6H2O, 0.5 mg Na2MoO4.2H2O, and 0.01% Tween 20 per liter] supplemented with 100 mg l−1 DEHP as the sole carbon source to enrich bacteria that can utilize DEHP for growth (Latorre et al. 2012). The enrichment cultures were incubated at 30 °C and agitated at 200 rpm. Two ml of the enrichments were used to inoculate fresh media once every three days. The enrichment process lasted for a month, and the final enrichment cultures were directly streaked on the MS media plates supplemented with 100 mg l−1 DEHP (MS-DEHP). The plates were incubated at 30 °C for 2 days until the colonies appeared.
Identification and sequencing of 16S rDNA gene
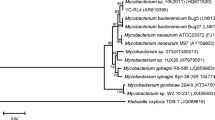
The genomic DNA of the single purified colony of the isolate from MS-DEHP media was extracted by Tissue & Cell Genomic DNA Purification kit (GeneMark, Taiwan). The 16S rDNA gene was PCR amplified using two universal primers, F8 (5′-AGAGTTTGATCCTGGCTCAG-3′) (Edwards et al. 1989) and R1492 (5′-GGTTACCTTGTTACGACTT-3′) (Stackebrandt and Liesack 1993). The PCR amplification was executed in a Thermal cycler (TProfessional thermocycler, Biometra, Germany). The PCR products were resolved in 1% agarose gel by electrophoresis and DNA fragments of 16S rDNA were purified with an innuPREP Gel Extraction kit (Analytikjena, Biometra, Germany). Sequencing of the purified PCR products was carried out at Genomics BioSci & Tech (Taipei, Taiwan) and the 16S rDNA sequence was submitted to GenBank with accession number KT306966. The sequencing results were analyzed using the NCBI BLAST. As compared with the known bacterial 16S rDNA gene sequences in GenBank, nine 16S rDNA sequences of the relative bacteria were retrieved and aligned with that of the isolated bacteria by the tool of MUSCLE in MEGA11 (Tamura et al. 2021). Phylogenetic trees were then constructed using the neighbor-joining method with MEGA11 software (Tamura et al. 2021). The bootstrap values were indicated at the nodes with 1000 replicates.
Effects of environmental factors on DEHP biodegradation by Burkholderia sp. SP4
The MS-DEHP liquid media at 35 °C, was used as a basic growth condition to determine the DEHP degradation ratio by Burkholderia sp. SP4. The growth condition of SP4 in MS-DEHP media was modified from the previous study (He et al. 2013). 100 ml MS media supplemented with 300 mg l−1 or 100 mg l−1 DEHP in a 250 ml Erlenmeyer flask, were used for batch culture. By centrifugation (8,000 rpm, 10 min), the overnight bacterial cultures in LB were collected and the cell pellets were rinsed by MS media twice followed by re-suspending in MS media. One mililiter of the washed overnight cultures was distributed into 100 ml fresh MS-DEHP media. At time 0 and every 24 h for 4 days, the bacterial growth in MS-DEHP was indicated by measuring OD600 and then 1 mL of the bacterial cultures in MS-DEHP (300 mg l−1) were harvested for DEHP extraction. Bacterial cells and cell debris in the supernatant were removed by centrifugation followed by filtration using a 0.22 μm syringe filter. For extraction of DEHP residuals, an aliquot (1 ml) of the resultant filtrates was mixed with an equal volume of ethyl acetate and the mixture was vigorously shaken for about 20 min. After centrifugation at 12,000 rpm for 10 min, the upper layer was collected and the rest solution was again extracted using an equal volume of ethyl acetate. The final collected top layer solution was evaporated and dried, which was then dissolved in methanol for HPLC analyses. The environmental features chosen to assess their effects on DEHP degradation in MS media with 100 mg l−1 DEHP at a 200 rpm shaking rate are temperature (15, 20, 25, 30, 35, 40 or 45 °C), initial pH value (3, 4, 5, 6, 7, 8, 9, 10 or 11), carbon or nitrogen source [glucose, peptone, yeast, sodium acetate or mannite by three various applications (0.5 g l−1, 1.5 g l−1 or 3.0 g l−1)] and surfactants [Sodium dodecyl sulfate (SDS), Triton X-100, Brij 35 and Tween 80] by three various applications [0.2%, 0.5% or 1.0% (v/v)]. After 48 h growth, the samples were collected as above for extraction of DEHP. The DEHP degradation ratio for each sample was determined by the following equation: DEHP degradation ratio (%) = [the initial concentration of DEHP—DEHP concentration of a single sample]/the initial concentration of DEHP × 100.
HPLC analytical methods
An aliquot of each sample was separately subjected to HPLC analysis. A Purospher® STAR RP-18e (250 × 4.6 mm, 5 μm) chromatography column was equipped in the machine. To detect DEHP, chromatography was executed under the following condition: temperature of 40 °C, a UV wavelength of 228 nm, the methanol: water (95:5, v/v) of mobile phase and 1 ml/min of a flow rate. The retention time for DEHP was 6 min. All measurements were accomplished on triplicate and the mean values and standard deviations were calculated for data analyses.
Degradation efficiency with different initial concentrations of DEHP and the identification of the intermediates during DEHP degradation
Various initial concentrations (100, 300, 500, 700 or 900 mg l−1) of DEHP in MS media were prepared for investigation of the consumption ability toward DEHP by SP4 using the procedures described above. To analyze the intermediate compounds during DEHP degradation by SP4, the final extracted samples were dissolved in methanol and the resulting suspension of each sample was applied to a high resolution HPLC-ESI-TOF–MS spectrometry device. Here, an HPLC (UltiMate 3000 Series system, DIONEX Technologies, Sunnyvale, CA, USA) equipped with a time-of-flight mass-spectrometer (Maxis, Bruker Daltonik, Bremen, Germany) was used for detection of the chemical compounds derived from degradation of DEHP by SP4. The detection procedures were modified according to the reference (Lee et al. 2015). Accordingly, a reverse phase column (Atlantis T3 Column, 2.1 mm × 100 mm, 3 µm particle size) was used for the analyses. The mobile phase was a gradient prepared from 0.1% formic acid in water (component A) and methanol (component B). The gradient program for the HPLC was as follows: 0–3 min, 10% B; 3–6 min 100% B; 6–6.5 min 100% B; 6.5–13 min 10% B, and the flow rate was 0.3 mL/min. The injection volume was 10 µl and the column temperature was 40 °C. Mass spectra in the m/z range 50–1500 were obtained by the use of electrospray ionization in the positive-ion mode. The mass spectrometric conditions were optimized as follows: gas temperature 180 °C, drying gas flow rate 9.0 l/min, nebulizer gas pressure 3.0 bar, and capillary 4500 V. The mass axis was calibrated using 0.01 N lithium formate as an internal calibration solution. Acquired data were analyzed by Bruker Compass Data Analysis (version 1.1).
Statistical analysis
Data shown in this study are mean ± standard deviation (SD) of three replicates from one of at least three independent experiments. Statistical analysis of degradation ratio for DEHP was determined by Student’s t-test, and the difference was considered significant when one-tailed tests resulted in a p-value less than 0.05.
Results
Identification of the DEHP-degrading bacterium isolated from agricultural soils
To isolate possible DEHP-degrading bacteria, soil samples were collected and inoculated in MS medium with DEHP as the primary carbon source. After a month of enrichment in which the culture was re-inoculated every three days, we isolated a bacterium that could grow on MS-DEHP media, but not on MS medium only. Identical colony morphology on MS-DEHP medium plates after serial transferring indicated that a single strain of bacterium was isolated. The 16S rDNA fragment of this bacterial strain was PCR-amplified and sequenced. We found that the isolated bacterium shared more than 95.3% 16S rDNA pairwise sequence similarities with bacteria from Burkholderia spp. Analyses of the neighbor-joining phylogenetic tree and the estimation of evolutionary discrepancy revealed that no definitive species match the isolated bacterium (Fig. S1). Hence, this bacterial strain was designated Burkholderia sp. SP4.
Degradation curve of DEHP and growth curve of SP4 strain in MS-DEHP medium
To explore whether SP4 directly degrades DEHP or not, the decreasing concentration of DEHP and the growth phase of SP4 were simultaneously measured in MS-DEHP medium at different time intervals. As shown in Fig. 1a, within 24 h, SP4 degraded about 73% of DEHP (with an initial concentration of 300 mg l−1) and within 48 h, approximately 99% of DEHP was consumed. Within 72 h, the concentration of DEHP decreased to less than 2 mg l−1. Meanwhile, faster growth of SP4 was observed when DEHP was degraded at a higher rate within 24 h, while bacterial growth slowed down when the DEHP-degradation rate dropped after 24 h. To verify that the decrease of DEHP was not due to exterior/denatured proteins of the inoculum bacteria, DEHP concentrations in MS media inoculated with sterilized SP4 were determined. As shown in Fig. 1b, DEHP contents did not decrease significantly in MS-DEHP medium with sterilized SP4 inoculation. In addition, since 0.01% Tween 20 was used in the initial DEHP stock to assist the solubilization of hydrophobic DEHP, the growth of SP4 in MS medium containing 0.01% Tween 20 was measured to verify whether SP4 could utilize this trace carbon source for substantial growth. As a result, the biomass of SP4 did not increase during the incubation time (data not shown). Therefore, 0.01% Tween 20 was not capable of supporting the growth of SP4.
Degradation of DEHP by Burkholderia sp. SP4. a Degradation curve of DEHP and growth curve of Burkholderia sp. SP4 in MS-DEHP (initial concentration as 300 mg l−1). The line with black circles represents the residual concentrations of DEHP in the presence of SP4 at different time points (0, 24, 48, and 72 h). The line with triangles represents the growth curve (OD600) of SP4. Error bars represent standard deviations of the means. (b) DEHP concentration (initial 300 mg l−1) in MS media inoculated with sterilized SP4 (100 °C boiling) was detected by HPLC-ESI-QTOF-MS on day 0 and day 4. Significant DEHP compound signals are indicated
Influences of temperature, pH, various carbon sources and surfactants on DEHP degradations
It is known that temperature and pH have effects on the degradation of phthalate by microorganisms (Ren et al. 2018). We assessed the degradation ability of SP4 with 100 mg l−1 of DEHP in MS medium at various temperatures and pH values. The degradation ratio of SP4 increased with increasing temperature between 15 °C and 35 °C and maintained above 50% between 20 °C to 35 °C. However, at 40 °C and 45 °C, the DEHP degradation ratio of SP4 declined significantly, and less than 30% of DEHP was degraded (Fig. 2a). In terms of the optimal pH, DEHP degradation ratios increased with increasing initial pH values and presented the highest with the initial pH of 6. When the initial pH value was equal or higher than 7, the degradation activity decreased (Fig. 2b). Based on these results, we performed subsequent experiments at 35 °C and at the initial pH of 6.
Effects of environmental factors on DEHP degradation by Burkholderia sp. SP4. Effects of different A temperatures, B pH values, C carbon sources: glucose, peptone, yeast, sodium acetate, or mannite (0.5 g l−1, 1.5 g l−1 or 3.0 g l−1), and D surfactants: SDS, TritonX-100, Brij35, or Tween 80 [0.2%, 0.5% or 1.0% (v/v)] on degradation of 100 mg l−1 DEHP by Burkholderia sp. SP4. The (*) symbol indicated that the data are significantly higher from the control, with p < 0.05 in the Student’s t-test. Error bars represent standard deviations of the means
A previous study has demonstrated that an addition of yeast extracts (1% w/v) as auxiliary materials in growth media is necessary for significant DEHP-degrading activities in bacterium B. pyrrocinia B1213 (Li et al., 2019). We therefore examined whether the supplementation of other carbon or nitrogen sources can enhance degradation activity of SP4 and found that glucose at a concentration of 3.0 g l−1 and peptone at a concentration of 0.5 g l−1 enhances DEHP degradation activity. However, other carbon sources seemed to have no significant effects (Fig. 2c). Since addition of surfactants could improve the solubility of DEHP in media, we also examined the effects of surfactants on DEHP degradation by SP4. DEHP degradation activity slightly increased in the presence of non-ionic surfactant Brij 35 and anionic surfactant 0.2% SDS; however, DEHP degradation activity decreased with increasing SDS concentration or with Triton X-100 and Tween 80 at all concentrations used (Fig. 2d). From our data, SP4 did not necessarily require additionally specific carbon or nitrogen sources or certain surfactants; its intrinsic DEHP degradation ability was already significant.
Effects of initial DEHP concentrations and biodegradation kinetics of DEHP by SP4 strain
To characterize the DEHP degradation ability of SP4, especially for high concentrations of DEHP, we prepared MS media supplemented with different initial concentrations of DEHP (100, 300, 500, 700, or 900 mg l−1) and determined residual DEHP concentrations by HPLC–ESI-QTOF-MS at different time points after incubation with SP4. As shown in Fig. 3, at all five initial concentrations, the residual DEHP concentration decreased rapidly with the incubation time. Without an apparent lag phase, DEHP with different initial concentrations were effectively consumed by SP4 before 72 h, including the one with the highest initial concentration of 900 mg l−1 (Fig. 3). Degradation kinetics for SP4 was determined to be a first-order reaction with the following equation: ln C = -Kt + A, in which C is the concentration of DEHP; K is the first-order rate constant; t is the reaction time; and A is a constant value, respectively. Half-life for DEHP degradation can then be determined by the formula: t1/2 = ln2/K, where t1/2 denotes the half-life. Table 1 represents the kinetic equations and the half-lives of DEHP degradation by SP4 against the various initial concentrations aforementioned. Compared with similar degradation kinetics by other DEHP-degrading bacteria, SP4 presented a better DEHP degradation ability. For example, Ochrobactrum anthropi L1-W degraded DEHP at 200 mg l−1 for 72 h (Nshimiyimana et al. 2020), but SP4 degraded the higher concentration of DEHP (300 mgl−1) within 48 h.
Identification of the diverse metabolites from degradations of DEHP by SP4 strain using HPLC–ESI-QTOF-MS analyses
The significant DEHP-degrading activity of SP4 may result from a special degradation pathway. To determine the pathway used by SP4 for DEHP degradation, we identified the degradation intermediates by HPLC-ESI-QTOF-MS. Data of mass measurements were listed in Table 2. First, MEHP (m/z 279.1599), generally considered to be the first product of DEHP degradation, was identified (Fig. 4a). MBP (m/z 223.0980), the novel intermediate recently identified in DEHP degradation of B. pyrrocinia B1213 and bacterial consortium (Li et al. 2018, 2019) was also identified (Fig. 4b), indicating that SP4 adopted this bio-conversion in which MEHP was first converted to MBP. Next, PA (m/z 167.0333) in the classic DEHP degradation pathway was identified (Fig. 4c). Notably, SA (m/z 139.0401), the intermediate that does not belong to the classic pathway of Gram-negative bacteria under aerobic conditions, was identified in the mass measurements (Fig. 4d). Finally, 4-oxo-hexanoic acid (m/z 131.0699), a late product of DEHP degradation was identified (Fig. 4e).
As noted, 4-oxo-hexanoic acid and SA are the intermediates of two distinct pathways. The coincidental yet significant appearance of both intermediates in DEHP-degradation of SP4 indicates that SP4 may use two routes for DEHP biodegradation: SP4 may convert PA into benzoate, which subsequently converts to SA and 4-hydroxybenzoate, in addition to the classic conversion of PA to PCA (Fig. 5). To demonstrate again the existence of SA and other intermediates, Thin-Layer-Chromatography (TLC) was used to isolate the intermediate compounds, MEHP and SA, in the samples after incubation of SP4 in MS-DEHP (1 g l−1) media (Fig. S2). Additionally, DEHP gradually decreased over time, while MEHP and SA both increased and accumulated. These results confirm that SA is indeed a key intermediate during DEHP degradation by SP4 and suggest that SP4 possesses dual pathways for DEHP degradation.
Discussion
One of the major ways of degrading PAE in the environment is through metabolic catabolism of PAE by microorganisms. Unlike di-ethyl phthalate (DEP) and di-butyl phthalate (DBP), which have smaller alkyl chains more susceptible to microbial degradation, DEHP with its extensive alkyl chain is difficult to degrade (O'Grady, et al. 1985; Wang et al. 2000; Chang, et al. 2004). Hence, DEHP prevails in the environment, leading to toxic accumulation that conveys reproductive toxicity in humans and animals (Talsness et al. 2009). In this study, we identified a new DEHP-degrading microorganism, Burkholderia sp. SP4, isolated from agricultural soils. We demonstrated that SP4 is highly effective in biodegrading DEHP, motivating us to characterize it further.
SP4 was isolated from soil samples. To ensure effective isolation (Gu 2018, 2021), MS medium with DEHP as the major carbon source was used for bacterial enrichment. The additional carbon source, if at all possible in this case, would be the Tween 20 that was added to aid the DEHP dissolution, and the overall concentration did not exceed 0.01%. In our growth test, 0.01% Tween 20 in MS medium could not support the growth of SP4 (data not shown); so DEHP in the medium should account for the sole carbon source. SP4 has been selectively enriched in this process for its potent DEHP-degrading capacity. On the other hand, the sterilized SP4 could not minimize the DEHP contents in MS-DEHP medium (Fig. 1b). Our results show that the decrease in DEHP contents observed in the medium with live SP4 was caused by the biodegradation and biotransformation of SP4 itself.
SP4 was determined to have maximum activity at 35 °C (Fig. 2a) and at pH range 5 to 6 (Fig. 2b); the former is similar to the previously identified toxic chemicals degrading microbes, including DEHP-degrader (Nomura et al. 1992; Jin et al. 2012; Nahurira et al. 2017). SP4 in the batch culture removed about 99% DEHP at an initial concentration of 300 mg l−1 within 48 h (Fig. 1). This degradation activity is better than that of the other bacteria. For example, Sphingomonas sp. DK4 and Corynebacterium sp. O18 only degraded 11.5 and 11.2% of DEHP (100 mgl−1), respectively, within 168 h (Chang et al. 2004). Although Rhizobium sp. LMB-1 and Acinetobacter sp. SN13 degraded 88% and 90% of DEHP (100 mgl−1), 120 h was required in both cases (Tang et al. 2016; Xu et al. 2017). Moreover, Ochrobactrum anthropi L1-W removed 98.7% of DEHP (200 mgl−1) but only at 72 h (Nshimiyimana et al. 2020). Finally, since Burkholderia sp. B1213 (Li et al., 2019) required yeast extracts for the biomass growth, it is not applicable for comparing the efficiency of DEHP degradation between SP4 and B1213. However, compared with the above other DEHP degraders, SP4 has an exceptional capability to metabolize DEHP. According to our result, SP4 could even degrade DEHP at concentration as high as 1gl−1 (Fig. S2).
To determine the degradation of DEHP at high concentrations, surfactants such as Tween 20 are added to ensure the DEHP solubility. If additional surfactants were added, low-concentration SDS and Brij 35 increased the degradation rate of SP4, whereas Triton X-100 and Tween 80 negatively impact the DEHP degradation activity (Fig. 2d). The negative effects of Triton X-100 and Tween 80 may be caused by their toxicity towards SP4, as SP4 grows less in such media. Assisted by the surfactant Tween 20, we were able to determine the kinetics of SP4 in degrading high concentrations of DEHP (from 100 to 900 mgL−1) (Fig. 3). In our results, SP4 used a first-order kinetic reaction for DEHP degradation (Fig. 3; Table 1), which might be one of the causes of its high degradation efficiency. In the example of B. pyrrocinia B1213, an apparent lag phase was observed in its biodegradation curve for DEHP (Li et al. 2019). Considering that B. pyrrocinia B1213 required yeast extract as auxiliary material in the medium, its more complex degradation pattern may reflect the fact that the bacteria need to digest yeast extracts for enrichment before degrading DEHP. In contrast, SP4 can degrade DEHP directly. The O. anthropi L1-W bacterium also uses the first-order kinetic reaction for DEHP degradation (Nshimiyimana et al. 2020). However, SP4 has a shorter half-life for degrading DEHP, at 8.84 h with an initial concentration of 300 mgl−1, compared to a half-life of 11.26 h for O. anthropi L1-W in degrading DEHP (Nshimiyimana et al. 2020).
Finally, we identified the intermediates derived from the degradation of DEHP by SP4 in order to characterize the degradation pathway. SP4 first degraded DEHP into MEHP (Fig. 4a) and, similar to B. pyrrocinia B1213 (Li et al., 2019), converted MEHP to MBP (Fig. 4b), likely based on the β-oxidation as well. Mass determination further identified PA (Fig. 4c) and the end product 4-oxo-hexanoic acid (Fig. 4e) of the classic bio-degradation pathway of Gram-negative bacteria under aerobic conditions. However, SA was identified as one of the intermediates (Fig. 4d), indicating that another pathway exists in the DEHP degradation of SP4. We speculate that SP4 can convert PA into benzoate, in addition to the conversion of PA to PCA (Fig. 5). Since the hydroxylation of benzoate in microorganisms favors the ortho- and para- positions (Omori and Yamada 1973), SP4-derived benzoate may be converted to para-hydroxybenzoate (4-hydroxybenzoate) and ortho-hydroxybenzoate (SA, 2-hydroxybenzoate). A dual pathway is then adopted: in one route, 4-hydroxybenzoate is converted to PCA; and in another route, SA may be transformed into catechol and subsequently cis-cis muconic acid. Our speculation is supported by our finding that SP4 can grow independently in MS medium supplemented with either phthalate, benzoate, or PCA (Table S1, compared to Escherichia coli BL21 that could not grow in the MS medium supplemented with these compounds), indicating its physiological ability to metabolize any of these compounds for biomass production. In one bacteria and two fungi, β-carboxy-cis, cis muconic acid or cis, cis-muconic acid, is ultimately cleaved to produce β-ketoadipate (Xu et al. 2017; Rocheleau et al. 2019; Lubbers et al. 2021) and then possibly converted into 4-oxo-hexanoic acid (also identified in reports by Xu et al. 2017 and Li et al. 2019) for entering the TCA cycle with the production of CO2 and H2O. Given the identification of 4-oxo-hexanoic acid (Fig. 4e), SP4 may also possess a similar conversion route. However, during the stepwise conversion of PA to 4-oxo-hexanoic acid, the aforementioned microbes all adopt a single pathway, whereas SP4 is highly likely to utilize the dual degradation pathway diverged from benzoate for its biomass growth.
In order to adopt the dual pathway, SP4 may possess enzymes such as 1-monooxygenase or 2, 3-dihydroxybenzoate decarboxylase, that convert SA or 2, 3-dihydroxybenzoate intermediate, respectively, into catechol (Rocheleau et al. 2019; Lubbers et al. 2021); a catechol 1, 2-dioxygenase that converts catechol to cis–cis muconic acid as reported in Burkholderia cepacia (Ngoc Thi et al., 2020); and likely enzymes similar to that found in Streptomyces niger, such as catechol 1, 2-dioxygenase (for degrading catechol compound derived from benzoate and SA) and protocatechuate 3, 4-dioxygenase (for degrading PCA compound derived from para-hydroxybenzoate) (Grund et al. 1990). Future studies will focus on discovering these enzymes in the DEHP-degradation pathways of SP4 to support its degradative mechanisms and the development of its application on environmental remediation. Given the dual pathway, SP4 may possess broad substrate specificity (Huang et al. 2019), which will also be confirmed by the identification of the degradative enzymes.
Conclusions
Our study identifies a DEHP-degrader bacterium, Burkholderia sp. SP4, which is newly isolated from agricultural soils. Compared to the previously isolated DEHP-degrading microbes, SP4 exhibits significantly higher activity for DEHP degradation. Moreover, SP4 can degrade DEHP without adding yeast extracts that are obligatory for other DEHP-degradable Burkholderia spp. Uniquely, the identification of intermediates proposed a novel dual pathway for DEHP degradation by SP4, which is very different from the single degradative pathway normally found in DEHP-degrading microbes. Our study not only demonstrates a highly effective DEHP-degradable bacterium that has potential for environmental remediation but also deepens the understanding of DEHP biodegradation of bacteria.
Data availability
The 16S rDNA sequence of SP4 was submitted to GenBank with accession number KT306966. The other datasets generated or analyzed during the study are available from the corresponding author on reasonable request.
References
Adeniyi AA, Okedeyi OO, Yusuf KA (2011) Flame ionization gas chromatographic determination of phthalate esters in water, surface sediments and fish species in the Ogun river catchments, Ketu, Lagos, Nigeria. Environ Monit Assess 172:561–569. https://doi.org/10.1007/s10661-010-1354-2
Bai N, Li S, Zhang J, Zhang H, Zhang H, Zheng X, Lv W (2020) Efficient biodegradation of DEHP by CM9 consortium and shifts in the bacterial community structure during bioremediation of contaminated soil. Environ Pollut 266:115112. https://doi.org/10.1016/j.envpol.2020.115112
Chang BV, Yang CM, Cheng CH, Yuan SY (2004) Biodegradation of phthalate esters by two bacteria strains. Chemosphere 55:533–538. https://doi.org/10.1016/j.chemosphere.2003.11.057
Chen JA, Li X, Li J, Cao J, Qiu Z, Zhao Q, Xu C, Shu W (2007) Degradation of environmental endocrine disruptor di-2-ethylhexyl phthalate by a newly discovered bacterium, Microbacterium sp. strain CQ0110Y. Appl Microbiol Biotechnol 74:676–682. https://doi.org/10.1007/s00253-006-0700-3
Chen Y-H, Chen L-L, Shang N-C (2009) Photocatalytic degradation of dimethyl phthalate in an aqueous solution with Pt-doped TiO 2-coated magnetic PMMA microspheres. J Hazard Mater 172:20–29
Clara M, Windhofer G, Hartl W, Braun K, Simon M, Gans O, Scheffknecht C, Chovanec A (2010) Occurrence of phthalates in surface runoff, untreated and treated wastewater and fate during wastewater treatment. Chemosphere 78:1078–1084. https://doi.org/10.1016/j.chemosphere.2009.12.052
Edwards U, Rogall T, Blocker H, Emde M, Bottger EC (1989) Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res 17:7843–7853. https://doi.org/10.1093/nar/17.19.7843
Grund E, Knorr C, Eichenlaub R (1990) Catabolism of benzoate and monohydroxylated benzoates by Amycolatopsis and Streptomyces spp. Appl Environ Microbiol 56:1459–1464. https://doi.org/10.1128/aem.56.5.1459-1464.1990
Gu J-D (2018) The endocrine-disrupting plasticizers will stay with us for a long time. Appl Environ Biotechnol 3:4. https://doi.org/10.26789/aeb.2018.01.008
Gu J-D (2021) Biodegradability of plastics: the issues, recent advances, and future perspectives. Environ Sci Pollut Res 28:1278–1282. https://doi.org/10.1007/s11356-020-11501-9
He Z, Xiao H, Tang L, Min H, Lu Z (2013) Biodegradation of di-n-butyl phthalate by a stable bacterial consortium, HD-1, enriched from activated sludge. Bioresour Technol 128:526–532. https://doi.org/10.1016/j.biortech.2012.10.107
Huang PC, Tien CJ, Sun YM, Hsieh CY, Lee CC (2008) Occurrence of phthalates in sediment and biota: relationship to aquatic factors and the biota-sediment accumulation factor. Chemosphere 73:539–544. https://doi.org/10.1016/j.chemosphere.2008.06.019
Huang H, Zhang XY, Chen TL, Zhao YL, Xu DS, Bai YP (2019) Biodegradation of structurally diverse phthalate esters by a newly identified esterase with catalytic activity toward Di(2-ethylhexyl) phthalate. J Agric Food Chem 67:8548–8558. https://doi.org/10.1021/acs.jafc.9b02655
Jin Q, Hu Z, Jin Z, Qiu L, Zhong W, Pan Z (2012) Biodegradation of aniline in an alkaline environment by a novel strain of the halophilic bacterium, Dietzia natronolimnaea JQ-AN. Bioresour Technol 117:148–154. https://doi.org/10.1016/j.biortech.2012.04.068
Kashyap D, Agarwal T (2018) Concentration and factors affecting the distribution of phthalates in the air and dust: a global scenario. Sci Total Environ 635:817–827. https://doi.org/10.1016/j.scitotenv.2018.04.158
Lamraoui I, Eltoukhy A, Wang J, Lamraoui M, Ahmed A, Jia Y, Lu T, Yan Y (2020) Biodegradation of Di (2-ethylhexyl) phthalate by a novel Enterobacter spp. strain YC-IL1 isolated from polluted soil, Mila, Algeria. Int J Environ Res Public Health 17:7501. https://doi.org/10.3390/ijerph17207501
Latorre I, Hwang S, Montalvo-Rodriguez R (2012) Isolation and molecular identification of landfill bacteria capable of growing on di-(2-ethylhexyl) phthalate and deteriorating PVC materials. J Environ Sci Health A 47:2254–2262. https://doi.org/10.1080/10934529.2012.707549
Lee MJ, Chung IM, Kim H, Jung MY (2015) High resolution LC-ESI-TOF-mass spectrometry method for fast separation, identification, and quantification of 12 isoflavones in soybeans and soybean products. Food Chem 176:254–262. https://doi.org/10.1016/j.foodchem.2014.12.073
Li F, Liu Y, Wang D, Zhang C, Yang Z, Lu S, Wang Y (2018) Biodegradation of di-(2-ethylhexyl) phthalate by a halotolerant consortium LF. PLoS ONE 13:e0204324. https://doi.org/10.1371/journal.pone.0204324
Li J, Zhang J, Yadav MP, Li X (2019) Biodegradability and biodegradation pathway of di-(2-ethylhexyl) phthalate by Burkholderia pyrrocinia B1213. Chemosphere 225:443–450. https://doi.org/10.1016/j.chemosphere.2019.02.194
Liao CS, Yen JH, Wang YS (2009) Growth inhibition in Chinese cabbage (Brassica rapa var. chinensis) growth exposed to di-n-butyl phthalate. J Hazard Mater 163:625–631. https://doi.org/10.1016/j.jhazmat.2008.07.025
Lubbers RJM, Dilokpimol A, Visser J, Hilden KS, Makela MR, de Vries RP (2021) Discovery and functional analysis of a salicylic acid hydroxylase from Aspergillus niger. Appl Environ Microbiol. https://doi.org/10.1128/AEM.02701-20
Magdouli S, Daghrir R, Brar SK, Drogui P, Tyagi RD (2013) Di 2-ethylhexylphtalate in the aquatic and terrestrial environment: a critical review. J Environ Manag 127:36–49. https://doi.org/10.1016/j.jenvman.2013.04.013
Meeker JD, Sathyanarayana S, Swan SH (2009) Phthalates and other additives in plastics: human exposure and associated health outcomes. Phil Trans r Soc b 364:2097–2113. https://doi.org/10.1098/rstb.2008.0268
Meng X, Niu G, Yang W, Cao X (2015) Di(2-ethylhexyl) phthalate biodegradation and denitrification by a Pseudoxanthomonas sp. strain. Bioresour Technol 180:356–359. https://doi.org/10.1016/j.biortech.2014.12.071
Nahurira R, Ren L, Song J, Jia Y, Wang J, Fan S, Wang H, Yan Y (2017) Degradation of di(2-ethylhexyl) phthalate by a novel Gordonia alkanivorans strain YC-RL2. Curr Microbiol 74:309–319. https://doi.org/10.1007/s00284-016-1159-9
Naito W, Gamo Y, Yoshida K (2006) Screening-level risk assessment of di(2-ethylhexyl) phthalate for aquatic organisms using monotoring data in Japan. Enviiron Monit Assess 115:451–471
Naumova RP, Zaripova SK, Usmanova LP (1986) Regulation of terephthalate catabolism in Rhodococcus rubropertinctus. Mikrobiologiia 55:918–923
Net S, Sempere R, Delmont A, Paluselli A, Ouddane B (2015) Occurrence, fate, behavior and ecotoxicological state of phthalates in different environmental matrices. Environ Sci Technol 49:4019–4035. https://doi.org/10.1021/es505233b
Ngoc Thi TV, Hoang Sinh DD, Ha Thanh LT, Huy ND, Tue NH, Shintani M, Kimbara K, Loc NH (2020) Cloning, expression and characterization of catechol 1,2-dioxygenase from Burkholderia cepacia. J Gen Appl Microbiol 66:188–194. https://doi.org/10.2323/jgam.2019.06.002
Nomura Y, Nakagawa M, Ogawa N, Harashima S, Oshima Y (1992) Genes in PHT plasmid encoding the initial degradation pathway of phthalate in Pseudomonas putida. J Ferment Bioeng 74:333–344
Nshimiyimana JB, Khadka S, Zou P, Adhikari S, Proshad R, Thapa A, Xiong L (2020) Study on biodegradation kinetics of di-2-ethylhexyl phthalate by newly isolated halotolerant Ochrobactrum anthropi strain L1-W. BMC Res Notes 13:252. https://doi.org/10.1186/s13104-020-05096-0
O’Grady DP, Howard PH, Werner AF (1985) Activated sludge biodegradation of 12 commercial phthalate esters. Appl Environ Microbiol 49:443–445
Omori T, Yamada K (1973) Relation between electronic structure and hydroxylation of aromatic compounds by microorganisms. Agric Biol Chem 37:1809–1811. https://doi.org/10.1080/00021369.1973.10860927
Orecchio S, Indelicato R, Barreca S (2013) The distribution of phthalate esters in indoor dust of Palermo (Italy). Environ Geochem Health 35:613–624. https://doi.org/10.1007/s10653-013-9544-9
Quintana-Belmares RO, Krais AM, Esfahani BK, Rosas-Pérez I, Mucs D, López-Marure R, Bergman Å, Alfaro-Moreno E (2018) Phthalate esters on urban airborne particles: levels in PM10 and PM2.5 from Mexico City and theoretical assessment of lung exposure. Environ Res 161:439–445. https://doi.org/10.1016/j.envres.2017.11.039
Ren L, Lin Z, Liu H, Hu H (2018) Bacteria-mediated phthalic acid esters degradation and related molecular mechanisms. Appl Microbiol Biotechnol 102:1085–1096. https://doi.org/10.1007/s00253-017-8687-5
Rhee GS, Kim SH, Kim SS, Sohn KH, Kwack SJ, Kim BH, Park KL (2002) Comparison of embryotoxicity of ESBO and phthalate esters using an in vitro battery system. Toxicol In Vitro 16:443–448
Rocheleau H, Al-Harthi R, Ouellet T (2019) Degradation of salicylic acid by Fusarium graminearum. Fungal Biol 123:77–86. https://doi.org/10.1016/j.funbio.2018.11.002
Stackebrandt E, Liesack W (1993) Nucleic acids and classification. In: Goodfellow M, O’Donnell AG (eds) Handbook of new bacterial systematics. Academic Press, London
Staples CA, Peterson DR, Parkerton TF, Adams WJ (1997) The environmental fate of phthalate esters: a literature review. Chemosphere 35:667–749
Staples CA, Parkerton TF, Peterson DR (2000) A risk assessment of selected phthalate esters in North American and Western European surface waters. Chemosphere 40:885–891
Talsness CE, Andrade AJM, Kuriyama SN, Taylor JA, vom Saal FS (2009) Components of plastic: experimental studies in animals and relevance for human health. Philos Trans R Soc b 364:2079–2096. https://doi.org/10.1098/rstb.2008.0281
Tamura K, Stecher G, Kumar S (2021) MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38:3022–3027. https://doi.org/10.1093/molbev/msab120
Tang WJ, Zhang LS, Fang Y, Zhou Y, Ye BC (2016) Biodegradation of phthalate esters by newly isolated Rhizobium sp. LMB-1 and its biochemical pathway of di-n-butyl phthalate. J Appl Microbiol 121:177–186. https://doi.org/10.1111/jam.13123
Tran BC, Teil MJ, Blanchard M, Alliot F, Chevreuil M (2015a) BPA and phthalate fate in a sewage network and an elementary river of France. Influence of hydroclimatic conditions. Chemosphere 119:43–51. https://doi.org/10.1016/j.chemosphere.2014.04.036
Tran BC, Teil MJ, Blanchard M, Alliot F, Chevreuil M (2015b) Fate of phthalates and BPA in agricultural and non-agricultural soils of the Paris area (France). Environ Sci Pollut Res Int 22:11118–11126. https://doi.org/10.1007/s11356-015-4178-3
Tran HT, Lin C, Bui XT, Itayama T, Dang BT, Cheruiyot NK, Hoang HG, Vu CT (2021) Bacterial community progression during food waste composting containing high dioctyl terephthalate (DOTP) concentration. Chemosphere 265:129064. https://doi.org/10.1016/j.chemosphere.2020.129064
Tran HT, Lin C, Bui X-T, Ky NM, Dan Thanh Cao N, Mukhtar H, Giang HH, Varjani S, Hao NH, Nghiem LD (2022) Phthalates in the environment: characteristics, fate and transport, and advanced wastewater treatment technologies. Biores Technol 344:126249. https://doi.org/10.1016/j.biortech.2021.126249
VanWezel AP, van Vlaardingen P, Posthumus R, Crommentuijn GH, Sijm DTHM (2000) Environmental risk limits for two phthalates, with special emphasis on endocrine disruptive properties. Ecotox Environ Saf 46:305–321
Wang J (2004) Effect of di-n-butyl phthalate (DBP) on activated sludge. Process Biochem 39:1831–1836
Wang JL, Chen LJ, Shi HC, Qian Y (2000) Microbial degrdation of phthalic acid esters under anaerobic digestion of sludge. Chemosphere 41:1245–1248
Wang Y, Zhu H, Kannan K (2019) A review of biomonitoring of phthalate exposures. Toxics. https://doi.org/10.3390/toxics7020021
Wen ZD, Gao DW, Wu WM (2014) Biodegradation and kinetic analysis of phthalates by an Arthrobacter strain isolated from constructed wetland soil. Appl Microbiol Biotechnol 98:4683–4690. https://doi.org/10.1007/s00253-014-5568-z
Xu J, Lu Q, de Toledo RA, Shim H (2017) Degradation of di-2ethylhexyl phthalate (DEHP) by an indigenous isolatae Acinetobacter sp. SN13. Int Biodeter Biodegr 117:205–214
Yang T, Ren L, Jia Y, Fan S, Wang J, Wang J, Nahurira R, Wang H, Yan Y (2018) Biodegradation of Di-(2-ethylhexyl) phthalate by Rhodococcus ruber YC-YT1 in contaminated water and soil. Int J Environ Res Public Health. https://doi.org/10.3390/ijerph15050964
Yuan SY, Liu C, Liao CS, Chang BV (2002) Occurrence and microbial degradation of phthalate esters in Taiwan river sediments. Chemosphere 49:1295–1299
Zhao HM, Hu RW, Chen XX, Chen XB, Lü H, Li YW, Li H, Mo CH, Cai QY, Wong MH (2018) Biodegradation pathway of di-(2-ethylhexyl) phthalate by a novel Rhodococcus pyridinivorans XB and its bioaugmentation for remediation of DEHP contaminated soil. Sci Total Environ 640–641:1121–1131. https://doi.org/10.1016/j.scitotenv.2018.05.334
Zheng Z, He PJ, Shao LM, Lee DJ (2007) Phthalic acid esters in dissolved fractions of landfill leachates. Water Res 41:4696–4702. https://doi.org/10.1016/j.watres.2007.06.040
Acknowledgements
We thank Dr. Pei-Jen Chen, and Dr. Nai-Chun Lin for technique help. We thank Dr. Yang-Hsin Shih for the suggestions. We also acknowledge the mass spectrometry technical research services from NTU consortia of Key Technologies.
Funding
This study was funded by the Ministry of Science and Technology, Taiwan, R.O.C (MOST-104-2320-B-002-038).
Author information
Authors and Affiliations
Contributions
YSH, YHL and WFW conceived and designed research. YSH, YHL and CHL conducted experiments. CHT and WFW wrote the manuscript. CHT and WFW analyzed and interpreted the data. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no financial or non-financial competing interests.
Research involving human participants or animals
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hsu, YS., Liu, YH., Lin, CH. et al. Dual bio-degradative pathways of di-2-ethylhexyl phthalate by a novel bacterium Burkholderia sp. SP4. World J Microbiol Biotechnol 39, 44 (2023). https://doi.org/10.1007/s11274-022-03490-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-022-03490-3