Abstract
The polyethylene terephthalate (PET) is one of the major plastics with a huge annual production. Alongside with its mass production and wide applications, PET pollution is threatening and damaging the environment and human health. Although mechanical or chemical methods can deal with PET, the process suffers from high cost and the hydrolyzed monomers will cause secondary pollution. Discovery of plastic-degrading microbes and the corresponding enzymes emerges new hope to cope with this issue. Combined with synthetic biology and metabolic engineering, microbial cell factories not only provide a promising approach to degrade PET, but also enable the conversion of its monomers, ethylene glycol (EG) and terephthalic acid (TPA), into value-added compounds. In this way, PET wastes can be handled in environment-friendly and more potentially cost-effective processes. While PET hydrolases have been extensively reviewed, this review focuses on the microbes and metabolic pathways for the degradation of PET monomers. In addition, recent advances in the biotransformation of TPA and EG into value-added compounds are discussed in detail.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
With the development of material science and chemical industry, polyesters have been widely used in almost every aspect of our lives, due to their perfect properties such as light weight, low maintenance requirements, weathering resistance, low toxicity, transparency, and low production costs (Geyer et al. 2017). Polyethylene terephthalate (PET), with ethylene glycol (EG) and terephthalic acid (TPA) as monomers, is one of the most-consumed synthetic polymers, with wide applications in manufacturing bottles, packaging, and textile industries. Due to its durability and stability, PET degradation is extremely slow in the natural environment, resulting in accumulation and pollution. PET materials are firstly activated by ultraviolet light, followed by the incorporation of oxygen molecules in the polymer (Venkatachalam et al. 2012), and then cracked into smaller debris including microplastics (< 5 mm in diameter) and nanoplastics (< 1 μm). These debris will exist in nature for decades, even hundreds of years (Sang et al. 2020), which can enter tissues and cells to adversely affect the physiological functions, such as provoking immune and stress responses and inducing reproductive and developmental toxicity (Blackburn and Green 2022). What’s more, additives or pollutants, such as di(2-ethyl-hexyl) phthalate (DEHP), can be released from the debris continuously, which has been found to be potentially carcinogenic and estrogenic in the long term (Li et al. 2021). Considering enormous PET waste leads to serious environmental pollution and has become a threat to global ecosystem and human health (Sinha et al. 2008; Rochman et al. 2013; Koelmans et al. 2014; Jambeck et al. 2015), there is an increasing demand for dealing with PET wastes.
Nowadays, PET wastes can be recycled by thermochemical or mechanical processes to generate recycled PET (rPET). Thermochemical process cleaves the ester linkage of PET to release a mixture of bis-(2-hydroxyethyl) terephthalate (BHET), monohydroxyethyl terephthalate (MHET), EG, and TPA at high temperature (≈ 450 °C) and high pressure (≈ 15 MPa) (Kim et al. 2021). Although monomers can be recycled theoretically, the cost for purification and downstream operations is rather high. Thus, its wide applications are hindered by the high processing cost (Sinha et al. 2008; Damayanti and Wu 2021). At the same time, toxic byproducts are generated during the process and result in secondary pollution (Webb et al. 2013; Dzięcioł and Trzeszczyński 2015). Mechanical process is another commonly employed recycling method (Khalid et al. 2022). Unfortunately, the plastic properties are significantly deteriorated after a number of processing cycles, leading to decreased commercial values and limited rPET applications (Sinha et al. 2008; Koelmans et al. 2014). Moreover, the temperatures used for both processes are often higher than 200oC, indicating high energy input and inhibitory cost. The relatively low cost of virgin PET stimulates the demand of PET production capacity and makes the recycling processes not economically competitive (Kubowicz and Booth 2017).
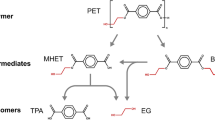
Although PET is rather stable and generally regarded as non-biodegradable, the accumulation of PET in the environment provides a unique niche for the evolution of microorganisms for the degradation of PET polymer and subsequent utilization of PET monomers for cell growth (Kawai et al. 2019; Perrin et al. 2020; Gambarini et al. 2021). In recent years, many PET-degrading microbes and enzymes have been isolated and studied in depth, bringing new hopes for dealing with PET accumulation (Roth et al. 2014; Wei et al. 2014; Yoshida et al. 2016). Müller et al. (2005) firstly reported a lipase, TfH from Thermobifida fusca, with the capacity to hydrolyze PET films, which opens a new era for PET biodegradation. In 2016, Yoshida et al. (2016) isolated a PET-degrading bacterial strain, Ideonella sakaiensis 201-F6, and characterized the corresponding PET. The PET hydrolases mainly act on the ester bond of PET, hydrolyzing it into BHET, MHET, TPA, and EG (Fig. 1). As incomplete hydrolyzation products, BHET can be further degraded by PET hydrolases, while MHET is hydrolyzed by MHETase to release TPA and EG. Compared with the traditional chemical and mechanical processes, biodegradation can be performed under mild and environment-friendly conditions with limited energy input, representing a promising approach for the complete degradation and even up-recycling of post-consumer PET materials in near future (Webb et al. 2013).
Schematic diagram for the degradation and biotransformation of PET. PET hydrolase catalyzes the hydrolytic cleavage of PET to produce BHET, MHET, and monomers (TPA and EG). The monomers could be used to produce high-value compounds, including GA (gallic acid), VA (vanillic acid), and PCA (protocatechuate)
Different with PET degrading enzymes that have been studied in detail (Sulaiman et al. 2012; Roth et al. 2014; Liu et al. 2019; Bollinger et al. 2020; Tournier et al. 2020), the utilization or degradation of EG and TPA is largely overlooked. As TPA and EG are not natural carbon and energy sources, they cannot be efficiently degraded and/or utilized by microorganisms. In other words, the degradation of PET polymer will result in the accumulation of TPA and EG, representing a secondary pollution to the environment. In recent years, metabolic engineering and directed evolution strategies have been performed to engineer strains for complete degradation of TPA and EG into harmless compounds, i.e. CO2 or CH4 (Lykidis et al. 2011; Mückschel et al. 2012; Wu et al. 2013; Nobu et al. 2015; Clarkson et al. 2017; Li et al. 2019; Pardo et al. 2020). While complete degradation is preferred for dealing with environmental PET wastes (particularly those micro-plastics in soil and ocean), there is a growing interest in converting PET monomers into value-added compounds using synthetic biology approaches. The idea of “turn trash to cash”, rather than complete degradation, is not only helpful to offset the processing cost, but also beneficial for the development of green economy (Wang et al. 2021).
While there have been plenty of reviews summarizing the discovery, characterization, and molecular engineering of PET hydrolases, this manuscript focuses on recent progress in the metabolism (including complete degradation and valorization) of PET monomers (Kawai et al. 2019; Mohanan et al. 2020; Urbanek et al. 2020; Amobonye et al. 2021; Gao et al. 2021). This review starts with the introduction of TPA and EG degrading microbes, followed by detailed discussion of the degradation pathways of PET monomers. Recent advances in the biotransformation of PET monomers into value-added compounds are also summarized. This review is concluded with future perspectives in PET biodegradation and valorization.
Microbes for the degradation of TPA and EG
TPA-degrading microbes
As early as 1970s, sludge has been employed to treat TPA-containing wastewater (Kleerebezem et al. 1997; Lykidis et al. 2011). Although the degradation rate is far from satisfactory, sludge becomes an important source for the isolation of microbes or microbial populations responsible for the degradation of phthalate isomers, including TPA. Wu et al. (2001) analyzed the microbial composition in a TPA-degrading granular sludge system and characterized novel δ-Proteobacterial group to be responsible for degrading terephthalate to acetate and hydrogen, which were finally degraded into methane via cooperation with the members of Methanobacteriaceae. A TPA-degrading bacterium was isolated from the enriched culture and designated as strain JT, belonging to Desulfotomaculum lineage I. Unfortunately, as a syntrophic bacterium depending strictly on the presence of hydrogen (and/or formate)-consuming partner organisms, strain JT could not work alone and should be cocultured with a hydrogenotrophic methanogen (e.g. Methanospirillum hungatei) to utilize TPA as well as other aromatic compounds (Qiu et al. 2004). Chen et al. (2004) also reported the similar TPA-degrading methanogenic consortium in an anaerobic hybrid reactor.
Although microbial community is capable of TPA degradation, the reaction rate is rather low and the corresponding mechanism is not clear. Thus, scientists have been devoted to isolating TPA-degrading strains from the TPA contaminated wastewater and polluted soil (Table 1). Karegoudar and Pujar (1985) reported a gram-positive bacterium with the ability to utilize terephthalic acid as sole carbon source. Comamonas testosteroni T-2 was reported to degrade TPA into 1,2-dihydroxy-3,5-cyclohexadiene-1,4-dicarboxylic acid (DCD) and subsequently to protocatechuate (PCA) (Schlafli et al. 1994). Afterwards, Comamonas sp. strain E6 was found to be able to utilize TPA as the sole carbon source (Sasoh et al. 2006).
Hara et al. (2007) confirmed that Rhodococcus sp. strain RHA1 degraded TPA via the PCA pathway and catechol pathway. Kenny et al. (2008) isolated three strains of the Pseudomonas genus that could utilize TPA to accumulate polyhydroxyalkanoate (PHA). Zhang et al. (2013) found that Arthrobacter sp. 0574, a gram-positive strain, was capable of growing aerobically on TPA as the sole carbon and energy source. Recently, Liu et al. (2018) isolated a strain WL-3, belonging to the Delftia genus, with the ability to hydrolyze diethyl terephthalate (DET) and subsequently degrade Narancic et al. (2021) sequenced and analyzed the genome of Pseudomonas umsongensis GO16, which could degrade PET-derived TPA into PCA and subsequently to medium and short chain length PHA.
EG-degrading microbes
Sludge is also a good source to isolate functional microbes for the degradation of EG (Table 1). As early as 1963, Gaston and Stadtman (1963) isolated an anaerobic organism, Clostridium glycolicum sp. N, from mud, which could utilize EG as the energy and carbon source. Dwyer and Tiedje reported the methanogenic enrichments, capable of converting EG to acetate, ethanol, and methane (Dwyer and Tiedje 1983). Another EG-fermenting anaerobe was isolated and identified as Acetobacterium woodii (strain Gra EG 12) (Schink and Stieb 1983), which was subsequently demonstrated to dehydrate EG to acetaldehyde and further disproportionated to ethanol and acetyl-CoA (Trifunović et al. 2016). Kataoka et al. (2001) screened microorganisms capable of oxidizing EG to glycolate, and obtained several yeasts and acetic acid bacteria with high efficiency. Among these isolates, Pichia naganishii AKU 4267 showed the accumulation of the highest concentration of glycolate (Kataoka et al. 2001). Another soil strain, Burkholderia sp. EG13 was also found to possess high EG-oxidizing activity to produce glycolate (Gao et al. 2014). Mückschel et al. (2012) investigated the metabolism of EG in Pseudomonas and found that P. putida JM37 grew rapidly with EG as a sole source of carbon and energy, while P. putida KT2440 could not grow within 2 days of incubation under the same conditions.
Degradation pathways of PET monomers
With the isolation of microbes for the degradation of TPA and EG, the corresponding degradation pathways have been gradually elucidated afterwards. Similar to the oxygen sensitivity of the PET monomer degradation microbes, aerobic and anaerobic degradation pathways are discovered for both TPA and EG,
TPA degradation pathways
As for the degradation of TPA under aerobic conditions, pathways in Comamonas have been studied in depth. Sasoh et al. (2006) isolated a gram-negative bacterium Comamonas sp. strain E6 with the capacity to utilize TPA as the sole carbon source and described the TPA degradation gene clusters. The pathway encoded by the tph genes converted TPA into PCA, a key metabolic intermediate for central metabolism. Two hydroxyl groups are added on positions 4 and 5 by TPA dioxygenase TphA1A2A3 to produce 1,2-dihydroxy-3,5-cyclohexadiene-1,4-dicarboxylate (DCD). Subsequently, the DCD dehydrogenase TphB removes the carboxyl group in position 6 (Fig. 2). In addition to these tph genes, TphR (transcriptional factor regulating the induction of TPA degradation pathway genes) and TphC (TPA binding protein) are also involved in TPA metabolism (Kasai et al. 2010; Hosaka et al. 2013). Subsequent studies showed that there are three characterized pathways for degrading PCA, including 4,5-cleavage pathway, 2,3-cleavage pathway and 3,4-cleavage pathway (Harwood et al. 1996; Masai et al. 2007; Kasai et al. 2009). Clarkson et al. (2017) constructed and optimized a heterologous 3,4-cleavage pathway for PCA catabolism in E. coli via rational and evolutionary approaches. The optimized strain showed significantly increased growth rate with PCA as the sole carbon source, serving as a platform for further reconstruction of catabolic pathways for aromatic compounds. A similar TPA degradation pathway as that of Comamonas has been described in the gram-positive bacteria Rhodococcus genus, with PCA as the key intermediate. Hara et al. (2007) performed transcriptomic analysis of Rhodococcus sp. Strain RHA1 and found that not only PCA pathway genes were induced by TPA, but also the catechol pathway. These results indicated that the catechol branch of 3-oxoadipate pathway is another route for the degradation of TPA (Fig. 2), which have been described in several other bacteria as well (Choi et al. 2005; Martínková et al. 2009). Recently, Pardo et al. (2020) introduced tphCA2A3BA1 and tpiBA from Comamonas sp. E6 to the PCA-utilization bacterium Acinetobacter baylyi ADP1, and finally obtained strains with the capability to utilize TPA as sole carbon source by combining with adaptive laboratory evolution.
Metabolic pathways for TPA degradation under anaerobic and aerobic conditions. Aerobic degradation begins with the conversion of TPA to protocatechuic acid (PCA). Blue gridline presents aerobic TPA degrading pathway in Rhodococcus sp. Strain RHA1 and Comamonas sp. strain E6. The orange gridline presents anaerobic degradation pathway in Syntrophorhabdus aromaticivorans strain UI and Pelotomaculum spp. TphA1A2A3, TPA dioxygenase; TphB, DCD dehydrogenase; CatA, catechol 1,2-dioxygenase; CatB, muconate cycloisomerase; CatC, muconolactone isomerase; PcaHG, protocatechuate 3,4-dioxygenase; PcaB, 3-carboxy-cis,cis-muconate cycloisomerase; PcaL, γ-carboxy-muconolactone decarboxylase/β-ketoadipate enol-lactone hydrolase; PcaIJ, β-ketoadipate succinyl-CoA transferase; PcaF, β-ketoadipyl CoA thiolase; BCR, benzoyl-CoA reductase; CH/3-HBD, crotonyl-CoA hydratase/(S)-3-hydroxy butyryl-CoA dehydrogenase; β-HBD, β-hydroxy butyryl-CoA dehydrogenase
In addition to TPA degradation enzymes, TPA transportation also attracts increasing attention. As the first key step, cellular uptake of TPA is vital for TPA degradation (Rosa et al. 2018). Though TPA could enter the cells by passive diffusion at high concentration, active transport is essential to improve the rate and efficiency (Sasoh et al. 2006; Hosaka et al. 2013). TphC, a TPA binding protein, plays an important role TPA uptake and metabolism (Kasai et al. 2010; Hosaka et al. 2013). It is predicted to be a solute-binding protein (SBP), which belongs to the tripartite tricarboxylate transporters (TTT) class of transporters (Sasoh et al. 2006; Rosa et al. 2018). Hosaka et al. (2013) demonstrated that TpiA-TpiB membrane transporter encoded by tpiBA together with TphC were essential for the uptake of TPA. Most recently, Gautom et al. (2021) carried out biochemical and structural characterization of TphC, demonstrating the narrow ligand specificity of TphC towards aromatic para-substituted dicarboxylates. In another work, the endogenous muconate transporter MucK rather than heterologous proteins from Comamonas sp. E6 mediated TPA transportation, indicating the TPA uptake system as a key point of metabolic engineering on TPA utilization (Pardo et al. 2020). P. putida KT2440 also possesses relevant uptake systems to utilize aromatic substrates. Wada et al. (2021) demonstrated that three transporters, PP_1376 (PcaK), PP_3349 (HcnK) and VanK, were aromatic acid/H+ symporter family transporters categorized into major facilitator superfamily, which have ability to take up aromatic substrates. PcaK, HcnK, and VanK were confirmed with ability to take up PCA, ferulate/4-coumarate, and vanillate/PCA, respectively.
On the other hand, TPA could also be metabolized through anaerobic methanogenic processes. Completely different with the aerobic pathways, the anaerobic pathway converted TPA to CH4 and/or CO2 as the final products (Kleerebezem and Lettinga 2000). In Syntrophorhabdus aromaticivorans UI, TPA is degraded via the benzoyl-CoA degradation pathway (Fig. 2) (Wu et al. 2013; Nobu et al. 2015). A series of genes encoding decarboxylases, hydratase, dehydrogenase, hydrolase, and acetyltransferase are involved in this process to achieve TPA complete biodegradation (Wu et al. 2013). In addition, microbial syntrophy could also degrade TPA under moderate condition. Jer-Horng Wu et al. (2001) proved that δ-Proteobacteria was presumed to be the primary population responsible for degrading TPA to acetate and hydrogen, and associated with different methanogenic populations to convert acetate, hydrogen, and carbon dioxide into methane. In 2013, Wu et al. (2013) reported that Pelotomaculum, Methanosaeta, and Methanolinea were predominant in TPA-degrading biofilms. Among three bacteria, Pelotomaculum spp. degraded TPA and intermediate by-products could support other microbial populations to grow. The intermediate by-products, including acetate, H2/CO2, and butyrate, were produced during the process, which could support the growth of methanogens and be used as substrates to produce CH4.
EG degradation pathways
Although EG has broad applications, its production and accumulation have caused huge problems for environment and health. On the other hand, EG is a potential carbon source to support the growth of microorganisms. Therefore, there is a growing interest in studying the degradation and utilization of EG as a carbon source.
In aerobic bacterial species, EG is degraded with the formation of glycolate as an important intermediate, which is subsequently incorporated into the TCA cycle for complete degradation. In the glycolate utilization microorganisms, such Pseudomonas and E. coli, glycolate is converted into intermediates of the central metabolism through different pathways. Mückschel et al. (2012) reported that P. putida strain JM37 could grow with EG rapidly to enable biomass formation, while another strain, P. putida KT2440, could only utilize EG very slowly. Currently, P. putida KT2440 has been considered as a good platform for EG degradation, and its genes involved in EG metabolism are also identified by genome analysis (Narancic et al. 2021). Two periplasmic quinoproteins PedE (PP_2674) and PedH (PP_2679) oxidize EG to glycolaldehyde, followed by cytoplasmic aldehyde dehydrogenases PedI (PP_2680) and PP_0545 to catalyze glycolaldehyde into glycolate (Vallon et al. 2015). Subsequently, membrane anchored oxidase GlcDEF oxidizes glycolate to glyoxylate, which flows into the catabolic pathways for complete degradation (Wehrmann et al. 2017; Narancic et al. 2021). In the conversion process, pyrroloquinoline quinone (PQQ) coupling with the electron transport chains is necessary (Duine and Jongejan 1989). Glyoxylate could be further metabolized by flowing into the TCA cycle (Fig. 3) (Franden et al. 2018). Blank et al. (2008) reported that aceA (isocitrate lyase) or glcB (malate synthase) of the glyoxylate shunt is responsible for further utilization of glyoxylate. In this shunt, C2 substrates flow into carbon conservation pathway to be metabolized at the level of acetyl-CoA. Alternatively, the glyoxylate carboligase (Gcl) enzyme of P. putida KT2440 could condense two molecules of glyoxylate into tartronate semialdehyde, followed by the conversion to glycerate and 2-phosphoglycerate, important glycolysis intermediates (Franden et al. 2018). Although P. putida possesses all the genes required for EG conversion, the carbon conservation pathway is not efficient enough for the utilization of glyoxylate as a sole carbon source. Therefore, Li et al. (2019) isolated P. putida KT2440 mutants which used EG as a sole source of carbon and energy through adaptive laboratory evolution. The authors revealed a central role of the transcriptional regulator, GclR, which represses the glyoxylate carboligase pathway, in EG and glyoxylate utilization. In addition, the growth ability of the evolved strains was further improved through the introduction of secondary mutations in a transcriptional regulator encoded by PP_2046 and a porin encoded by PP_2662. Finally, an efficient EG utilization strain was constructed by reverse engineering.
Metabolic pathways for EG degradation under anaerobic and aerobic conditions. The red pathway presents the aerobic degradation pathway, while the blue for anaerobic pathway. The gray gridline presents the possible routes of glyoxylate metabolism. PedE and PedF, PQQ-dependent methanol/ethanol family dehydrogenase; PedI and PP_0545, cytoplasmic aldehyde dehydrogenases; GlcDEF, glycolate dehydrogenase; MS, malate synthase; Gcl, glyoxylate carboligase; GlxR, tartronate semialdehyde reductase; Hyi, hydroxypyruvate isomerase; PduCDE, diol dehydratase; PduP, propionaldehyde dehydrogenase; ALDH, alcohol dehydrogenase
As E. coli is considered as preferred chassis for both fundamental studies and biotechnological applications, there is a growing interest in engineering E. coli for EG utilization. Unfortunately, wild-type E. coli strains could not utilize EG directly. Boronat et al. (1983) obtained strains of E. coli K-12 with capacity to grow on EG as a sole source of carbon and energy via adaptive evolution. The evolved strains converted EG to glycolaldehyde and glycolate via increasing activities of propanediol oxidoreductase and glycolaldehyde dehydrogenase. Afterwards, glycolate was catalyzed by glycolate oxidase to generate glyoxylate, which is further catalyzed by malate synthase or glyoxylate carboligase for complete degradation (Fig. 3) (Barkulis and Krakow 1956). On the other hand, although EG is not a natural substrate for E. coli, 1,2-propanediol oxidoreductase mutant (fucO) could catalyze the oxidation of EG. An engineered E. coli with fucO mutant (I7L/L8V) and aldA can utilize EG to produce glycolate (Pandit et al. 2021).
The anaerobic acetogenic bacteria are another class of important microorganisms to use EG as carbon source, whose metabolic pathway is different from those mentioned above (Schink and Stieb 1983). However, the metabolism of EG in acetogens has not been well studied until recently (Eichler and Schink 1984,1985). Trifunović et al. (2016) elucidated the pathway for EG utilization in A. woodii. EG is initially dehydrated to acetaldehyde, and then is disproportionated to ethanol and acetyl coenzyme A (acetyl-CoA), respectively. Ethanol is in part further oxidized and the reducing equivalents are recycled by reducing CO2 to acetate in the Wood-Ljungdahl pathway. The pdu gene cluster (Awo_c25780 to Awo_c25910) expresses propanediol dehydratase (PduCDE) and CoA-dependent propionaldehyde dehydrogenase (PduP) during growth on EG. The PduC subunit of PduCDE is the only dehydratase produced in EG-growing cells (Schuchmann et al. 2015). PduP, a CoA-dependent aldehyde dehydrogenase, probably oxidizes acetaldehyde to acetyl-CoA for complete degradation (Bertsch et al. 2016). Moreover, genes encoding bacterial microcompartments as part of the pdu gene cluster are also expressed during growth on EG, indicating a dual function (degradation of both propanediol and EG) of the Pdu microcompartment system (Trifunović et al. 2016).
Biotransformation and valorization of PET monomers
Due to the low cost in PET production, there is a lack of motivation in PET recycling (López et al. 2014). The discovery and engineering of PET hydrolases bring new approaches and new hope to initiate PET biotransformation and valorization. Combined with synthetic biology and metabolic engineering, the hydrolyzed PET monomers can be converted to high-value compounds to offset the high cost in the treatment of PET. The PET monomers derived compounds could be used as important components for perfumes, pharmaceuticals, cosmetics, and animal feeds (Fig. 4).
The monomers of PET hydrolysis could be converted into high-value compounds. TPA can be converted to vanillic acid (VA), gallic acid (GA), pyrogallol, and muconic acid (MA) through the intermediate PCA by metabolically engineered microbes. MA could be further converted into different compounds, including polyhydroxyalkanoate (PHA), polyhydroxybutyrate (PHB), hydroxyalkanoyloxy-alkanoate (HAA) β-ketoadipic acid (βKA), and Nylon. EG can be converted to glycolate, glyoxylate, malate, and fumarate. OMT, O-methyltransferase; AroY, protocatechuic acid decarboxylase; CatA, catechol dioxygenase; PobA, para-hydroxybenzoic acid hydroxylase; LpdC, gallate decarboxylase; PhKLMNOPQ, phenol hydroxylase operon
Biotransformation and valorization of TPA
Although TPA is often treated with anaerobic methanogenic processes to produce CH4 and CO2, this process is considered as time-consuming and low value-added (Wu et al. 2001; Lykidis et al. 2011). Even worse, both CH4 and CO2 are greenhouse gases, bringing in secondary environmental concerns. Through metabolically engineered microbes, TPA could be biotransformed into other added-value products, by various enzymes or pathways (Furtwengler and Avérous 2018; Salvador et al. 2019). Kenny et al. (2008) isolated three strains of the Pseudomonas genus, which could utilize TPA for growth and meanwhile accumulate large amount of PHA, a class of renewable and biodegradable polymer. After subsequent treatment, these Pseudomonas species were found to convert the TPA fraction to PHA with a maximal production rate at approximately 8.4 mg·L− 1·h− 1. In following studies, the authors supplied TPA and glycerol waste to P. putida GO16 and the production rate of PHA reached approximately 108.8 mg·L− 1·h− 1 (Kenny et al. 2012).
Kim et al. (2019) engineered E. coli to express necessary metabolic enzymes for converting TPA to PCA, the key precursor metabolite of various high-value aromatic products. Then, by additionally introducing PobAMut and LpdC or AroY and PhKLMNOPQ, pyrogallol, an antioxidant in the oil industry, was biosynthesized via gallic acid (GA) or catechol from TPA with a molar yield of ~ 40% or ~ 20%, respectively. GA is also an important chemical intermediate, which can be further processed into propyl gallate, an antioxidant and anticancer agent (Ng et al. 1994; Saxena et al. 2008; Badhani et al. 2015; Reverón et al. 2017; Brückner et al. 2018). In addition, muconic acid (MA), the precursor of adipic acid and vanillin, was produced from catechol by CatA or from PCA by HsOMTHis, respectively (Fig. 4) (Kim et al. 2019). Sadler and Wallace (2021) designed a 5-step enzymatic pathway in E. coli MG1665 RARE to convert TPA into value-added vanillin (VA). In order to improve TPA uptake, n-butanol was used to increase E. coli cell membrane permeability towards small molecules. Then process optimization was performed to achieve 79% conversion of TPA to VA, representing a 157-fold improvement over initial conditions. Werner et al. (2021) engineered P. putida KT2440 for BHET catabolism and the TPA monomer was converted into PCA. The deletion of pcaIJ resulted in the production of β-ketoadipic acid (βKA) with a titer of 15.1 g/Land a molar yield of 76% from BHET. βKA can react with hexamethyl diamine to produce a polyamide with increased melting temperature and crystallinity while decreased water uptake than nylon 6.6 (Sudarsan et al. 2016; Dissanayake and Jayakody 2021). Tiso et al. 2021) modified P. umsongensis GO16 for the conversion of TPA and EG into the extracellular building block hydroxyalkanoyloxy-alkanoate (HAA) and native intracellular polymer PHA. HAA was used as platform molecule to further produce a novel bio-based poly(amide urethane). Liu et al. (2021) designed a co-cultivation system with PET-degrading and TPA-utilization microorganisms, and then introduced a recombinant plasmid containing the phbCAB operon to achieve polyhydroxybutyrate (PHB) production. Up to now, the conversion of TPA to value-added compounds is still limited to a few examples and there is an urgent need to construct and optimize efficient cell factories for TPA valorization.
Biotransformation and valorization of EG
EG is the second component of PET and used widely in many fields (Pereira et al. 2015). EG released from hydrolysis could be oxidized to glycolaldehyde firstly, and then converted into glycolate or glyoxylate, both of which are important α-C2 carboxylic acids with broad applications (Yue et al. 2012).
Glycolate is an industrial valuable compound, which could be used in chemical industry, food processing, and cosmetics, and its demand has been increasing continuously. Some microbes have been reported to oxidize EG to glycolate, such as P. naganishii and G. oxydans (Kataoka et al. 2001; Wei et al. 2009). Meanwhile, although glycolate is a potential carbon and energy source for microbes, it is not an easy-to-use substrate and only a few microbes could grow on glycolate directly (Hansen and Hayashi 1962; Kornberg and Morris 1965). On the other hand, it is beneficial for its accumulation during EG metabolism. The oxidase-producing strain Burkholderia sp. EG13 could convert EG to glycolate with high efficiency (98.8% yield), which was potential for further investigation and industrial application (Gao et al. 2014). As a model and preferred host for industrial applications, E. coli strains could not grow on glycolate without adaptation (Krakow et al. 1961; Hansen and Hayashi 1962), indicating the potential of E. coli for the production and accumulation of glycolate (Kataoka et al. 2001; Wei et al. 2009; Gao et al. 2014; Deng et al. 2018; Hua et al. 2018). Recently, an engineered E. coli MG1655 can consume EG and produce glycolate by overexpressing endogenous genes fucO and aldA (Pandit et al. 2021). They obtained a yield of 0.8 g/g from EG and productivity of 0.1 g·L− 1·h− 1 during the production stage. The strain, Gluconobacter oxydans, without glycolate downstream pathways, was employed as a whole-cell biocatalyst to produce glycolate from EG (Hua et al. 2018, 2019; Kim et al. 2019).
In addition to glycolate, other intermediates in TCA are the target compounds to shunt EG metabolism. Succinate is a valuable and multipurpose platform chemical, which could be produced by microorganisms from renewable carbon sources (Thakker et al. 2012). It has been listed as one of the twelve platform chemicals from biomass by the US Department of Energy (DOE) (McKinlay et al. 2007). Succinate production from EG is a novel approach to deal with and more importantly valorize the PET monomer. E. coli, an ideal microbial cell factory, has been engineered to produce succinate from different carbon sources (Unden and Bongaerts 1997; Dharmadi et al. 2006; Blankschien et al. 2010; Thakker et al. 2012; Soellner et al. 2013; Kim et al. 2018; Chiang et al. 2021; Zhou et al. 2021). In addition, L-malate with broad applications in beverage, chemical, agricultural, food, and pharmaceutical industries, is also one of the most promising platform chemicals (Rosenberg et al. 1999; Bressler et al. 2002). Similar with succinate, some microorganisms have been developed and engineered to produce L-malate with the fermentation and genetic strategies (Sauer et al. 2008; Zhang et al. 2011; Dong et al. 2017; Gao et al. 2018; Martinez et al. 2018; Jiang et al. 2020).
Summaries and perspectives
Huge PET waste accumulation and inefficient recycling methods result in extensive and global crisis. The discovery of PET hydrolysis and degradation microbes is a great event and brings in new hope to deal with PET waste. As a growing number of PET-degrading enzymes and microorganisms have been isolated, biodegradation not only would contribute to creating a path beyond current recycling methods, but also provide a suitable measure for environment-friendly applications.
Although PET could be degraded and biotransformed by enzymes and microbes, there are still grand challenges to be addressed before practical applications. As for PET biodegradation, low hydrolysis activity results in partial hydrolyzation and oligomer accumulation (Sang et al. 2020). The high crystallinity and surface hydrophobicity of PET hamper enzyme accessibility, leading to preferential degradation of the amorphous regions. In addition, due to the unsatisfactory thermal stability, PET hydrolases perform poorly at glassy transition temperature (~ 72oC). Meanwhile, the degradation and valorization of PET monomers suffer from low rate and efficiency, concerning the issues with TPA uptake and enzymatic cofactor regeneration (Kasai et al. 2010; Hosaka et al. 2013). While there have been a few reports on the conversion of PET monomers to value-added compounds, the successful examples are still rather limited and economic analysis and life cycle analysis on PET waste valorization have not been performed yet. Nevertheless, synthesis biology and metabolic engineering are expected to play increasingly important roles in addressing bottlenecks in microbial degradation and valorization of PET wastes (Keasling 2012). For example, protein engineering combined with computer science will provide strong support to obtain ideal properties of PET hydrolases, monomer transporters, and key biotransformation enzymes to meet industrial expectations. Well-characterized genetic components together with engineered enzymes and transcription factors can be employed to design genetic circuits to construct customized and highly efficient microbial cell factories (Xu et al. 2021; Yu et al. 2021). Moreover, considering the dominance of microbial consortia to perform complex tasks in nature, artificial microbial consortium is another challenging yet promising metabolic engineering strategy for complete degradation of PET and its monomers (Qi et al. 2021; Su et al. 2021).
With deeper understanding of polymer hydrolases, metabolic pathways, and microbial strains, the established system can also be broadened to degrade polymers with similar structures to create revenue while also reducing its release into the environment. In other words, the biodegradation and valorization strategies are not only restricted to PET, but also other polymers. With the development of metabolic engineering and synthetic biology tools, it is expected to observe more successful examples on microbial degradation and valorization of PET and other polymers.
References
Amobonye A, Bhagwat P, Singh S, Pillai S (2021) Plastic biodegradation: Frontline microbes and their enzymes. Sci Total Environ 759:143536. https://doi.org/10.1016/j.scitotenv.2020.143536
Bache R, Pfennig N (1981) Selective isolation of Acetobacterium woodii on methoxylated aromatic acids and determination of growth yields. Arch Microbiol 130(3):255–261. https://doi.org/10.1007/BF00459530
Badhani B, Sharma N, Kakkar R (2015) Gallic acid: a versatile antioxidant with promising therapeutic and industrial applications. RSC Adv 5(35):27540–27557. https://doi.org/10.1039/c5ra01911g
Barkulis SS, Krakow G (1956) Conversion of glyoxylate to hydroxypyruvate by extracts of Escherichia coli. Biochim Biophys Acta Biomembr 21(3):593–594. https://doi.org/10.1016/0006-3002(56)90208-6
Bertsch J, Siemund AL, Kremp F, Müller V (2016) A novel route for ethanol oxidation in the acetogenic bacterium Acetobacterium woodii: the acetaldehyde/ethanol dehydrogenase pathway. Environ Microbiol 18(9):2913–2922. https://doi.org/10.1111/1462-2920.13082
Blackburn K, Green D (2022) The potential effects of microplastics on human health: What is known and what is unknown. Ambio 51(3):518–530. https://doi.org/10.1007/s13280-021-01589-9
Blank LM, Ionidis G, Ebert BE, Bühler B, Schmid A (2008) Metabolic response of Pseudomonas putida during redox biocatalysis in the presence of a second octanol phase. Febs j 275(20):5173–5190. https://doi.org/10.1111/j.1742-4658.2008.06648.x
Blankschien MD, Clomburg JM, Gonzalez R (2010) Metabolic engineering of Escherichia coli for the production of succinate from glycerol. Metab Eng 12(5):409–419. https://doi.org/10.1016/j.ymben.2010.06.002
Bollinger A, Thies S, Knieps-Grunhagen E, Gertzen C, Kobus S, Hoppner A, Ferrer M, Gohlke H, Smits SHJ, Jaeger KE (2020) A novel polyester hydrolase from the marine bacterium Pseudomonas aestusnigri - structural and functional insights. Front Microbiol 11:114. https://doi.org/10.3389/fmicb.2020.00114
Boronat A, Caballero E, Aguilar J (1983) Experimental evolution of a metabolic pathway for ethylene glycol utilization by Escherichia coli. J Bacteriol 153(1):134–139. https://doi.org/10.1128/jb.153.1.134-139.1983
Bressler E, Pines O, Goldberg I, Braun S (2002) Conversion of fumaric acid to L-malic by sol-gel immobilized Saccharomyces cerevisiae in a supported liquid membrane bioreactor. Biotechnol Prog 18(3):445–450. https://doi.org/10.1021/bp010139t
Brückner C, Oreb M, Kunze G, Boles E, Tripp J (2018) An expanded enzyme toolbox for production of cis, cis-muconic acid and other shikimate pathway derivatives in Saccharomyces cerevisia. FEMS Yeast Res 18(2). https://doi.org/10.1093/femsyr/foy017
Chen CL, Macarie H, Ramirez I, Olmos A, Ong SL, Monroy O, Liu W (2004) Microbial community structure in a thermophilic anaerobic hybrid reactor degrading terephthalate. Microbiology 150(10):3429–3440
Chiang CJ, Hu RC, Huang ZC, Chao YP (2021) Production of Succinic Acid from Amino Acids in Escherichia coli. J Agric Food Chem 69(29):8172–8178. https://doi.org/10.1021/acs.jafc.1c02958
Choi KY, Kim D, Sul WJ, Chae JC, Zylstra GJ, Kim YM, Kim E (2005) Molecular and biochemical analysis of phthalate and terephthalate degradation by Rhodococcus sp. strain DK17. FEMS Microbiol Lett 252(2):207–213. https://doi.org/10.1016/j.femsle.2005.08.045
Clarkson SM, Giannone RJ, Kridelbaugh DM, Elkins JG, Guss AM, Michener JK (2017) Construction and optimization of a heterologous pathway for protocatechuate catabolism in Escherichia coli enables bioconversion of model aromatic compounds. Appl Environ Microbiol 83(18):e01313–01317. https://doi.org/10.1128/aem.01313-17
Damayanti, Wu HS (2021) Strategic possibility routes of recycled PET. Polym (Basel) 13(9). https://doi.org/10.3390/polym13091475
Deng Y, Ma N, Zhu K, Mao Y, Wei X, Zhao Y (2018) Balancing the carbon flux distributions between the TCA cycle and glyoxylate shunt to produce glycolate at high yield and titer in Escherichia coli. Metab Eng 46:28–34
Dharmadi Y, Murarka A, Gonzalez R (2006) Anaerobic fermentation of glycerol by Escherichia coli: a new platform for metabolic engineering. Biotechnol Bioeng 94(5):821–829. https://doi.org/10.1002/bit.21025
Dissanayake L, Jayakody LN (2021) Engineering microbes to bio-upcycle polyethylene terephthalate. Front Bioeng Biotechnol 9:656465. https://doi.org/10.3389/fbioe.2021.656465
Dong X, Chen X, Qian Y, Wang Y, Wang L, Qiao W, Liu L (2017) Metabolic engineering of Escherichia coli W3110 to produce L-malate. Biotechnol Bioeng 114(3):656–664. https://doi.org/10.1002/bit.26190
Duine JA, Jongejan JA (1989) Quinoproteins, enzymes with pyrrolo-quinoline quinone as cofactor. Annu Rev Biochem 58:403–426
Dwyer DF, Tiedje JM (1983) Degradation of ethylene glycol and polyethylene glycols by methanogenic consortia. Appl Environ Microbiol 46(1):185–190
Dzięcioł M, Trzeszczyński J (2015) Volatile products of poly(ethylene terephthalate) thermal degradation in nitrogen atmosphere. J Appl Polym Sci 77(9):1894–1901
Eichler B, Schink B (1984) Oxidation of primary aliphatic alcohols by Acetobacterium carbinolicum sp. nov., a homoacetogenic anaerobe. Arch Microbiol 140(2–3):147–152
Eichler B, Schink B (1985) Fermentation of primary alcohols and diols and pure culture of syntrophically alcohol-oxidizing anaerobes. Arch Microbiol 143(1):60–66. https://doi.org/10.1007/BF00414769
Franden MA, Jayakody LN, Li WJ, Wagner NJ, Cleveland NS, Michener WE, Hauer B, Blank LM, Wierckx N, Klebensberger J, Beckham GT (2018) Engineering Pseudomonas putida KT2440 for efficient ethylene glycol utilization. Metab Eng 48:197–207. https://doi.org/10.1016/j.ymben.2018.06.003
Furtwengler P, Avérous L (2018) Renewable polyols for advanced polyurethane foams from diverse biomass resources. Polym Chem 9(32):4258–4287. https://doi.org/10.1039/c8py00827b
Gambarini V, Pantos O, Kingsbury JM, Weaver L, Handley KM, Lear G (2021) Phylogenetic distribution of plastic-degrading microorganisms. mSystems 6(1):e01112–01120. https://doi.org/10.1128/mSystems.01112-20
Gao C, Wang S, Hu G, Guo L, Chen X, Xu P, Liu L (2018) Engineering Escherichia coli for malate production by integrating modular pathway characterization with CRISPRi-guided multiplexed metabolic tuning. Biotechnol Bioeng 115(3):661–672. https://doi.org/10.1002/bit.26486
Gao R, Pan H, Lian J (2021) Recent advances in the discovery, characterization, and engineering of poly(ethylene terephthalate) (PET) hydrolases. Enzyme Microb Technol 150:109868. https://doi.org/10.1016/j.enzmictec.2021.109868
Gao X, Ma Z, Yang L, Ma J (2014) Enhanced bioconversion of ethylene glycol to glycolic acid by a newly isolated Burkholderia sp. EG13. Appl Biochem Biotechnol 174(4):1572–1580. https://doi.org/10.1007/s12010-014-1114-9
Gaston LW, Stadtman ER (1963) Fermentation of ethylene glycol by Clostridium glycolicum, sp. n. J Bacteriol 85(2):356. https://doi.org/10.1111/j.1365-2672.1963.tb04803.x
Gautom T, Dheeman D, Levy C, Butterfield T, Alvarez Gonzalez G, Le Roy P, Caiger L, Fisher K, Johannissen L, Dixon N (2021) Structural basis of terephthalate recognition by solute binding protein TphC. Nat Commun 12(1):6244. https://doi.org/10.1038/s41467-021-26508-0
Geyer R, Jambeck JR, Law KL (2017) Production, use, and fate of all plastics ever made. Sci Adv 3(7):e1700782. https://doi.org/10.1126/sciadv.1700782
Hansen RW, Hayashi JA (1962) Glycolate metabolism in Escherichia coli. J Bacteriol 83(3):679–687. https://doi.org/10.1128/jb.83.3.679-687.1962
Hara H, Eltis LD, Davies JE, Mohn WW (2007) Transcriptomic analysis reveals a bifurcated terephthalate degradation pathway in Rhodococcus sp. strain RHA1. J Bacteriol 189(5):1641–1647. https://doi.org/10.1128/JB.01322-06
Harwood CS, And, Parales RE (1996) The β-ketoadipate pathway and the biology of self-identity. Annu Rev Microbiol 50:553–590. https://doi.org/10.1146/annurev.micro.50.1.553
Hosaka M, Kamimura N, Toribami S, Mori K, Kasai D, Fukuda M, Masai E (2013) Novel tripartite aromatic acid transporter essential for terephthalate uptake in Comamonas sp. strain E6. Appl Environ Microbiol 79(19):6148–6155. https://doi.org/10.1128/AEM.01600-13
Hua X, Du G, Xu Y (2019) Cost-practical of glycolic acid bioproduction by immobilized whole-cell catalysis accompanied with compressed oxygen supplied to enhance mass transfer. Bioresour Technol 283:326–331. https://doi.org/10.1016/j.biortech.2019.03.094
Hua X, Zhou X, Xu Y (2018) Improving techno-economics of bioproduct glycolic acid by successive recycled-cell catalysis of ethylene glycol with Gluconobacter oxydans. Bioprocess Biosyst Eng 41(10):1–5
Jambeck JR, Geyer R, Wilcox C, Siegler TR, Perryman M, Andrady A, Narayan R, Law KL (2015) Plastic waste inputs from land into the ocean. Science 347(6223):768–771
Jiang Y, Zheng T, Ye X, Xin F, Zhang W, Dong W, Ma J, Jiang M (2020) Metabolic engineering of Escherichia coli for L-malate production anaerobically. Microb Cell Fact 19(1):165. https://doi.org/10.1186/s12934-020-01422-0
Karegoudar TB, Pujar BG (1985) Degradation of terephthalic acid by a Bacillus species. FEMS Microbiol Lett 30(1–2):217–220
Kasai D, Fujinami T, Abe T, Mase K, Katayama Y, Fukuda M, Masai E (2009) Uncovering the protocatechuate 2,3-cleavage pathway genes. J Bacteriol 191(21):6758–6768
Kasai D, Kitajima M, Fukuda M, Masai E (2010) Transcriptional regulation of the terephthalate catabolism operon in Comamonas sp. strain E6. Appl Environ Microbiol 76(18):6047–6055. https://doi.org/10.1128/aem.00742-10
Kataoka M, Sasaki M, Hidalgo A R G D, Nakano M, Shimizu S (2001) Glycolic acid production using ethylene glycol-oxidizing microorganisms. J Agricultural Chem Soc Japan 65(10):2265–2270
Kawai F, Kawabata T, Oda M (2019) Current knowledge on enzymatic PET degradation and its possible application to waste stream management and other fields. Appl Microbiol Biotechnol 103(11):4253–4268. https://doi.org/10.1007/s00253-019-09717-y
Keasling JD (2012) Synthetic biology and the development of tools for metabolic engineering. Metab Eng 14(3):189–195. https://doi.org/10.1016/j.ymben.2012.01.004
Kenny ST, Runic JN, Kaminsky W, Woods T, Babu RP, Keely CM, Blau W, O’connor KE (2008) Up-cycling of PET (polyethylene terephthalate) to the biodegradable plastic PHA (polyhydroxyalkanoate). Environ Sci Technol 42(20):7696–7701
Kenny ST, Runic JN, Kaminsky W, Woods T, Babu RP, O’connor KE (2012) Development of a bioprocess to convert PET derived terephthalic acid and biodiesel derived glycerol to medium chain length polyhydroxyalkanoate. Appl Microbiol Biotechnol 95(3):623–633. https://doi.org/10.1007/s00253-012-4058-4
Khalid MY, Arif ZU, Ahmed W, Arshad H (2022) Recent trends in recycling and reusing techniques of different plastic polymers and their composite materials. Sustainable Mater Technol 31. https://doi.org/10.1016/j.susmat.2021.e00382
Kim HT, Hee Ryu M, Jung YJ, Lim S, Song HM, Park J, Hwang SY, Lee HS, Yeon YJ, Sung BH, Bornscheuer UT, Park SJ, Joo JC, Oh DX (2021) Chemo-biological upcycling of poly(ethylene terephthalate) to multifunctional coating materials. Chemsuschem 14(19):4251–4259. https://doi.org/10.1002/cssc.202100909
Kim HT, Kim JK, Cha HG, Kang MJ, Lee HS, Khang TU, Yun EJ, Lee D-H, Song BK, Park SJ, Joo JC, Kim KH (2019) Biological valorization of poly(ethylene terephthalate) monomers for upcycling waste PET. ACS Sustain Chem & Eng 7(24):19396–19406. https://doi.org/10.1021/acssuschemeng.9b03908
Kim NY, Kim SN, Kim OB (2018) Long-term adaptation of Escherichia coli to methanogenic co-culture enhanced succinate production from crude glycerol. J Ind Microbiol Biotechnol 45(1):71–76. https://doi.org/10.1007/s10295-017-1994-0
Kleerebezem R, Lettinga G (2000) High-rate anaerobic treatment of purified terephthalic acid wastewater. WATER SCI TECHNOL 42(5–6):259–268
Kleerebezem R, Mortier J, Hulshoff Pol LW, Lettinga G (1997) Anaerobic pre-treatment of petrochemical effluents: terephthalic acid wastewater. WATER SCI TECHNOL 36(2–3):237–248
Koelmans AA, Gouin T, Thompson R, Wallace N, Arthur C (2014) Plastics in the marine environment. Environ Toxicol Chem 33(1):5–10. https://doi.org/10.1002/etc.2426
Kornberg HL, Morris JG (1965) The utilization of glycollate by Micrococcus denitrificans: the p-hydroxyaspartate pathway. Biochem J 95(3):577–586. https://doi.org/10.1042/bj0950577
Krakow G, Barkulis SS, Hayashi JA (1961) Glyoxylic acid carboligase: an enzyme present in glycolate-grown Escherichia coli. J Bacteriol 81(4):509–518. https://doi.org/10.1128/jb.81.4.509-518.1961
Kubowicz S, Booth AM (2017) Biodegradability of plastics: challenges and misconceptions. Environ Sci Technol 51(21):12058–12060. https://doi.org/10.1021/acs.est.7b04051
Li WJ, Jayakody LN, Franden MA, Wehrmann M, Daun T, Hauer B, Blank LM, Beckham GT, Klebensberger J, Wierckx N (2019) Laboratory evolution reveals the metabolic and regulatory basis of ethylene glycol metabolism by Pseudomonas putida KT2440. Environ Microbiol 21(10):3669–3682. https://doi.org/10.1111/1462-2920.14703
Li X, Han X, Vogt RD, Zhou J, Zheng B, Song Y, Lu X (2021) Distributions, temporal trends and ecological risks of polyethylene terephthalate (PET) and di-(2-ethylhexyl) phthalate (DEHP) in sediments of Jiaozhou Bay, China. Mar Pollut Bull 165:112176. https://doi.org/10.1016/j.marpolbul.2021.112176
Liu C, Shi C, Zhu S, Wei R, Yin CC (2019) Structural and functional characterization of polyethylene terephthalate hydrolase from Ideonella sakaiensis. Biochem Biophys Res Commun 508(1):289–294. https://doi.org/10.1016/j.bbrc.2018.11.148
Liu J, Xu G, Dong W, Xu N, Xin F, Ma J, Fang Y, Zhou J, Jiang M (2018) Biodegradation of diethyl terephthalate and polyethylene terephthalate by a novel identified degrader Delftia sp. WL-3 and its proposed metabolic pathway. Lett Appl Microbiol 67(3):254–261. https://doi.org/10.1111/lam.13014
Liu P, Zhang T, Zheng Y, Li Q, Su T, Qi Q (2021) Potential one-step strategy for PET degradation and PHB biosynthesis through co-cultivation of two engineered microorganisms. Eng Microbiol 1:100003. https://doi.org/10.1016/j.engmic.2021.100003
López M, Pernas AA, López MA, Latorre AL, Vilariño JL, Rodríguez MG (2014) Assessing changes on poly(ethylene terephthalate) properties after recycling: Mechanical recycling in laboratory versus postconsumer recycled material. Mater Chem Phys 147(3):884–894
Lykidis A, Chen CL, Tringe SG, Mchardy AC, Copeland A, Kyrpides NC, Hugenholtz P, Macarie H, Olmos A, Monroy O, Liu WT (2011) Multiple syntrophic interactions in a terephthalate-degrading methanogenic consortium. Isme j 5(1):122–130. https://doi.org/10.1038/ismej.2010.125
Martinez I, Gao H, Bennett GN, San KY (2018) High yield production of four-carbon dicarboxylic acids by metabolically engineered Escherichia coli. J Ind Microbiol Biotechnol 45(1):53–60. https://doi.org/10.1007/s10295-017-1991-3
Martínková L, Uhnáková B, Pátek M, Nesvera J, Kren V (2009) Biodegradation potential of the genus Rhodococcus. Environ Int 35(1):162–177. https://doi.org/10.1016/j.envint.2008.07.018
Masai E, Katayama Y, Fukuda M (2007) Genetic and biochemical investigations on bacterial catabolic pathways for lignin-derived aromatic compounds. Biosci Biotech Bioch 71(1):1–15
Mckinlay JB, Vieille C, Zeikus JG (2007) Prospects for a bio-based succinate industry. Appl Microbiol Biotechnol 76(4):727–740. https://doi.org/10.1007/s00253-007-1057-y
Mohanan N, Montazer Z, Sharma PK, Levin DB (2020) Microbial and enzymatic degradation of synthetic plastics. Front Microbiol 11:580709. https://doi.org/10.3389/fmicb.2020.580709
Mückschel B, Simon O, Klebensberger J, Graf N, Rosche B, Altenbuchner J, Pfannstiel J, Huber A, Hauer B (2012) Ethylene glycol metabolism by Pseudomonas putida. Appl Environ Microbiol 78(24):8531–8539. https://doi.org/10.1128/aem.02062-12
Müller R, Schrader H, Profe J, Dresler K, Deckwer W (2005) Enzymatic degradation of poly(ethylene terephthalate): rapid hydrolyse using a hydrolase from T. fusca. Macromol Rapid Commun 26(17):1400–1405
Narancic T, Salvador M, Hughes GM, Beagan N, Abdulmutalib U, Kenny ST, Wu H, Saccomanno M, Um J, O’connor K, E, Jiménez JI (2021) Genome analysis of the metabolically versatile Pseudomonas umsongensis GO16: the genetic basis for PET monomer upcycling into polyhydroxyalkanoates. Microb Biotechnol 14(6):2463–2480. https://doi.org/10.1111/1751-7915.13712
Ng LC, Shingler V, Sze CC, Poh CL (1994) Cloning and sequences of the first eight genes of the chromosomally encoded (methyl) phenol degradation pathway from Pseudomonas putida P35X. Gene 151(1–2):29–36. https://doi.org/10.1016/0378-1119(94)90629-7
Nobu MK, Narihiro T, Hideyuki T, Qiu YL, Sekiguchi Y, Woyke T, Goodwin L, Davenport KW, Kamagata Y, Liu WT (2015) The genome of Syntrophorhabdus aromaticivorans strain UI provides new insights for syntrophic aromatic compound metabolism and electron flow. Environ Microbiol 17(12):4861–4872. https://doi.org/10.1111/1462-2920.12444
Pandit AV, Harrison E, Mahadevan R (2021) Engineering Escherichia coli for the utilization of ethylene glycol. Microb Cell Fact 20(1):22. https://doi.org/10.1186/s12934-021-01509-2
Pardo I, Jha RK, Bermel RE, Bratti F, Gaddis M, Mcintyre E, Michener W, Neidle EL, Dale T, Beckham GT, Johnson CW (2020) Gene amplification, laboratory evolution, and biosensor screening reveal MucK as a terephthalic acid transporter in Acinetobacter baylyi ADP1. Metab Eng 62:260–274. https://doi.org/10.1016/j.ymben.2020.09.009
Pereira B, Zhang H, Mey MD, Lim CG, Li ZJ, Stephanopoulos G (2015) Engineering a novel biosynthetic pathway in Escherichia coli for production of renewable ethylene glycol. Biotechnol Bioeng 113(2):376–383
Perrin E, Ghini V, Giovannini M, Di Patti F, Cardazzo B, Carraro L, Fagorzi C, Turano P, Fani R, Fondi M (2020) Diauxie and co-utilization of carbon sources can coexist during bacterial growth in nutritionally complex environments. Nat Commun 11(1):3135. https://doi.org/10.1038/s41467-020-16872-8
Qi X, Ma Y, Chang H, Li B, Ding M, Yuan Y (2021) Evaluation of PET degradation using artificial microbial consortia. Front Microbiol 12:778828. https://doi.org/10.3389/fmicb.2021.778828
Qiu YL, Sekiguchi Y, Imachi H, Kamagata Y, Tseng IC, Cheng SS, Ohashi A, Harada H (2004) Identification and isolation of anaerobic, syntrophic phthalate isomer-degrading microbes from methanogenic sludges treating wastewater from terephthalate manufacturing. Appl Environ Microbiol 70(3):1617–1626
Reverón I, Jiménez N, Curiel JA, Peñas E, De López F, De Las Rivas B, Muñoz R (2017) Differential gene expression by Lactobacillus plantarum WCFS1 in response to phenolic compounds reveals new genes involved in tannin degradation. Appl Environ Microbiol 83(7):e03387–e03316. https://doi.org/10.1128/aem.03387-16
Rochman CM, Browne MA, Halpern BS, Hentschel BT, Hoh E, Karapanagioti HK, Rios-Mendoza LM, Takada H, Teh S, Thompson RC (2013) Policy: Classify plastic waste as hazardous. Nature 494(7436):169–171. https://doi.org/10.1038/494169a
Rosa LT, Bianconi ME, Thomas GH, Kelly DJ (2018) Tripartite ATP-independent periplasmic (TRAP) transporters and tripartite tricarboxylate transporters (TTT): from uptake to pathogenicity. Front Cell Infect Microbiol 8:33. https://doi.org/10.3389/fcimb.2018.00033
Rosenberg M, Miková H, Kristofíková L (1999) Formation of L-malic acid by yeasts of the genus Dipodascus. Lett Appl Microbiol 29(4):221–223. https://doi.org/10.1046/j.1365-2672.1999.00601.x
Roth C, Wei R, Oeser T, Then J, Follner C, Zimmermann W, Strater N (2014) Structural and functional studies on a thermostable polyethylene terephthalate degrading hydrolase from Thermobifida fusca. Appl Microbiol Biotechnol 98(18):7815–7823. https://doi.org/10.1007/s00253-014-5672-0
Sadler JC, Wallace S (2021) Microbial synthesis of vanillin from waste poly(ethylene terephthalate). Green Chem 23(13):4665–4672. https://doi.org/10.1039/d1gc00931a
Salvador M, Abdulmutalib U, Gonzalez J, Kim J, Smith AA, Faulon J-L, Wei R, Zimmermann W, Jimenez JI (2019) Microbial genes for a circular and sustainable bio-PET economy. Genes 10(5):373. https://doi.org/10.3390/genes10050373
Sang T, Wallis CJ, Hill G, Britovsek GJP (2020) Polyethylene terephthalate degradation under natural and accelerated weathering conditions. Eur Polymer J 136. https://doi.org/10.1016/j.eurpolymj.2020.109873
Sasoh M, Masai E, Ishibashi S, Hara H, Kamimura N, Miyauchi K, Fukuda M (2006) Characterization of the terephthalate degradation genes of Comamonas sp. strain E6. Appl Environ Microbiol 72(3):1825–1832. https://doi.org/10.1128/aem.72.3.1825-1832.2006
Sauer M, Porro D, Mattanovich D, Branduardi P (2008) Microbial production of organic acids: expanding the markets. Trends Biotechnol 26(2):100–108. https://doi.org/10.1016/j.tibtech.2007.11.006
Saxena HO, Faridi U, Srivastava S, Kumar JK, Darokar MP, Luqman S, Chanotiya CS, Krishna V, Negi AS, Khanuja SP (2008) Gallic acid-based indanone derivatives as anticancer agents. Bioorg Med Chem Lett 18(14):3914–3918. https://doi.org/10.1016/j.bmcl.2008.06.039
Schink B, Stieb M (1983) Fermentative degradation of polyethylene glycol by a strictly anaerobic, gram-negative, nonsporeforming bacterium, Pelobacter venetianus sp. nov. Appl Environ Microbiol 45(6):1905–1913. https://doi.org/10.1128/aem.45.6.1905-1913.1983
Schlafli HR, Weiss MA, Leisinger T, Cook AM (1994) Terephthalate 1,2-dioxygenase system from Comamonas testosteroni T-2: purification and some properties of the oxygenase component. J Bacteriol 176(21):6644–6652. https://doi.org/10.1128/jb.176.21.6644-6652.1994
Schuchmann K, Schmidt S, Martinez Lopez A, Kaberline C, Kuhns M, Lorenzen W, Bode HB, Joos F, Müller V (2015) Nonacetogenic growth of the acetogen Acetobacterium woodii on 1,2-propanediol. J Bacteriol 197(2):382–391. https://doi.org/10.1128/jb.02383-14
Sinha V, Patel MR, Patel JV (2008) PET Waste management by chemical recycling: a review. J Polym Environ 18(1):8–25. https://doi.org/10.1007/s10924-008-0106-7
Soellner S, Rahnert M, Siemann-Herzberg M, Takors R, Altenbuchner J (2013) Evolution of pyruvate kinase-deficient Escherichia coli mutants enables glycerol-based cell growth and succinate production. J Appl Microbiol 115(6):1368–1378. https://doi.org/10.1111/jam.12333
Su H, Wei H, Liu J, Wang S, Liu Y (2021) Design and progress of synthetic consortia: a new frontier in synthetic biology. Synth Biology J 2(4):635–650. https://doi.org/10.12211/2096-8280.2021-031
Sudarsan S, Blank LM, Dietrich A, Vielhauer O, Takors R, Schmid A, Reuss M (2016) Dynamics of benzoate metabolism in Pseudomonas putida KT2440. Metab Eng Commun 3:97–110. https://doi.org/10.1016/j.meteno.2016.03.005
Sulaiman S, Yamato S, Kanaya E, Kim JJ, Koga Y, Takano K, Kanaya S (2012) Isolation of a novel cutinase homolog with polyethylene terephthalate-degrading activity from leaf-branch compost by using a metagenomic approach. Appl Environ Microbiol 78(5):1556–1562. https://doi.org/10.1128/AEM.06725-11
Thakker C, Martinez I, San KY, Bennett GN (2012) Succinate production in Escherichia coli. Biotechnol J 7(2):213–224. https://doi.org/10.1002/biot.201100061
Tiso T, Narancic T, Wei R, Pollet E, Beagan N, Schroder K, Honak A, Jiang M, Kenny ST, Wierckx N, Perrin R, Averous L, Zimmermann W, O’connor K, Blank LM (2021) Towards bio-upcycling of polyethylene terephthalate. Metab Eng 66:167–178. https://doi.org/10.1016/j.ymben.2021.03.011
Tournier V, Topham CM, Gilles A, David B, Folgoas C, Moya-Leclair E, Kamionka E, Desrousseaux ML, Texier H, Gavalda S, Cot M, Guemard E, Dalibey M, Nomme J, Cioci G, Barbe S, Chateau M, Andre I, Duquesne S, Marty A (2020) An engineered PET depolymerase to break down and recycle plastic bottles. Nature 580(7802):216–219. https://doi.org/10.1038/s41586-020-2149-4
Trifunović D, Schuchmann K, Mueller V (2016) Ethylene glycol metabolism in the acetogen Acetobacterium woodii. J Bacteriol 198(7):1058–1065
Unden G, Bongaerts J (1997) Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim Biophys Acta 1320(3):217–234. https://doi.org/10.1016/s0005-2728(97)00034-0
Urbanek AK, Mironczuk AM, Garcia-Martin A, Saborido A, De La Mata I, Arroyo M (2020) Biochemical properties and biotechnological applications of microbial enzymes involved in the degradation of polyester-type plastics. Biochim Biophys Acta Proteins Proteom 1868(2):140315. https://doi.org/10.1016/j.bbapap.2019.140315
Vallon T, Simon O, Rendgen-Heugle B, Frana S, Mückschel B, Broicher A, Siemann-Herzberg M, Pfannenstiel J, Hauer B, Huber A, Breuer M, Takors R (2015) Applying systems biology tools to studyn-butanol degradation in Pseudomonas putida KT2440. Eng Life Sci 15(8):760–771. https://doi.org/10.1002/elsc.201400051
Venkatachalam S, Nayak GS, Labde JV, Gharal PR, Rao K, Kelkar AK (2012) Degradation and recyclability of poly. Ethylene Terephthalate), Polyester
Wada A, Prates ÉT, Hirano R, Werner AZ, Kamimura N, Jacobson DA, Beckham GT, Masai E (2021) Characterization of aromatic acid/proton symporters in Pseudomonas putida KT2440 toward efficient microbial conversion of lignin-related aromatics. Metab Eng 64:167–179. https://doi.org/10.1016/j.ymben.2021.01.013
Wang Q, Zeng Y, Zhang Y (2021) Advances in synthetic biomanufacturing. Synth Biology J 2(2):145–160. https://doi.org/10.12211/2096-8280.2020-052
Webb HK, Arnott J, Crawford RJ, Ivanova EP (2013) Plastic degradation and its environmental implications with special reference to poly(ethylene terephthalate). Polymers 5:1–18. https://doi.org/10.3390/polym5010001
Wehrmann M, Billard P, Martin-Meriadec A, Zegeye A, Klebensberger J (2017) Functional role of lanthanides in enzymatic activity and transcriptional regulation of pyrroloquinoline quinone-dependent alcohol dehydrogenases in Pseudomonas putida KT2440. mBio 8(3):e00570–e00517. https://doi.org/10.1128/mBio.00570-17
Wei G, Yang X, Gan T, Zhou W, Lin J, Wei D (2009) High cell density fermentation of Gluconobacter oxydans DSM 2003 for glycolic acid production. J Ind Microbiol Biotechnol 36(8):1029–1034
Wei R, Oeser T, Zimmermann W (2014) Synthetic polyester-hydrolyzing enzymes from thermophilic actinomycetes. Adv Appl Microbiol 89:267–305. https://doi.org/10.1016/B978-0-12-800259-9.00007-X
Werner AZ, Clare R, Mand TD, Pardo I, Ramirez KJ, Haugen SJ, Bratti F, Dexter GN, Elmore JR, Huenemann JD, Peabody GLT, Johnson CW, Rorrer NA, Salvachúa D, Guss AM, Beckham GT (2021) Tandem chemical deconstruction and biological upcycling of poly(ethylene terephthalate) to β-ketoadipic acid by Pseudomonas putida KT2440. Metab Eng 67:250–261. https://doi.org/10.1016/j.ymben.2021.07.005
Wu J-H, Liu W-T, Tseng I-C, Cheng S-S (2001) Characterization of microbial consortia in a terephthalate-degrading anaerobic granular sludge system. Microbiol (Reading) 147(Pt 2):373–382
Wu JH, Wu FY, Chuang HP, Chen WY, Huang HJ, Chen SH, Liu WT (2013) Community and proteomic analysis of methanogenic consortia degrading terephthalate. Appl Environ Microbiol 79(1):105–112. https://doi.org/10.1128/AEM.02327-12
Xu K, Zheng R, Wan Y, Zheng Y (2021) In vitro biosynthesis of chemicals: pathway design, component assembly and applications-a review. Synth Biology J 2(6):886–901. https://doi.org/10.12211/2096-8280.2021-019
Yoshida S, Hiraga K, Takehana T, Taniguchi I, Yamaji H, Maeda Y, Toyohara K, Miyamoto K, Kimura Y, Oda K (2016) A bacterium that degrades and assimilates poly(ethylene terephthalate). Science 351(6278):1196–1199. https://doi.org/10.1126/science.aad6359
Yu H, Wang M, Du Y, Liang Y, Zheng Y (2021) Microbial promoter engineering strategies in synthetic biology. Synth Biology J 2(4):598–611. https://doi.org/10.12211/2096-8280.2020-092
Yue H, Zhao Y, Ma X, Gong J (2012) Ethylene glycol: properties, synthesis, and applications. Chem Soc Rev 41(11):4218–4244. https://doi.org/10.1039/c2cs15359a
Zhang X, Wang X, Shanmugam KT, Ingram LO (2011) L-malate production by metabolically engineered Escherichia coli. Appl Environ Microbiol 77(2):427–434. https://doi.org/10.1128/aem.01971-10
Zhang YM, Sun YQ, Wang ZJ, Zhang J (2013) Degradation of terephthalic acid by a newly isolated strain of Arthrobacter sp.0574. S Afr J Sci 109(7/8):1–4. https://doi.org/10.1590/sajs.2013/20120019
Zhou L, Wang Y, Han L, Wang Q, Liu H, Cheng P, Li R, Guo X, Zhou Z (2021) Enhancement of patchoulol production in Escherichia coli via multiple engineering strategies. J Agric Food Chem 69(27):7572–7580. https://doi.org/10.1021/acs.jafc.1c02399
Funding
This work was financially supported by the Natural Science Foundation of Zhejiang Province (LR20B060003), the Natural Science Foundation of China (21808199), and the National Key Research and Development Program of China (2018YFA0901800 & 2021YFC2103200).
Author information
Authors and Affiliations
Contributions
All the authors were involved in the concept and design of this review. RG and HP wrote the article; JL read and revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gao, R., Pan, H., Kai, L. et al. Microbial degradation and valorization of poly(ethylene terephthalate) (PET) monomers. World J Microbiol Biotechnol 38, 89 (2022). https://doi.org/10.1007/s11274-022-03270-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-022-03270-z