Abstract
The microbial communities responsible for the degradation of poly(lactic acid)/poly(3-hydroxybutyrate) (PLA/PHB) blend foils were investigated in 1 year long laboratory soil burial experiments. Different PLA/PHB foils were tested: (a) PLA/PHB original transparent foil, (b) PLA/PHB carbon black filled foil and (c) PLA/PHB black foil previously exposed for 90 days to sun light. The microbiome diversity of these three types of foil was compared with that identified from soil/perlite sample at the beginning of experiment and that developed on a cellulose mat. Culture-dependent and culture-independent (DGGE-cloning) approaches together with PLA, PHB and PLA/PHB degradation plate assays were employed. The cultivation strategy combined with degradation tests permitted the isolation and evaluation of several PLA/PHB blend degrading microorganisms such as members of the genera Bacillus, Paenibacillus, Streptomyces, Rhodococcus, Saccharothrix, Arthrobacter, Aureobasidium, Mortierella, Absidia, Actinomucor, Bjerkandera, Fusarium, Trichoderma and Penicillium. The DGGE-cloning investigation increased the information about the microbial communities occurring during bioplastic degradation detecting several bacterial and fungal taxa and some of them (members of the orders Anaerolineales, Selenomonadales, Thelephorales and of the genera Pseudogymnoascus and Pseudeurotium) were revealed here for the first time. This survey showed the microbiome colonizing PLA/PHB blend foils and permitted the isolation of several microorganisms able to degrade the tested polymeric blends.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The biopolymers for the production of mulching foils are considered, with respect to their ecologically friendly properties, as a valid alternative to other kind of plastic polymers, as for example those derived from fossil fuels (Rudnik and Briassoulis 2011; Brodhagen et al. 2015; Dharmalingam et al. 2015; Muthuraj et al. 2017).
Two different families of biopolymers are represented by polylactic acid (PLA) and polyhydroxyalkanoates (PHAs). PLA is a thermoplastic produced from natural resources which combines biocompatibility, biodegradability and an excellent processability (Corneillie and Smet 2015). However, PLA also has some disadvantages as for example high crystallinity and brittleness, and slow biodegradability in soil (Shah et al. 2008; Corneillie and Smet 2015).
PHAs are a group of biopolyesters produced by several kinds of microorganisms as intracellular inclusions. There are different types of PHAs, the simplest and generally the most frequently studied PHA is poly(3-hydroxybutyrate) (PHB). The disadvantages of the PHB include brittleness, slow crystallization, poor thermal stability and poor melt processability (Możejko-Ciesielska and Kiewisz 2016).
In order to improve the mechanical properties of mulching foils is a normal practice to prepare blends composed by different types of polymers. One solution is the production of blends combining PLA and PHB. In this way also the biodegradation characteristics of the new mulching foil can be modified and improved (Brodhagen et al. 2015).
Previously, many studies were aimed at elucidating the biodegradation properties of several PLA/PHB blends. The experimentation included respiratometric test (Abdelwahab et al. 2012), composting conditions (Arrieta et al. 2016) and soil environment (Weng et al. 2013). Although, all these investigations produced important data regarding the changes of the properties of blends after a period of incubation in soil buried conditions, they didn’t indicate which kind of microorganisms are the main actors during the degradation of PLA/PHB blends.
In literature, several studies identified the cultivable soil microbial community responsible for bioplastic degradation. They were oriented either to PLA (Karamanlioglu et al. 2014) or PHB polymer (Mergaert et al. 1993; Boyandin et al. 2013; Volova et al. 2017).
To our knowledge the microbial communities that contribute to the degradation of PLA/PHB blends in soil were not studied until now and also no culture-independent strategies were applied to have a better view of such microbial populations. The aim of this study was the investigation of bacterial and fungal assemblages, through culture-dependent and culture-independent approaches, responsible for the degradation of different kinds of PLA/PHB mulching foils (T8—PLA/PHB foil white; T12—PLA/PHB foil black; T21—PLA/PHB foil black exposed 90 days to sunlight) before and after 1 year of incubation in respirometric reactors.
Materials and methods
Polymeric foils
Commercial blend PLA/PHB Nonoilen® pellets were provided by Panara s.r.o. (Nitra, Slovakia; Nonoilen contained PLA, PHB polymers and citrate ester as plasticizer). Delivered pellets were converted into transparent film and 1 wt% containing carbon black (CB, N220, supplied by Corax®) composite film using film blowing technology as is described in details by Mosnáčková et al. (2017). The samples named as cellulose (T6) for cellulose foil (used as control), PLA/PHB_w (T8) for original transparent foil, PLA/PHB_b (T12) for original carbon black filled foil and PLA/PHB_b 90D sun (T21) for outdoor weathering foil exposed 90 days to sun light.
Soil samples and isolation of microorganisms
Soil was collected in an agricultural region at South Slovakia near Danube River. The pH of the soil was determined according to the standard STN ISO 10390 and was 7.5. Its moisture hold capacity (MHC) represents the moisture in soil that keeps inside against the gravity and was established on 0.85. Expanded Perlite EP 200 for agricultural purposes has grain diameter 0.5/4 mm and was purchased from LB Minerals, a.s. (Košice, Slovakia).
The sieved agriculture soil was mixed with perlite in equivalent amounts (SSP; totally 200 g). This soil mix was using in order to bury polymeric foils in several respirometric reactors as schematically showed in Fig. 1.
10 g (wet weight) portion of this soil mixture (SSP), at the beginning of experimentation were mixed in sterile 250 ml Erlenmeyer flasks with 90 ml of a 0.9% (w/v) NaCl solution and incubated at room temperature 24–26 °C in a shaker at 90 rpm for 2 h. The obtained suspension were filtered through Whatman 1 filter paper (Merck, Darmstadt, Germany), and these filtered soil suspension were inoculated directly onto different agar media for the isolation of bacteria and fungi.
After 1 year of incubation, in respirometric reactors, the polymeric foils (T6, T8, T12 and T21) were taken off and immersed in 3 ml of physiological solution in 50 ml Erlenmeyer flasks. In this way the soil and dust attached to foils were spread in the solution. Each suspension was serially diluted and inoculated in specific agar plate for the growth of bacteria and fungi.
For the isolation of bacteria SM1 agar (Tan et al. 2006), Bennett’s Agar (BA; Himedia; Mumbai, India), Actinomycete Isolation Agar (AIA; Himedia) and LB10 agar (peptone 1 g l−1, yeast extract 0.5 g l−1, NaCl 1 g l−1, agar 15 g l−1; Grivalský et al. 2016) were used. The fungi were isolated on Potato Dextrose Agar (PDA; Himedia) and Sabouraud agar (SAB; Himedia). Both bacteria and fungi were also isolated on Filtered Soil Suspension agar (FSS) which was prepared using 1000 g (wet weight) of the same agriculture soil mixed with 2 l of a 50 mM NaOH solution and incubated overnight at laboratory temperature. Then, this mixture was filtered through Whatman 1 filter paper (Merck) and centrifuged 60 min at 18,000 rpm. The pH value of filtered soil suspension supernatant was adjusted to 7–7.5. For the preparation of FSS medium 1000 ml of filtered soil suspension and 1.5 g of agar were mixed and boiled for three times at 100 °C during 24 h.
In order to inhibit the growth of fungi and bacteria some media were supplemented either with 80 mg l−1 cycloheximide (LB10 and FSS for bacteria) or with 100 mg l−1 chloramphenicol (PDA, SAB and FSS for fungi), respectively.
The pure bacterial colonies were maintained on LB10 or on AIA plates, while the fungi on SAB slants.
Mineralization test under laboratory controlled conditions
Mineralization test was carried out according to standards ASTM D5988-12 and STN 17556-2012 in soil under aerobic conditions in the reactor schematically showed in Fig. 1. Organic carbon content in tested PLA/PHB foil determined by elemental analysis was 52.15%. Polymer foil with size 7 cm × 7 cm with thickness 0.1 mm (weight approximately 0.45 g) was buried in mixed perlite and soil equivalent amount (totally 200 g) in 500 ml Erlenmayer flask. Flask was closed by perforated glass tube fulfilled with 20 ml of KOH 0.5 M solution and equipped by rubber stopcock. CO2 produced by mineralization was absorbed in the KOH solution. Samples of the trap solution were opened periodically at least every 10 days for 1 h to tend free respiration of microorganisms in the soil and to establish the amount of absorbed CO2. 1 ml of KOH solution from the trap was titrated with 0.05 M HCl with phenolphthalein as indicator. For new period the new 20 ml of KOH was added to the trap. All samples were processed by triplicate. Flasks with only perlite and soil were used as control and empty flasks were used as blank, respectively, to measure production of the CO2 by the microorganisms itself and to measure CO2 present in the air, respectively.
DNA extraction of isolated microorganisms, clustering and identification
Bacterial chromosomal DNA was isolated using the InstaGene Matrix (Biorad, Hercules, CA, USA) following the instructions of the manufacturer. The isolated strains were clustered by ITS PCR. The internal transcribed spacer (ITS) between 16S and 23S rRNA genes was amplified and the produced amplicons were separated by Qiaxcel electrophoresis (Qiagen, Hilden, Germany) according to Bučková et al. (2018). Bacterial representatives of each ITS cluster were identified by the sequencing of their 16S rRNA genes using the primers 27F and 685R (Puškárová et al. 2016). The final PCR mixture (25 µl) consisted 50 pmol of each primer, 200 µmol l−1 of dNTP, 1.5 U BioTherm Taq DNA polymerase (Genecraft, Köln, Germany), 1× PCR buffer, and 6 µl of template. The PCR program consisted of initial denaturation at 95 °C for 1 min, 30 cycles (95 °C for 1 min, 54 °C for 1 min, 72 °C for 1 min 30 s) and a final polymerization step at 72 °C for 8 min.
The purified fungal isolates were selected on the basis of the different morphology and polymer degradation properties (“PLA/PHB blend, PHB and PLA plate degradation assays” section). For DNA extraction from fungal isolates, the isolates were inoculated in SAB broth at 28 °C until growth; they were then separated from the broth by filtration through sterile filter paper, followed by DNA extraction with Ron’s fungal DNA mini kit (Bioron, Ludwigshafen, Germany), according to the instructions of the manufacturer. The ITS region of rDNA was amplified with the primers ITS1 and ITS4 (Sclocchi et al. 2017). The 25 µl PCR mixture contained 50 pmol of each primer, 200 µmol l−1 of dNTP, 1.5 U HotStar Taq plus DNA polymerase (Qiagen), 1× PCR buffer and 3 µl of the extracted DNA. The PCR program consisted of an initial denaturation at 94 °C for 5 min, followed by 30 cycles (denaturation at 94 °C for 30 s, annealing at 54 °C for 45 s, extension at 72 °C for 1 min) and a final polymerization step at 72 °C for 10 min.
The resulting PCR products from both fungal and bacterial isolates were purified using ExoSAP-IT (Affymetrix, Cleveland, Ohio, USA) and sequenced at a commercial facility (GATC-Biotech, Konstanz, Germany). The resulting sequences were directly compared with those in GenBank using the BLAST program (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and were subsequently deposited in GenBank under the accession numbers MH155488–MH155548 (bacterial isolates) and MH161229–MH161273 (fungal isolates).
Direct DNA extraction from soil and polymer foil samples
One and half milliliters of soil suspension used to inoculate the agar plates (SSP) and also of the polymer foil suspensions of the respirometric reactors T6, T8, T12, T21 were used to extract the DNA by the PowerSoil DNA Isolation Kit (MO BIO Laboratories, Carlsbad, USA), according to the protocol of the manufacturer and repeatedly eluted to a final volume of 2 × 30 µl of TE buffer.
Culture-independent analysis: denaturing gradient gel electrophoresis and clone library
First PCR amplification
Bacterial 16S rDNA and eukaryotic ITS fragment were amplified by two steps, a portion of the PCR product of the first step being used for the construction of clone libraries, and another portion in the second amplification step, a semi-nested PCR facilitating fingerprint analysis based on denaturing gradient gel electrophoresis (DGGE).
The first step involved PCR with primers 27F and 685R (Lane 1991) oriented to 16S rRNA gene. For amplification of ITS region of yeasts and fungi, primers ITS1 and ITS4 were used (White et al. 1990). The PCR mixture (25 µl) contained 50 pmol of each primer, 200 µmol l−1 of dNTP, 1.5 U SuperHot-Taq DNA polymerase (Bioron) and 1× PCR buffer. 3 µl of extracted DNA were used as a template in the first amplification. The temperature programme consisted of initial denaturation at 94 °C for 5 min, 30 cycles (94 °C for 30 s, 54 °C for 45 s, 72 °C for 1 min) and a final polymerization step at 72 °C for 10 min. For each DNA target (16S rDNA and ITS), four reactions of 25 µl (100 µl altogether) were produced. The four reactions of each DNA target were mixed together and the specificity of amplification was checked by agarose gel electrophoresis.
Semi-nested PCR and DGGE fingerprinting
The PCR product of the first step (2 µl) was used as a template in the second amplification, a semi-nested PCR for each DNA target. The 16S rDNA was re-amplified with primers 518f and 685R-GC (Puškárová et al. 2016). Primers ITS1f-GC and ITS2 were used for the semi-nested amplification of ITS fragment (Pangallo et al. 2014; Puškárová et al. 2016). The PCR conditions were the same as above. Four semi-nested PCR products (four reactions) for each DNA target were pooled, checked by electrophoresis in agarose gel, and precipitated with 96% ethanol, resuspended in 20 µl H2O and the precipitate (10 µl) was analysed by DGGE in 8% polyacrylamide gel (acryl amide-bisacrylamide 37.5:1) with the denaturation gradient of 25–55% for separation of 16S rDNA amplicons and 20–50% for separation of ITS amplicons (100% denaturant contained 7 mol l−1 urea and 40% (v/v) formamide). DGGE was run on DCode System (Bio-Rad) in 0.5 × TAE (20 mmol l−1 Tris, 10 mmol l−1 acetate, 0.5 mmol l−1 Na2 EDTA; pH 8.0) at 200 V and 60 °C for 3 h for bacteria, or for 5 h for fungi.
Construction of clone libraries and sequencing
The rest of the PCR products from the first amplifications were used for the construction of bacterial 16S rDNA and eukaryotic ITS clone libraries. Briefly, the PCR products were purified by QIAquick PCR purification kit (Qiagen), ligated to pGEM-T Easy vector (Promega, Madison, Wisconsin, USA), transformed to E. coli XLI-Blue, and spread to LB plates with ampicillin (100 µg ml−1), X-Gal (0.1 mmol l−1) and IPTG (0.2 mmol l−1). A number of about 120 white colonies from each clone library was checked by vector-specific PCR with primers SP6 (5′-ATT TAG GTG ACA CTA TAG AAT AC-3′) and T7 (5′-TAA TAC GAC TCA CTA TAG GG-3′).
One hundred positive clones of each library were analysed by DGGE at conditions described above, using bacterial primers 518f and 685R-GC, or ITS primers ITS1f-GC and ITS2. Profiles of individual clones were compared with each other and with the profile of the whole community. Clones with different profiles were sequenced using primers SP6 and T7. The obtained sequences were compared with those present in the GenBank database (nucleotide collection database and, in the case of sequences identified as uncultured bacterium or fungus, the 16S ribosomal RNA sequences database or the exclusion of the uncultured/environmental sample sequences, respectively) by BLAST search (National Center for Biotechnology Information, Bethesda, Maryland, USA). The sequences were deposited in GenBank under accession numbers MH155549–MH155592 (bacterial clones) and MH161274–MH161325 (eukaryotic clones).
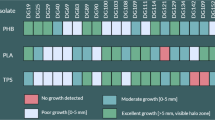
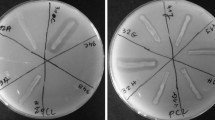
PLA/PHB blend, PHB and PLA plate degradation assays
In order to assess the degradation abilities of isolated strains for biodegradable polymers three different media containing respectively PLA/PHB blend, PHB and PLA (provided by Panara s.r.o. and Polymer Institute, SAS) were prepared. One gram of PLA/PHB, PHB or PLA granules were dissolved in 30 ml of dichloromethane. To each solution were added 150 ml of a basal medium composed of 1 g l−1 KH2PO4, 1 g l−1 (NH4)2SO4, 0.2 g l−1 MgSO4·7H2O, 0.01 g l−1 FeCl3, 0.05 g l−1 NaCl, 0.05 g l−1 CaCl2, 0.25 g l−1 Yeast extract, 0.1 g l−1 Triton X and 15 g l−1 of agar according to Grivalský et al. (2017). Then, media were stirred at 70 °C until dichloromethane was evaporated, which takes approximately 20 h. The media were sterilized by autoclaving and poured on Petri dishes. The assays were performed for each isolate in triplicate and the positive reaction was displayed as a zone of clearance around the assayed microbial colonies.
Results
Mineralization of PLA/PHB (CB) blends before and after outdoor weathering
Cellulose mat (T6) was used as reference material in order to check the microbial activity of the soil. It can be seen in Fig. 2 that cellulose sample started to mineralize almost immediately, it grew linearly and reached approximately 70% of mineralization in a year. In mineralization tests hydrolytic degradation of the PLA/PHB foils can not be excluded, but it can assumed that measured evolved CO2 can be produced exclusively by microorganisms, which use this polymer for feeding. In this context, initial pre-degradation by sunlight could affect velocity of followed mineralization. According to Fig. 2 all polymer samples undergo mineralization faster compared to control soil sample without polymer foil, suggesting that polymer samples increase microbial activity. However, this difference was small and mineralization was slow for all investigated polymers including cellulose.
PLA/PHB_w (T8) polymer sample exhibited slightly higher mineralization compared to PLA/PHB_b (T12) polymer composite. The mineralization seemed to be slightly faster for the sunlight exposed sample PLA/PHB_b 90D (T21), but generally for all of the samples the difference could be considered to be within the experimental error (Fig. 2).
Surprisingly to this promising results, after withdrawal samples lose only approximately 8–12% of its original weight (Fig. 3). Only cellulose sample exhibit approximately 50% weight loss. This is in contradiction to the mineralization tests, which showed five times overestimation. While disintegration of polymer foils was expected at such degree of mineralization, all withdrawal foils were still self-holding. Only some of the samples exhibited visible cracks, which are usually a cause of loss of mechanical properties such as decreasing of elongation at break and increasing of brittleness of polymer foils. These cracks were most visible for PLA/PHB_b 90D (T21; Fig. 4).
Cultivation analysis
Bacteria belonging to three phyla (Firmicutes, Actinobacteria and Proteobacteria) were isolated from soil sample (SSP) at the beginning of the mineralization process and from the cellulose foil (T6) after mineralization test. Members of only Firmicutes and Actinobacteria were recovered from the different PLA/PHB foils (T8, T12 and T21) after 1 year of incubation in the respirometric reactors. The most isolated bacteria were the members of the genus Bacillus. They were present already in the SSP sample and repeatedly isolated from all polymeric foils (Fig. 5; Table S1).
The bacterial community isolated from transparent foil (PLA/PHB_w; T8) was dominated mainly by Bacillales and Streptomycetales members. Bacteria recovered from PLA/PHB black foils were represented also by different kinds of Actinobacteria: Streptomyces, Rhodococcus and Arthrobacter (Fig. 5; Table S1). For the isolation several types of culture media were used (SM1, BA, AIA, FSS and LB10), but in our case the best isolation media were represented by LB10, FSS and AIA. We were not able to isolate any microorganisms from polymeric samples using BA and SM1 media. In addition the specificity of SM1, BA and AIA for the isolation of Actinobacteria was partially respected, in fact only by SM1 medium this group of bacteria was exclusively isolated. In the other media (AIA and BA) also members of other bacterial groups, mainly Bacillus, grew very well.
The fungal community isolated from soil (SSP) was composed by few more taxa (9 genera) compared to the bacterial community (7 genera). Members of the genera Phialophora, Cladosporium and Trichoderma were the prevalent fungi isolated from soil. Penicillium spp. were isolated on all the three agar media used.
The fungal analysis after 1 year of incubation in the respirometric reactors showed different changes in the composition of community depending on polymeric substrate. In fact, from the cellulose mat were isolated only Fusarium and Penicillium (Fig. 6; Table S2). The Fusarium were isolated from all PLA/PHB foils together with members of Mucorales (Absidia and Actinomucor). The largest fungal diversity, constituted by Fusarium, Penicillium, Aspergillus, Actinomucor, Bjerkandera and Mortierella, was found in the PLA/PHB foil previously exposed to sun light for 90 days (Fig. 6; Table S2).
Polymers degradation abilities of isolated strains
The hydrolytic agar media assays were performed in order to evaluate the degradation power of bacterial and fungal isolates.
The 68% of bacteria recovered from soil were able to degrade at least one of the tested polymeric substrates (PHB, PLA/PHB blend and PLA). This percentage increased when the bacteria from polymeric foils were assayed: 86% from cellulose mat, 94% from film T8 and 79% from T12. The bacteria from sample T21 showed at least one degradation property (Table S1).
Several isolates of the genus Bacillus demonstrated to be valuable polymers degraders. They were isolated mainly from soil. Among bacteria isolated from the different polymeric foils the best degradation abilities were exhibited by Actinobacteria of the genus Streptomyces, Rhodococcus, Saccharothrix and the species Arthrobacter oryzae (Table 1; Table S1). Many bacterial isolates displayed very rapid and extensive positive reaction for each assayed substrate (Table 1).
Each selected and identified fungal isolate expressed at least positive reaction for two of the tested substrates. Numerous fungal isolates displayed high PLA/PHB blend degradation ability often associated with a relevant hydrolytic activity for PHB and PLA. The best fungal degraders encompassed Cladosporium subcinereum, Trichoderma harzianum, Aureobasidium pullulans, Absidia sp., Penicillium sp., Bjerkandera adusta, Actinomucor elegans, Aspergillus fumigatus and several members of the genera Fusarium and Mortierella (Table 1; Table S2).
Culture-independent analysis
The DGGE-cloning approach permitted a more complete analysis of the bacterial and fungal microbiomes in soil and associated to polymeric foils.
Twenty-seven bacterial orders were revealed, the biggest number of taxa were detected from soil sample, with 12 orders comprising 14 genera, followed by the cellulose mat, with 10 orders including 13 genera. The number of taxa detected from PLA/PHB foils was very similar (Fig. 7; Table S3).
The operational taxonomic units (OTUs) of the order Acidobacteria (subdivision 6) were the most spread, indeed they were detected in each analyzed samples (Fig. 7). The second most diffuse OTUs belonged to the orders Cytophagales (absent only in the T8 sample) and Anaerolineales (not detected in soil; Fig. 7). The OTUs of the orders Gemmatimonadales and Burkholderiales occurred in soil, on cellulose mat and PLA/PHB black foil (T12). The two PLA/PHB black foils (T12 and T21) displayed quite similar bacterial microbiomes. By contrast, the bacterial community of PLA/PHB transparent foil (T8) was different and composed by characteristic OTUs belonging to the orders Pseudonocardiales, Bacillales, Pseudomonadales, Methylococcales and Desulfuromonadales, that were not detected on the black foils (Fig. 7, Table S3).
Twelve different fungal orders together with four orders (Ulvales, Chlamydomonadales, Sporadotrichida and Stichotrichida) belonging to other eukaryotic organisms were identified by DGGE-cloning strategy. The highest number of fungal genera (9) of 8 different orders were identified in soil samples. From the polymeric samples the number of detected taxa sharply decreased, in fact only four orders were identified for each of investigated foils (Fig. 8; Table S4).
The eukaryotic community of cellulose mat was composed mostly by fungi, the other eukaryotic organisms representing only the 2%. The most detected OTUs belonged to the orders Agaricales (44%) and Leotiomycetes incertae sedis (37%). The latter order was common to all analyzed samples (Fig. 8). The OTUs of Pleosporales order were the second most diffused, although they reached lower percentages with respect to Leotiomycetes. They were detected in all samples except the PLA/PHB transparent foil (T8). The foil T8 showed a different fungal microbiome in comparison with the black foil samples. OTUs of the order Hypocreales were identified only here (Fig. 8).
The two black foils (T12 and T21) showed very similar fungal microbiomes composed by the orders Pleosporales, Mortierellales and Leotiomycetes. The Leotiomycetes community of the sample T12 exhibited also OTUs belonging to the genus Pseudogymnoascus, which was not detected on foil T21 (Table S4). The differences between the fungal microbiomes of the PLA/PHB black foils regarded also the orders Pezizales (2%) and Thelephorales (37%) identified on foil T12 and T21, respectively (Fig. 8).
Discussion
Our investigation showed the change of the microbial community responsible for the biodegradation of different polymeric foils (one cellulose mat and 3 PLA/PHB blend films) after 1 year of incubation into respirometric reactors.
Culture-dependent and culture-independent strategies were combined together with different agar degradation assays in order to characterize the degrading microbiome.
Both types of analysis showed how the microbiome changed on the basis of available substrate. The cellulose degrading microbiome is different with respect to the microbial communities isolated and detected from the PLA/PHB foils. Moreover, the microbiomes of the two black foils (T12 and T21) have more affinity between them than with the microbiota of transparent film (T8).
Many Bacillus, Paenibacillus, Streptomyces, Rhodococcus, Saccharothrix and Arthrobacter isolates displayed excellent abilities to degrade the three types of PLA/PHB foils. In the literature, it has been reported that some of these bacteria contributed to the degradation of bioplastics or mulching foils (Akutsu-Shigeno et al. 2003; Tokiwa et al. 2009; Brodhagen et al. 2015), but in these studies, only PLA or PHB were tested and not a PLA/PHB blend.
As far as fungal isolates are concerned, several of them were able to degrade PLA/PHB blend. This confirmed the findings of previous studies (Kim and Rhee 2003; Ghosh et al. 2013; Brodhagen et al. 2015) where Fusarium, Trichoderma, Cladosporium, Penicillium and Aspergillus isolates hydrolyzed either PLA or PHB. To our knowledge, in this study we have demonstrated for the first time the ability of various members of the genera Aureobasidium, Mortierella, Absidia, Actinomucor and Bjerkandera to degrade PLA, PHB and PLA/PHB blend polymers.
One of the novelty of our study included also the use of a culture-independent method (DGGE-cloning approach) to study the bacterial and eukaryotic microbiota responsible for PLA/PHB blend degradation. Until now only few attempts were tried for studying the PLA and PHB microbial degraders by culture-independent methods and such attempts regarded very different environments (Kamiya et al. 2007; Yagi et al. 2014; Qiu et al. 2017). Moreover, they were oriented either to fungal or to bacterial community.
Our DGGE-cloning analysis revealed a complex microbiome complementary to that identified by the cultivation strategy. A low correspondence was observed between isolated microorganisms and OTUs; the analogies were related only to limited bacterial and fungal orders. Both methods detected the presence of Bacillales and Pseudonocardiales on transparent PLA/PHB foil (T8). Importantly, the Bacillales members detected by both strategies were Bacillus and Paenibacillus, which again confirms that these two genera are implicated in the degradation of the PLA/PHB blend. On the other hand, the Pseudonocardiales species identified were different: Saccharothrix were isolated by cultivation and Pseudonocardia were identified by cloning.
The fungal analogies between culture-dependent and culture-independent analyses confirmed also the importance, during the biodegradation of PLA/PHB foils, of the members belonging to the genus Fusarium, detected by both strategies on sample T8, and Mortierella species revealed by the two identification approaches only on both black foils (T12 and T21). A previous study evidenced the ability of the fungus Mortierella subtilissima to degrade polyethylene films (Nowak et al. 2011).
Pleosporales is another taxon identified by both methods, mainly by DGGE-cloning, although the coincidence between the isolated and detected species did not match. Members of this order were associated in the degradation of PLA mulches in soil environment (Moore-Kucera et al. 2014).
We believe that other taxonomic orders detected in high percentages by DGGE-cloning, such as Acidobacteria (subdivision 6), Cytophagales, Burkholderiales, Anaerolineales and Selenomonadales, Leotiomycetes (incertae sedis), Agaricales and Thelephorales, also have a major role in PLA/PHB blend degradation. The bacterial orders Acidobacteria, Cytophagales and Burkholderiales have already shown their abilities to degrade different bioplastic mixtures, but not the PLA/PHB blend, in specific denitrification systems (Horiba et al. 2005; Shen et al. 2013; Chu and Wang 2017; Xu and Chai 2017). The orders Anaerolineales and Selenomonadales have so far demonstrated their capacity to colonize PLA/PHB foils only in this study.
Regarding the fungal taxa, it seems that only the genera Coprinus (order Agaricales) and Geomyces (Leotiomycetes incertae sedis) previously showed an ability to degrade nylon (Nomura et al. 2001) and a biodegradable mulching film (Moore-Kucera et al. 2014), respectively. We could find no information in the literature about the bioplastic degradation activities of the genera Pseudogymnoascus and Pseudeurotium (Leotiomycetes incertae sedis) or the order Thelephorales.
With our PLA/PHB foils experimental trial we did not assess the optimal conditions for reaching a mineralization rate of 60–70% (STN EN ISO 17556; Gómez and Michel 2013) that would reasonably represent complete biodegradation, but evidence suggests that 50% values are sufficient to prove that biodegradation was successful (Bayer and Lamed 1992; Weytjens et al. 1994; Solaro et al. 1998). In addition, our microbiological analysis (including culture-dependent and culture-independent approaches, and biodegradation plate assays) permitted the identification of PLA/PHB degraders. The obtained data showed the main actors of the PLA/PHB blend degradation and produced information for future studies focused on the development and modulation of new PLA/PHB foils with improved biodegradability.
References
Abdelwahab MA, Flynn A, Chiou BS, Imam S, Orts W, Chiellini E (2012) Thermal, mechanical and morphological characterization of plasticized PLA–PHB blends. Polym Degrad Stab 97:1822–1828
Akutsu-Shigeno Y, Teeraphatpornchai T, Teamtisong K, Nomura N, Uchiyama H, Nakahara T, Nakajima-Kambe T (2003) Cloning and sequencing of a poly (DL-lactic acid) depolymerase gene from Paenibacillus amylolyticus strain TB-13 and its functional expression in Escherichia coli. Appl Environ Microbiol 69:2498–2504
Arrieta MP, López J, López D, Kenny JM, Peponi L (2016) Biodegradable electrospun bionanocomposite fibers based on plasticized PLA–PHB blends reinforced with cellulose nanocrystals. Ind Crops Prod 93:290–301
ASTM D5988-12 (2012) Standard test method for determining aerobic biodegradation of plastic materials in soil. https://doi.org/10.1520/D5988-12
Bayer EA, Lamed R (1992) The cellulose paradox: pollutant par excellence and/or a reclaimable natural resource? Biodegradation 3:171–188
Boyandin AN, Prudnikova SV, Karpov VA, Ivonin VN, Đỗ NL, Nguyễn TH, Lê TMH, Filichev NL, Levin AL, Filipenko ML, Volova TG (2013) Microbial degradation of polyhydroxyalkanoates in tropical soils. Int Biodeterior Biodegrad 83:77–84
Brodhagen M, Peyron M, Miles C, Inglis DA (2015) Biodegradable plastic agricultural mulches and key features of microbial degradation. Appl Microbiol Biotechnol 99:1039–1056
Bučková M, Puškárová A, Ženišová K, Kraková L, Piknová Ľ, Kuchta T, Pangallo D (2018) Novel insights into microbial community dynamics during the fermentation of Central European ice wine. Int J Food Microbiol 266:42–51
Chu L, Wang J (2017) Denitrification of groundwater using a biodegradable polymer as a carbon source: long-term performance and microbial diversity. RSC Adv 7:53454–53462
Corneillie S, Smet M (2015) PLA architectures: the role of branching. Polym Chem 6:850–867
Dharmalingam S, Hayes DG, Wadsworth LC, Dunlap RN, DeBruyn JM, Lee J, Wszelaki AL (2015) Soil degradation of polylactic acid/polyhydroxyalkanoate-based nonwoven mulches. J Polym Environ 23:302–315
Ghosh SK, Pal S, Ray S (2013) Study of microbes having potentiality for biodegradation of plastics. Environ Sci Pollut Res Int 20:4339–4355
Gómez EF, Michel FC (2013) Biodegradability of conventional and bio-based plastics and natural fiber composites during composting, anaerobic digestion and long-term soil incubation. Polym Degrad Stab 98:2583–2591
Grivalský T, Bučková M, Puškárová A, Kraková L, Pangallo D (2016) Water-related environments: a multistep procedure to assess the diversity and enzymatic properties of cultivable bacteria. World J Microbiol Biotechnol 32:42
Grivalský T, Rychlý J, Rychlá L, Bučková M, Kraková L, Puškárová A, Orovčík Ľ, Pangallo D (2017) Aerobic biodegradation of aromatic aliphatic copolyester induced by bacteria obtained from different environments. J Polym Environ. https://doi.org/10.1007/s10924-017-0980-y
Horiba Y, Khan ST, Hiraishi A (2005) Characterization of the microbial community and culturable denitrifying bacteria in a solid-phase denitrification process using poly (ε-caprolactone) as the carbon and energy source. Microbes Environ 20:25–33
Kamiya M, Asakawa S, Kimura M (2007) Molecular analysis of fungal communities of biodegradable plastics in two Japanese soils. Soil Sci Plant Nutr 53:568–574
Karamanlioglu M, Houlden A, Robson GD (2014) Isolation and characterisation of fungal communities associated with degradation and growth on the surface of poly (lactic) acid (PLA) in soil and compost. Int Biodeterior Biodegrad 95:301–310
Kim DY, Rhee YH (2003) Biodegradation of microbial and synthetic polyesters by fungi. Appl Microbiol Biotechnol 61:300–308
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackenbrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, New York, pp 115–148
Mergaert J, Webb A, Anderson C, Wouters A, Swings J (1993) Microbial degradation of poly (3-hydroxybutyrate) and poly (3-hydroxybutyrate-co-3-hydroxyvalerate) in soils. Appl Environ Microbiol 59:3233–3238
Moore-Kucera J, Cox SB, Peyron M, Bailes G, Kinloch K, Karich K, Miles C, Inglis DA, Brodhagen M (2014) Native soil fungi associated with compostable plastics in three contrasting agricultural settings. Appl Microbiol Biotechnol 98:6467–6485
Mosnáčková K, Danko M, Šišková A, Falco LM, Janigová I, Chmela Š, Vanovčanová Z, Omaníková L, Chodák I, Mosnáček J (2017) Complex study of the physical properties of a poly (lactic acid)/poly (3-hydroxybutyrate) blend and its carbon black composite during various outdoor and laboratory ageing conditions. RSC Adv 7:47132–47142
Możejko-Ciesielska J, Kiewisz R (2016) Bacterial polyhydroxyalkanoates: still fabulous? Microbiol Res 192:271–282
Muthuraj R, Misra M, Mohanty AK (2017) Biodegradable compatibilized polymer blends for packaging applications: a literature review. J Appl Polym Sci. https://doi.org/10.1002/app.45726
Nomura N, Deguchi T, Shigeno-Akutsu Y, Nakajima-Kambe T, Nakahara T (2001) Gene structures and catalytic mechanisms of microbial enzymes able to biodegrade the synthetic solid polymers nylon and polyester polyurethane. Biotechnol Genet Eng Rev 18:125–147
Nowak B, Pająk J, Drozd-Bratkowicz M, Rymarz G (2011) Microorganisms participating in the biodegradation of modified polyethylene films in different soils under laboratory conditions. Int Biodeterior Biodegrad 65:757–767
Pangallo D, Šaková N, Koreňová J, Puškárová A, Kraková L, Valík L, Kuchta T (2014) Microbial diversity and dynamics during the production of May bryndza cheese. Int J Food Microbiol 170:38–43
Puškárová A, Bučková M, Habalová B, Kraková L, Maková A, Pangallo D (2016) Microbial communities affecting albumen photography heritage: a methodological survey. Sci Rep 6:20810
Qiu T, Xu Y, Gao M, Han M, Wang X (2017) Bacterial community dynamics in a biodenitrification reactor packed with polylactic acid/poly (3-hydroxybutyrate-co-3-hydroxyvalerate) blend as the carbon source and biofilm carrier. J Biosci Bioeng 123:606–612
Rudnik E, Briassoulis D (2011) Comparative biodegradation in soil behaviour of two biodegradable polymers based on renewable resources. J Polym Environ 19:18–39
Sclocchi MC, Kraková L, Pinzari F, Colaizzi P, Bicchieri M, Šaková N, Pangallo D (2017) Microbial life and death in a foxing stain: a suggested mechanism of photographic prints defacement. Microb Ecol 73:815–826
Shah AA, Hasan F, Hameed A, Ahmed S (2008) Biological degradation of plastics: a comprehensive review. Biotechnol Adv 26:246–265
Shen Z, Zhou Y, Wang J (2013) Comparison of denitrification performance and microbial diversity using starch/polylactic acid blends and ethanol as electron donor for nitrate removal. Bioresour Technol 131:33–39
Solaro R, Corti A, Chiellini E (1998) A new respirometric test simulating soil burial conditions for the evaluation of polymer biodegradation. J Polym Environ 6:203–208
STN EN ISO 17556 (2012) Plastics—determination of the ultimate aerobic biodegradability of plastic materials in soil by measuring the oxygen demand in a respirometer or the amount of carbon dioxide evolved. https://www.sutn.sk/eshop/public/standard_detail.aspx?id=116602. Accessed 7 Mar 2017
STN ISO 10390 (2005) Kvalita pôdy. Stanovenie pH [Soil quality. Measuring pH]. SÚTN, Bratislava (in Slovak)
Tan GYA, Ward AC, Goodfellow M (2006) Exploration of Amycolatopsis diversity in soil using genus-specific primers and novel selective media. Syst Appl Microbiol 29:557–569
Tokiwa Y, Calabia BP, Ugwu CU, Aiba S (2009) Biodegradability of plastics. Int J Mol Sci 10:3722–3742
Volova TG, Prudnikova SV, Vinogradova ON, Syrvacheva DA, Shishatskaya EI (2017) Microbial degradation of polyhydroxyalkanoates with different chemical compositions and their biodegradability. Microb Ecol 73:353–367
Weng YX, Wang L, Zhang M, Wang XL, Wang YZ (2013) Biodegradation behavior of P (3HB, 4HB)/PLA blends in real soil environments. Polym Test 32:60–70
Weytjens D, Van Ginneken I, Painter HA (1994) The recovery of carbon dioxide in the Sturm test for ready biodegradability. Chemosphere 28:801–812
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, New York, pp 315–321
Xu Z, Chai X (2017) Effect of weight ratios of PHBV/PLA polymer blends on nitrate removal efficiency and microbial community during solid-phase denitrification. Int Biodeterior Biodegrad 116:175–183
Yagi H, Ninomiya F, Funabashi M, Kunioka M (2014) Mesophilic anaerobic biodegradation test and analysis of eubacteria and archaea involved in anaerobic biodegradation of four specified biodegradable polyesters. Polym Degrad Stab 110:278–283
Acknowledgements
The work was mainly supported by the grant APVV-15-0528 “Modified polymers from renewable resources and their degradation”. This contribution is also the result of the projects VEGA 2/0158/17 and ITMS-26240220010 in the frame of the support program Research and Development of the European Regional Development Fund.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jeszeová, L., Puškárová, A., Bučková, M. et al. Microbial communities responsible for the degradation of poly(lactic acid)/poly(3-hydroxybutyrate) blend mulches in soil burial respirometric tests. World J Microbiol Biotechnol 34, 101 (2018). https://doi.org/10.1007/s11274-018-2483-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-018-2483-y