Abstract
Sulfur dioxide which is released from petroleum oil combustion causes pollution over the atmosphere and the soil. Biodesulfurization can be used as a complementary method of hydrodesulfurization, the common method of petroleum desulfurization in refineries. Many studies have been carried out to develop biological desulfurization of dibenzothiophene (DBT) with bacterial biocatalysts. However, fungi are capable to metabolize a wide range of aromatic hydrocarbons through cytochrome P450 and their extracellular enzymes. The aim of the present work was isolation and identification of fungi biocatalysts capable for DBT utilization as sulfur source and production of novel metabolites. DBT consumption and the related produced metabolites were analyzed by HPLC and GC–MS respectively. One of the isolated fungi that could utilize DBT as sole sulfur source was identified by both traditional and molecular experiments and registered in NCBI as Exophiala spinifera FM strain (accession no. KC952672). This strain could desulfurize 99 % of DBT (0.3 mM) as sulfur source by co-metabolism reaction with other carbon sources through the same pathway as 4S and produced 2-hydroxy biphenyl (2-HBP) during 7 days of incubation at 30 °C and 180 rpm shaking. However, the isolate was able to transform 2-HBP to 1,3-benzenediol, 5-hexyl. While biphenyl compounds are toxic to leaving cells, biotransformation of them can reduce their toxicity and the fungi will be more tolerant to the final product. These data are the first report about the desulfurization of DBT comparable to 4S-pathway and production of innovative metabolite by E. spinifera FM strain.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Oil is one of the most important energy sources which contain variety of organosulfur compounds (Soleimani et al. 2007) that are combustible. It can produce sulfur oxide during combusting which will cause pollution over the atmosphere and the soil (Van Hamme et al. 2003). Therefore, the reduction of sulfur in the oil through hydro-desulfurization (HDS), which is the most common method, is of crucial importance (Ma et al. 2007). However, HDS can’t be applied to many aromatic sulfur compounds such as dibenzothiophene (DBT) that are resistant to this process (Stoner et al. 1990) and also this process requires high temperature (200–450 °C) and pressure (150–250 atm) (Ma et al. 2007). So as an alternative or a complementary method, biotechnological processes such as bio-desulfurization can be used instead (Van Hamme et al. 2003). Microorganisms need to sulfur for growth and vital activities, because they can utilize sulfur from organosulfur compounds such as DBT and reduce sulfur content in fossil fuels (Torkamani et al. 2008). Many studies have been carried out to develop a biological biodesulfurization by using DBT as a model cyclic sulfur compounds in fossil fuels (Lin et al. 2010). Several microorganisms have been isolated that utilize DBT and its alkylated derivatives as sulfur source (Rashidi et al. 2006) which mainly belong to Gram—positive actinobacteria from genus Rhodococcus (Denis-Laros et al. 1997). Other actinobacteria such as Oreskovia sp. present in soil, have potential as biodesulfurization catalysts (Khedkar and Shanker 2014). The Gram-negative bacteria such as Klebsiella sp. 13T and Pantoea agglomerans were also reported for thermophilic and mesophilic biodesulfurization of DBT, respectively from different petroleum oils (Bhatia and Sharma 2010a, 2012). Most of these bacteria desulfurize DBT to 2-hydroxybiphenyl (2-HBP) during 4S-pathway (DBT-specific pathway) (Torkamani et al. 2008). The study of desulfurization ability of novel bacteria was performed by Bhatia and Sharma (2010b) based on in silico search by the protein sequences of Dsz proteins of R. erythropolis IGTS8.
Thus most of these studies are related to bacterial strains and nearly none of them have been focused over fungi strains (Bezalel et al. 1996; Campos-Martin et al. 2010). However, the use of fungi as a method of bioremediation has drawn little attention in the past two decades since these researches have focused on the use of bacteria. But, recently the application of fungi in bioremediation of different pollutants has received considerable attention, which is attributed to the potential enzymes they produce (Husaini et al. 2008). The advantage of fungi over the bacterial biocatalysts include their ability to metabolize a wide range of chemical compounds and multi ring aromatic hydrocarbons through cytochrome P-450 and their potential extracellular enzymes (Van Hamme et al. 2003; Etemadifar et al. 2006; Husaini et al. 2008). These enzymes have wide specificity and ability to metabolize high-molecular weight substrates and can use as biocatalysts (Van Hamme et al. 2003). Thus, fungi can be appropriate candidates for DBT desulfurization. Among fungi, Cunninghamella elegans has been reported that metabolize DBT to DBT-5-oxide and DBT-5-dioxide but do not produce biphenyl (Baldi et al. 2003) and Paecylomyces sp. desulfurize DBT by producing 2-di-hydroxy biphenyl using sulfur-specific oxidation pathway (Fasion et al. 1991). But no fungus have been reported to use 4S-pathway for DBT desulfurization, so the aim of present work was isolation and molecular identification of fungi which are able to bioremediate DBT as a sulfur source using 4S-pathway or pathway with thiophenic C–S bond break for application in petroleum desulfurization.
Materials and methods
Chemical materials
Methanol (HPLC grade), DBT, ethyl acetate, Gibb’s reagent (2,6-dichloroquinon 4-chloroimide) and all materials for basal salt medium used in this study were manufactured by Merck. 2-hydroxybiphenyl (2-HBP) was obtained from Fluka Chemika Co.
Culture medium
Basal salt medium (BSM) containing (g/l): 8 NaH2PO4·7.0 H2O, 4.0 KH2PO4, 2.0 NH4Cl, 2.0 MgCl2·6H20, 0.001 CaCl2·2H20, 0.001 FeCl3·6H20 in double distilled deionized water (DDW) or de-ionized water (DIW) was used for isolation of desulfurizing fungi. Glucose was added to BSM by 10 g/l final concentration as the carbon source, after autoclaving at 110 °C for 15–20 min, DBT was added from a stock solution of ethanol (100 mM) with final concentration of 5.4 mM as sulfur source.
Enrichment and isolation of DBT-desulfurizing fungi
Oil contaminated soil samples were collected from different regions of Khuzestan province of Iran and stored at low temperature for further studies. One gram of each soil sample was suspended in 50 ml BSM broth supplemented with 1 % glucose and 5.4 mM DBT as carbon and sulfur sources respectively in 250 ml Erlenmeyer flasks and incubated for 6 days at 30 °C and 180 rpm in an incubator shaker. After incubation, 5 ml enrichment culture was re-inoculated in 45 ml of freshly BSM containing 1 % glucose and 5.4 mM DBT. Every 6 days for three times, 1 % inoculum from the previous culture was transferred to the fresh medium and incubated under conditions as noted earlier. Thereafter for fungi isolation, 80 µl of the culture was streaked on potato dextrose agar (PDA) with chloramphenicol and incubated at 30 °C. Finally single colony of fungi was streaked on a new PDA medium and screened for DBT desulfurization and metabolite production.
Molecular identification of selected strain
DNA preparation
Fungal biomass (0.3 g) was transferred into a 1.5 ml microcentrifuge tube for DNA extraction by DNA extraction kit (Cinaa Gen Inc., Iran) based on manufacturer’s instructions.
Polymerase chain reaction (PCR)
Isolated fungi was identified using internal transcribed spacer region (ITS rDNA) that was amplified with universal primers ITS1 (5-TCCGTAGGTGAACCTGCGG-3) and ITS4 (5-TCCTCCGCTTATTGATATGC-3). ITS1 and ITS4 hybridize at the end of 18S rDNA and at the beginning of 28S rDNA respectively (Ferrer et al. 2001). PCR reaction was done in a 25 µl volume which was prepared by adding 2.5 µl of template DNA, 2.5 µl of each primer and 5 µl of DDW to 12.5 µl of master reaction mixture containing 2.5 µl 10X buffer, 0.7 µl MgCl2, 0.5 µl dNTP and 0.5 µl Taq DNA polymerase. Negative control included all master reaction material except template DNA. Finally amplification of DNA product was conducted using a thermal cycler that started with an initial denaturation at 95 °C for 5 min and was followed by 30 cycles of denaturing at 95 °C for 30 s, annealing at 55 °C for 30 s, extension at 72 °C for 2 min and a final extension at 72 °C for 10 min. DNA fragment was observed using ethidium bromide staining after electrophoresis on a 1 % agarose gel and then sequencing for fungi identification was conducted by FazaBiotech Corp.
Morphological and physiological characterization
Initial identification of isolated strain was based on macroscopic and microscopic features. For macroscopic observation, fungi were cultured on PDA and malt extract agar (MEA) at 30 °C for 2 weeks. Morphological and microscopical observations were carried out with slide culture technique on PDA and mounted in lactic acid or lactophenol cotton blue. PDA medium is mainly used for slide culture because represses aerial hyphal growth and induces sporulation (Bahuguna et al. 2011).
Phylogenetic analysis
ITS sequence of the isolated strain from oil-contaminated soil was blasted with GenBank databases by the Blastn algorithm and aligned by using CLC Genomic Workbench 6.6.2. Finally, phylogenetic tree was constructed with CLC Genomic Workbench 6.6.2.
Fungal inoculum preparation
Conidial suspensions was prepared using the harvest fungal colonies from PDA culture medium with sterile swab and was dissolved in approximately 5 ml of sterile BSM. Then vortexed for 15–30 s to make a suitable suspension and allowed the suspension to stand for a 5–10 min to larger hyphal particles were settled then top homogenous suspension was removed and its turbidity adjusted to A600 = 0.5 (Torkamani et al. 2008).
Growth curve of Exophiala spinifera FM strain and primary analysis of DBT utilization
To study the kinetic of DBT utilization, 1 ml of fungal suspension (A600 = 0.5) of isolated strain was inoculated to 50 ml BSM broth supplemented with 1 % (w/v) glucose and 0.3 mM DBT as carbon and sulfur sources, respectively. During different times of incubation, reaction products in medium were extracted with equal volume of ethyl acetate after acidification to pH 2.0 using HCL 6 N. Extracted DBT was assessed using UV-spectrophotometric analysis and specific absorption of DBT at 323 nm. DBT calibration curve was used for calculation of remained DBT concentration (Gai et al. 2007; Papizadeh et al. 2011). To confirm the presence of ending metabolite of DBT desulfurization (such as 2-HBP) Gibb’s assay was performed, which has been usually applied for determination of phenol ring (Quintana et al. 1997). This assay was carried out as follows: 1 ml of the supernatant from fungi culture on BSM medium was transferred to a Eppendorf tube containing 200 µl sodium bicarbonate (Na2CO3) (1 M, pH 8), then 20 µl Gibb’s reagent from stock solution in ethanol (1 µg/µl) was added. The reaction mixture was stirred for 30 min at 30 °C to form a blue colored complex (Stoner et al. 1990) and the absorbance of the complex was measured at 610 nm (Etemadifar et al. 2008).
DBT degradation by isolated strain
To determine DBT degradation, 1 ml of conidia suspension was inoculated into 250 ml Erlenmeyer flasks containing 50 ml BSM supplemented with 0.3 mM DBT as carbon and sulfur source and incubated at 30 °C and 180 rpm in a rotary shaker for 7 days. DBT was extracted and analyzed by UV-spectrophotometer at 24 h intervals as explained above.
Analaysis of DBT-desulfurization pathway and its metabolites
Measurement of DBT concentration and confirming the presence of 2-HBP (as product) and new peak, because of specific metabolites production and accumulation in medium, was performed using high-performance liquid chromatograph (HPLC) analysis. For this assay supernatant of fungal culture exposed to DBT was extracted by ethyl acetate during 4 and 7 days incubation periods and 20 µl ethyl acetate extracts was injected to a HPLC (Germany, Smartline Pump 1000, UV–Vis detector 2600) equipped with a perfectsil target C18 column (5 cm × 6.4 mm I.D.) (5 µm particles stationary phase diameters). Mobile phase consisted of 90 % methanol in water (V/V) with a flow rate of 1 ml/min and detection wavelength of 254 nm. All experiment steps were performed at room temperature (25–27 °C) and under the condition mentioned, retention time of DBT and 2HBP were 5.62 and 3.11 min, respectively. DBT concentration from extracted sample was calculated by using standard curve which was plotted through relation between known amounts of DBT and peak area.
Finally the molecular structures of produced metabolites were analyzed using Gas chromatography-mass spectrometry (GC–MS) equipped with a HP-5MS column [30 m by 0.25 µm (inner diameter) and 0.25 µm (film thickness)]. Helium gas at a rate 2.1 ml/min was entered to column and in order to isolate, the oven temperature program was as followed by 50 °C for 3 min with 8 °C increase per minute to 200 °C and finally to 290 °C with a rate of 12 °C increase per minute and 3 min at this temperature was kept constant.
Results
Isolation and characterization of a DBT-desulfurizing fungi
Among all fungi isolated from oil-contaminated soil on selective media one strain, designated FM strain, capable to consume DBT as the sulfur source was selected for further experiments. Initial identification of isolated FM strain was based on physiological and morphological characters. This fungus forms black, smooth, waxy and yeast-like colonies, and after 2-weeks was converted to downy and velvety in texture because of development of aerial hyphae (Fig. 1a, b). Hyphae were simple or branched with thin walls (Fig. 1c). Conidiogenous and conidiophores were tubular and spine-like, respectively. Conidia were sub-spherical to elliptical shapes and one-celled that were found in clusters at both sides of the conidiophores or accumulated at the tip of conidiogenous (Fig. 1d, e). Yeast-like cells were also seen in isolated strain (Fig. 1f).
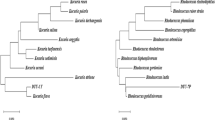
PCR product of ITS rDNA gave sharp band in 600 bp on agarose gel (Fig. 2). ITS sequence of isolated FM strain was compared with other sequences in the NCBI database using the BLAST algorithm. The BLAST result showed that FM strain was closely to the genus Exophiala spinifera with 98 % similarity to the ITS rDNA sequence of E. spinifera strain R68E3 in NCBI GenBank database. Finally phylogenetic tree was made using CLC Genomic Workbench 6.6.2. (Fig. 3). In this tree, FM strain and E. spinifera strain BMU are located in a branch of tree. The ITS sequence of the isolated strain was recorded in the NCBI database as E. spinifera strain FM with the accession number: KC952672.
Assessment of DBT desulfurization and growth of isolated strain FM
UV spectrophotometric and HPLC studies (Fig. 4) indicated that E. spinifera FM strain causing 99 % reduction (from 0.3 to 0.003 mM) of DBT after 7 days incubation at 30 °C and 180 rpm in an incubator shaker. The result (Fig. 5) showed that exponential phase began after 84 h long lag phase and after 180 h was transmitted to stationary phase. DBT utilization was started at the beginning of log phase, during this phase DBT concentration was utilized by 99 %. Maximum DBT utilization was during 168 h of cultivation and its concentration reduced to 0.003 mM.
Observation of blue-color in Gibb’s assay and the picks in HPLC analysis (Fig. 6a) indicated that 2-HBP was produced during DBT metabolism. So the result confirmed that DBT was desulfurized by isolated E. spinifera. Moreover, a new peak other than 2HBP presented in the extraction sample at t96. The new peak increased and 2-HBP peak deleted in extraction sample at the time of t168 (Fig. 6b).
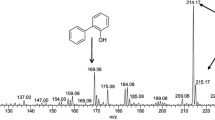
For characterization of observed new peak, a GC-mass equipped with a HP-5MS column detector was used and GC-mass analysis indicated that new peak was 1,3-benzenediol, 5-hexyl compound (Fig. 7).
The results implied that isolated fungi desulfurized DBT by passing through a way similar to 4S pathway, because 2-HBP peak observed on HPLC chromatogram and also peak related to DBT-sulfone observed on GC-mass analysis which is one of the intermediate metabolites from 4S-pathway. In fact, isolated fungus was able to transform the end product of this pathway (2-HBP) to 1,3-benzenediol, 5-hexyl which was less toxic. It is hypothesized that 2-HBP convert to di-hydroxy biphenyl by 2-Hydroxybiphenyl 3-monooxygenase enzyme that is an inducible flavoenzyme involved in the degradation of the toxic 2-hydroxybiphenyl produced by Exophiala and O2 through an oxidative meta-cleavage pathway, as shown below:

Then bioconversion of di-hydroxy biphenyl to 1,3-benzenediol, 5-hexyl as final product was occurred as following reaction:

Because of this transformation, 2-HBP peak reduced and 2,3-benzenediol, 5-hexyl appeared on extracted sample at t196. This phenomena and new metabolite proposed new biodesulfurization pathway for isolated strain FM, which is demonstrated in Fig. 8.
Discussion
Microorganisms require sulfur for their growth and biological activities. They are dependent on their enzymes and metabolic pathway so can obtain sulfur from different sources such as organosulfur compounds by using their enzymes (Stoner et al. 1990). Microorganisms may use hydrocarbons of oil as carbon source and this process reduces the oil fuel value (Gupta et al. 2005) thus, investigators were studying the bacteria do not consume petroleum hydrocarbons as carbon source and only use from sulfur of these compounds (Li and Jiang 2013). Accordingly, desulfurization activities by the organisms are divided into two groups: desulfurization without destroying the C–C bonds of organic sulfur compound and desulfurization with destroying the molecular structure (Soleimani et al. 2007).
Among metabolic researches, two major pathways for DBT consumption have been studied in bacteria: (1) Kudama pathway in Pseudomonas, that in this way DBT convert to 2-formyl, 3-hydroxyl banzothiophene by using cleavage on aromatic ring, (2) 4S pathway, which was identified in Rhodococcus erythropolis IGTS8 and some of the isolated strains (Yu et al. 2006). In 4S metabolic pathway the breakage does not happen on aromatic ring and 2-HBP and sulfate are its products (Gilbert et al. 1998; Arez et al. 2014). Sequencing and sub-molecular cloning tests on strain IGTS8 have revealed the desulfurization pathway is composed of three genes: dszA, dszB, dszC (Piddington et al. 1995). DBT convert to 2-HBP by using a multi-enzymatic pathway which consists of two mono-oxygenase and one di-oxygenase (Yu et al. 2006; Grossman et al. 2001). But the main known DBT desulfurization pathway is 4S. There is not a report for existence of 4S-pathway in DBT utilization by fungi.
Aranda et al. showed that white rot fungus Agrocybe is capable to convert DBT to eight different metabolites during 8 days. The metabolites are mainly products of the oxidation process (DBT-sulfone and DBT-sulfoxide) (Aranda et al. 2009). Conversion of DBT to DBT-sulfone by Bjerkandera sp. BOS55 with peroxidase-manganase enzyme followed by ring cleavage and production of 4-methoxy benzonic acid has been reported by Eibes et al. (2006). Crawford and Gupta (1990) reported that the fungus C. elegans converted 99 % of DBT to DBT-5-oxide and DBT-sulfone after 7 days and this DBT metabolism occurred by the P-450 cytochrome. No further metabolism of DBT by studied fungi was seen, because of the lack of appropriate enzymes.
In this study, one fungal strain was isolated from oil-contaminated soils. This fungus was capable of 99 % desulfurization of DBT in 7 days. GC–MS and HPLC techniques showed that this strain produced 2-hydroxy biphenyl and then convert it to 1,3-benzenediol, 5-hexyl. Among the studies on DBT utilization by the fungus, Fasion et al. reported that fungi Paecilomyces sp. TLI converted DBT to 2-2, dihydroxybiphenyl with an oxidation pathway (Fasion et al. 1991). Paecilomyces sp. TL1 was not able to metabolize hydroxyl biphenyl. Also oxidation of biphenyl to the hydroxylated derivatives 4,4′-dihydroxybiphenyl, 3,4-dihydroxybiphenyl, 2-hydroxybiphenyl and ring cleavage product 4-phenyl-2-pyrone-6-carboxylic acid by filamentous fungus Talaromyces helices was reported by Pinedo-Rivilla et al. (2009). Gesell et al. (2001) reported that fungi were able to biotransform and hydroxylate biphenyl compounds, also they could transform the hydroxylate biphenyl compounds and this lead to breakage or opening the aromatic rings (Gesell et al. 2001). Isolated strain FM in this study could be able to transform hydroxy biphenyl to 1,3-benzenediol, 5-hexyl as final product. In previous studies the investigators found that di-hydroxylate biphenyl production, induce breakage on aromatic rings and reduce toxicity of biphenyl compounds, so increased the fungal tolerance to these compounds (Romero et al. 2005).
Many metabolites determined by Zhang et al. (1997) during incubation of the antihistamine cyproheptadine hydrochloride with the zygomycete fungus C. elegans in liquid culture. Within 72 h cyproheptadine was extensively biotransformed to at least eight oxidative phase-1 metabolites primarily via aromatic hydroxylation metabolic pathways. Cyproheptadine was biotransformed predominantly to 2-hydroxy cyproheptadine. We are also subjected that our transformation is via hydroxylation of hydroxyl biphenyl.
Produced metabolites and end-products during the DBT metabolism by isolated strain FM, concluded this strain desulfurize DBT by passing through a way similar to 4S pathway. It is the first report about desulfurization of DBT through a pathway similar to 4S-pathway by fungus E. spinifera FM strain.
1,3-benzenediol, 4-Hexyl is slightly red insoluble powder which has anthelmintic and topical antiseptic usages; this is synthesize by chemical reaction. Most fungi and cyanobacteria produce metabolites with anthelmintic, anticancer, antibacterial activities or promote plant growth. Most studies so far have identified aromatic and polypeptide in fungi with antibacterial activities. For the first time in this study, it was shown that 1, 3 benzendiol, 5-hexyle is produced during DBT desulfurization. Considering the isomer structure of this to 1,3-benzendiol, 4-hexyl could be an important step for production of anti bacterial and anthelmintic drug. On the other hand the purity of final product tends to produce high yield drug and easy recovery by an organic phase extraction after fungal treatment (removal by filter). Thus, this paper not only demonstrates new pathway in desulfurization, but also detects this by-product and looked as new antibacterial drug. E. spinifera FM strain, in particular, is a preferred choice for this purpose.
References
Aranda E, Kinne M, Kluge M, Ullrich R, Hofrichter M (2009) Conversion of dibenzothiophene by the mushrooms Agrocybeae gerita and Coprinellus radians and their extracellular peroxygenases. Appl Microbiol Biotechnol 82:1066–1414
Arez BF, Alves L, Paixão SM (2014) Production and characterization of a novel yeast extracellular invertase activity towards improved dibenzothiophene biodesulfurization. Appl Biochem Biotechnol 174(6):2048–2057
Bahuguna A, Lily MK, Munjal A, Singh RN, Dangwal K (2011) Desulfurization of dibenzothiophene (DBT) by a novel strain Lysinibacillus sphaericus DMT-7 isolated from diesel contaminated soil. Environ Sci 23(6):975–982
Baldi F, Pepi M, Fava F (2003) Growth of Rhodosporidium toruloides strain DBVPG 6662 on dibenzothiophene crystals and orimulsion. Appl Environ Microbiol 69(8):4689–4696
Bezalel L, Hadar Y, Fu PP, Fremman JP, Cerniglia CJ (1996) Metabolism of phenanthrene by the White Rot Fungus Pleurotus ostreatus. Appl Environ Microbiol 62(7):2547–2553
Bhatia S, Sharma DK (2010a) Biodesulfurization of dibenzothiophene, its alkylated derivatives and crude oil by a newly isolated strain Pantoea agglomerans D23W3. Biochem Eng J 50(3):104–109
Bhatia S, Sharma DK (2010b) Mining of genomic databases to identify novel biodesulfurizing microorganisms. J Ind Microbiol Biotechnol 37(4):425–429
Bhatia S, Sharma DK (2012) Thermophilic desulfurization of dibenzothiophene and different petroleum oils by Klebsiella sp. 13T. Environ Sci Pollut Res 19(8):3491–3497
Campos-Martin JM, Capel-Sanchez MC, Perez-Presas K, Fierro JL (2010) Oxidative processes of desulfurization of liquid fuels. Chem Technol Biotechnol 85:879–890
Crawford DL, Gupta RK (1990) Oxidation of dibenzothiophene by Cunninghamella elegans. Curr Microbiol 21:229–231
Denis- Laros C, Labbe D, Bergeron H, Jones AM, Greer CW, Al-Hawari J, Grossman J, Sankey BM, Lau PC (1997) Conservation of plasmid- encoded dibenzothiophene desulfurization genes in several Rhodococcus. Appl Environ Microbiol 63:2915–2919
Eibes G, Cajthaml T, Moreira MT, Feijoo G, Lema JM (2006) Enzymatic degradation of anthracene, dibenzothiophene and pyrene by manganese peroxidase in media containing acetone. Chemosphere 64:408
Etemadifar Z, Emtiazi G, Peimanfar S (2006) Removal of dibenzothiophene, biphenyl and phenol from waste by Trichosporon sp. Sci Res Essays 1(3):072–076
Etemadifar Z, Emtiazi G, Christofi N (2008) Enhanced desulfurization activity in protoplast transformed Rhodococcus erythropolis. Am Eurasian J Agric Environ Sci 3(5):795–801
Fasion BD, Clark TM, Lewis SN, Ma CY, Sharkey DM, Woodward CA (1991) Degradation of organic sulfur compounds by a coal-solubilizing fungus. Appl Biochem Biotechnol 28:237–251
Ferrer C, Colom F, Frasés S, Mulet E, Abad JL, Alio JL (2001) Detection and identification of fungal pathogens by PCR and by ITS2 and 5.8S ribosomal DNA typing in ocular infections. Clin Microbiol 39(8):2873–2879
Gai Z, Yu B, Li L, Wang Y, Ma C, Feng J, Deng Z, Xu P (2007) Cometabolic degradation of dibenzofuran and dibenzothiophene by a newly isolated carbazole-degradaing Sphingomonas sp. strain. Appl Environ Microbiol 73(9):2832–2838
Gesell M, Hammer E, Specht M, Francke W, Schauer F (2001) Biotransformation of biphenyl by Paecilomyces lilacinus and characterization of ring cleavage products. Appl Environ Microbiol 67(4):1551–1557
Gilbert SC, Morton J, Buchanan S, Oldfield C, McRoberts A (1998) Isolation of a unique benzothiophene-desulfurizing bacterium, Gordonia sp. strain 213E (NCIMB 40816), and characterization of the desulfurization pathway. Microbiology 144(9):2545–2553
Grossman M, Lee M, Prince R, Minak-Bernero V, George G, Pickering I (2001) Deep desulfurization of extensively hydrodesulfurized middle distillate oil by Rhodococcus sp strain ECRD-1. Appl Environ Microbiol 67(4):1949–1952
Gupta N, Roychoudhury PK, Deb JK (2005) Biotechnology of desulfurization of diesel: prospects and challenges. Appl Environ Microbiol 66:356–366
Husaini A, Roslan HA, Hii KSY, Ang CH (2008) Biodegradation of aliphatic hydrocarbon by indigenous fungi isolated from used motor oil contaminated sites. World J Microb Biotechnol 24(12):2789–2797
Khedkar S, Shanker R (2014) Isolation and classification of a soil actinomycete capable of sulphur specific biotransformation of dibenzothiophene, benzothiophene and thianthrene. J Appl Microbiol 118(1):62–74
Li W, Jiang X (2013) Enhancement of bunker oil biodesulfurization by adding surfactant. World J Microbiol Biotechnol 29(1):103–108
Lin L, Hong L, Jianhua Q, Jinjuan X (2010) Progress in the technology for desulfurization of crude oil. China Pet Process PE 12(4):1–6
Ma CQ, Feng JH, Zeng YY, Cai XF, Sun BP, Zhang ZB, Blankespoor HD, Xu P (2007) Methods for the preparation of a biodesulfurization biocatalyst using Rhodococcus sp. Chemosphere 65:165–169
Papizadeh M, Roayaei Ardakani M, Motamedi H, Rasoli I, Zarei M (2011) C-S targeted biodegradation of dibenzothiophene by Stenotrophomonas sp. NISOC-04. Appl Biochem Biotechnol 165:938–948
Piddington CS, Kovacevich BR, Rambosek J (1995) Sequence and molecular characterization of a DNA region encoding the dibenzothiophene desulfurization operon of Rhodococcus sp. strain IGTS8. Appl Environ Microbiol 61:468–475
Pinedo-Rivilla C, Aleu J, Collado IG (2009) Pollutants biodegradation by fungi. Curr Org Chem 13(12):1194–1214
Quintana MG, Didion C, Dalton H (1997) Colorimetric method for a rapid detection of oxygenated aromatic. Biotechnol Techmol 11(8):585–587
Rashidi L, Mohebali G, Towfighi J, Rasekh B (2006) Biodesulfurization of dibenzothiophene and its alkylated derivaties through the sulfur-specific pathway by the bacterium RIPI-S81. Biotechnology 5:351–356
Romero MC, Hammer E, Hanschke R, Arambarri AM, Schauer F (2005) Biotransformation of biphenyl by the filamentous fungus Talaromyces helices. Microbiol Biotechnol 21:101–106
Soleimani M, Bassi A, Margaritis A (2007) Biodesulfurization of refractory organic sulfur compounds in fossil fuels. Biotechnol Adv 25:570–596
Stoner DL, Wey JE, Barrett KB, Jolley JG, Wright RB, Dugan PR (1990) Modification of water- soluble coal- derived products by dibenzothiophene-degrading microorganisms. Appl Environ Microbiol 56(9):2667–2676
Torkamani S, Shayegan J, Yaghmaei S, Alemzadeh I (2008) Study of the first isolated fungus capable of heavy crude oil biodesulfurization. Ind Eng Chem Res 47:7476–7482
Van Hamme JD, Wong ET, Dettman H, Gray MR, Pickard MA (2003) Dibenzyl sulfide metabolism by White Rot Fungi. Appl Environ Microbiol 69(2):1320–1324
Yu B, Xu P, Shi Q, Ma C (2006) Deep desulfurization of diesel oil and crude oils by a newly isolated Rhodococcus erythropolis strain. Appl Environ Microbiol 72(1):54–58
Zhang D, Hansen EB, Deck J, Heinze TM, Henderson A, Korfmacher W, Cerniglia C (1997) Fungal transformations of antihistamines: metabolism of cyproheptadine hydrochloride by Cunninghamella elegans. Xenobiotica 27(3):301–315
Acknowledgments
We are appreciated from Dr. Balali (Professor Associate in the University of Isfahan) for helping us in identification of fungal strain. Also we thank from the University of Isfahan for financial support of this work.
Conflict of interest
There is no conflict of interest between the authors of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Elmi, F., Etemadifar, Z. & Emtiazi, G. A novel metabolite (1,3-benzenediol, 5-hexyl) production by Exophiala spinifera strain FM through dibenzothiophene desulfurization. World J Microbiol Biotechnol 31, 813–821 (2015). https://doi.org/10.1007/s11274-015-1835-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11274-015-1835-0