Abstract
Excessive nitrogen (N) loading has had severe consequences in coastal zones around the world. Denitrification and anammox are major microbial pathways for removing N in aquatic environments before it is exported to the coast. To assess two processes in eutrophic riverine systems, the denitrification and anammox and their bacterial participants were investigated in sediments of the Xiaoqing (XQ) River and Jiaolai (JL) River in Northeast China. By combining the evidence from N15 isotope tracing experiment and functional gene-based analysis, it was found that denitrification and anammox are ubiquitous along the investigated riverine sediments. The denitrification varied from 39.38 to 1433.01 nmol N2 m−2 h−1. Moreover, the anammox rates were in the range of 15.91 to 1209.97 nmol N2 m−2 h−1. Quantitative PCR results revealed that the nirK and nirS genes were in the order of 104–106 copies g−1 and 103–105 copies g−1, respectively, in both river sediments, while the hzsA was in the order of 106–105 copies g−1 in XQ at approximately two orders of magnitude compared with JL. The phylogenetic analysis of functional genes revealed the high diversity of the denitrifier and low diversity of anammox bacteria. Variance partitioning analyses verified that the grain particle characteristics were the major factor group determined the N removal efficiency. The denitrification and anammox processes were estimated to have removed 16.1% of the inorganic nitrogen inputs before being exported to Laizhou Bay, which highlights that a more extensive understanding of the regularity of the N removal processes is important in the technical remediation of eutrophication problems.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Substances containing N that leak into the environment as a result of inefficient fertilizer usage have led to aquatic eutrophication problems around the world, particularly in estuarine and coastal zones (Painting et al. 2007; van Wijnen et al. 2015). The application of N fertilizer has increased the crop production to levels that enable the feeding of 40% of the world’s population, while 50–70% of the applied fertilizer have been lost into the water and air environment over the past century (Masclaux-Daubresse et al. 2010). The negative effects of the increased nitrate input are visible in coastal environments and include biodiversity losses, eutrophication, harmful algal blooms, and in more severe cases as hypoxia and even dead zones (Diaz and Rosenberg 2008; Qu and Kroeze 2010). Considering the growing population and increased use of irrigation, fertilizer usage is predicted to increase (Liu et al. 2005). Hence, the N eutrophication of aquatic environment is expected to severely situation (Strokal et al. 2014).
The biochemical removal of N in natural rivers reduces the delivery of N to coastal ecosystems and controls the development of marine eutrophication and dead zones (Donner and Kucharik 2008). It is estimated that 40 to 80% of N can be removed through assimilatory and dissimilatory processes before being exported to oceans (Breemen et al. 2002). Denitrification and anammox are the only two known microbiological pathways that can permanently remove N from aquatic systems by the end products of gaseous N2 or N2O. Denitrification is a step-wise anaerobic reduction from nitrate to nitrite, nitric oxide, nitrous oxide, and ultimately N2, through a complete reaction, or to N2O through an incomplete reaction. Several environmental conditions have been identified as key factors in controlling the denitrification rates, including temperature, nitrate concentrations, oxygen supply, water content, and even aquatic plants (Alldred and Baines 2016, Liu et al. 2015, Small et al. 2016, Speir et al. 2017). Anammox is also an anaerobic process converting ammonium with nitrite to N2 as the end product. The rates of anammox may vary depending on the available ammonium and nitrite competing with nitrification and denitrification. Previous studies on N removal in natural aquatic environments have suggested that denitrification and anammox are ubiquitous in anaerobic environments; however, their contributions may exhibit spatial and temporal variations. For example, anammox has been found to account for approximately 7% of N2 production in clay sediments, and for up to 58% in sand sediments at the Hampshire Avon Catchment, UK (Lansdown et al. 2016). Regarding the importance of denitrification and anammox in the efficiency of N removal in aquatic ecosystems, the available knowledge is still inadequate in terms of their responses and contributions under eutrophic constrains (Tang et al. 2018).
It is commonly assumed that microbial-mediated processes should be affected by the characteristics of their functional community, including abundance and diversity (Rocca et al. 2015). Both the nirK and nirS genes are frequently targeted to investigate denitrifier communities because they encode nitrite reductase which catalyzes the rate-limiting step of denitrification (Petersen et al. 2012). The hzsA gene, which encodes hydrazine synthase, has been proposed by studies that investigated anammox bacteria (Harhangi et al. 2012). Moreover, previous studies have reported that the functional gene abundance is positively related to the denitrification rate in riverine channels and coastal sediments (Guentzel et al. 2014; Zhang et al. 2018; Fozia et al. 2020). However, inconsistent trends have also been observed, which indicates that the environmental conditions, rather than the bacterial communities, mainly influence the denitrification rates (Lindemann et al. 2016; Temasek et al. 2017; Liu et al. 2018). It has been suggested that the denitrifier quantities may be the rate-limiting factor of the denitrification rates in oligotrophic environments, such that the nir gene abundances may appear to influence the denitrification rates. However, varying physicochemical conditions can restrict the denitrifier capacities in eutrophic environments, which may weaken the correlations between the genes and rates (Liu et al. 2018). Anammox bacterial communities and their biogeochemical activities have been observed in diverse environments (Crowe et al. 2017), but the relationship between the microbial communities and the anammox rates has rarely been reported, and the associations between the gene abundance and the process rates remain largely uncertain.
For semi-enclosed bays, N overloading can cause more severe problems on water quality owing to the weak water exchange. Laizhou Bay is a typical semi-enclosed inner sea in the southern Bohai Gulf of North China. According to the Chinese Ocean Environmental Quality Bulletin in 2013, more than one-fourth of Laizhou Bay failed to satisfy the grade IV national sea water quality standard, and N was found to be the primary pollutant leading to the deterioration of water quality and algal bloom onsets. Riverine inputs have been identified as the main source of N eutrophication, carrying approximately 13 thousand tons of dissolved inorganic nutrients (DIN) into Laizhou Bay every year (Zhang et al. 2006). Among these rivers surrounding Laizhou Bay, Xiaoqing (XQ) River and Jiaolai (JL) River are major contributors to eutrophication and their DIN discharges during 2013 have been reported as high as 3213 tons. This study investigated the denitrification and anammox patterns in the removal of N within the XQ and JL sediments with the objective of clarifying the biotic and abiotic variables affecting these two processes. Previous studies doubted that the denitrification and anammox processes would be accelerated under the impacts of the amount of substrate supply and abundance of functional microbes in eutrophic rivers (Kim et al. 2016; Lin et al. 2020). The denitrification and anammox rates along the XQ and JL Rivers were determined using the isotope pairing method (IPM). Additionally, DNA-based analyses were carried out to verify the distribution of functional bacteria and estimate their relationship with the N removal rates. This study had the following objectives: (1) verify the occurrence of denitrification and anammox processes in the sediments of two rivers; (2) investigate the control of environmental parameters and functional communities on the process rates and identify their relationships; (3) clarify the mechanism of N removal processes under eutrophic conditions and their contribution to the N loading of Laizhou Bay from the two eutrophic rivers.
2 Materials and Methods
2.1 Study Site and Sampling Procedure
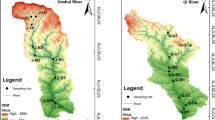
XQ and JL are located north of Shandong Peninsula in the south coastal area of Laizhou Bay. The mean annual rainfall of the river basin is approximately 740 mm, with 70–75% of precipitation occurring in the wet season from July to September (Liu et al. 2007). XQ is the second largest freshwater input to Laizhou Bay, estimated at 1.9 × 109 m3/year and flowing from the southwestern coast into Laizhou Bay. From the river’s 216-km length, four sites were selected with regard to their runoff and eutrophication from the estuary to the upstream 78-km meandering area, which is close to the petrochemical manufacturing base. In the southeastern part of Laizhou Bay, JL mainly flows through agricultural areas. As a smaller river, it dries up occasionally during the dry season. Four more sites were selected in the 50-km downstream area from JL’s total length of 130 km. The study areas of the eight sampling sites are shown in Fig. S1.
Sampling was carried out during the low tide from the riparian zone in April, August, and October of 2012 to account for seasonal variations. However, sampling was not conducted in winter because the river was frozen. All samples were labeled with site names followed by the month of collection. Sediment samples were collected in triplicate from the 5-cm surface of each site using a 5-ml syringe with the top removed for molecular microbial analysis. The samples were immediately stored in liquid N and transported to the laboratory. Then, the samples were stored at − 80 °C prior to being used for DNA extraction. Additional sediment samples were collected in triplicate using a 20-ml syringe to measure the geochemical variables, and were stored in a cooler at 4 °C before analysis. For all samples, six sediment cores (depth of 5 cm) were collected in perspex tubes (internal diameter of 3.4 cm, length of 0.2 m) and stored in a refrigerator at 4 °C for denitrification and anammox rate measurement. The overlying water was filtered through a 0.2-μm membrane and also collected to measure the rate in the two processes.
2.2 Investigation of Geochemical Variables
The physicochemical properties of the overlying water, including the temperature, pH, and salinity, were measured in situ using model 556MPS YSI. The interstitial water nutrients and sediment characteristics, including the particle diameter, water content, organic carbon (OC), and total N (TN), were determined from the sediment cores collected in triplicate. For nutrient analysis, interstitial water was obtained by centrifugation (10 min at 5000×g). The nutrient concentrations, including NH4+-N, NO2−-N, NO3−-N, and PO43—P, were measured using standard colorimetric methods in an AA3 segmented flow analyzer (Seal Analytical GmbH, Germany). The grain-size parameter of the mean diameter was investigated using a laser diffraction instrument (Malvern Mastersizer 2000, Malvern, UK). The water content percentage was determined by dividing the loss weight by the wet weight of the sediment. Freeze-dried sediment samples were used to measure the OC and TN in an elemental analyzer (Elementar, Vario Micro).
The wet weight of the sediment was recorded, and the sediment was subsequently freeze-dried for 48 h to determine the dry weight. The sediment volume was 15.71 cm3, as calculated by multiplying the cross-sectional area of the syringe by the depth of 5 cm. The wet bulk density was determined by dividing the wet weight by the sediment’s volume.
2.3 Determination of Denitrification and Anammox Rates
The sediment cores were overlaid with filtered-sterilized water collected from the sampling site and equilibrated overnight in a bath at the in situ temperature. Na15NO3 solution (abundance of 15N: 99.3%, Sigma) was added to every tube with 12.5 μmol or 25.0 μmol to obtain two sets of cores with different nitrate amendments (four replicate cores in each set). Thus, the denitrification and anammox could be measured using the isotope pairing technique (Dong et al. 2002; Risgaard-Petersen et al. 2003). The cores were immediately sealed with a rubber plug and re-equilibrated for 30 min to homogenize the distribution of the added 15NO3−, and then incubated at the in situ temperature for 3 h. The reference cores of the two concentration sets were sacrificed at the beginning of incubation by adding 0.5 ml of ZnCl2 solution (50%, wt/vol). At the end of the incubation, 0.5 ml of ZnCl2 solution was added to terminate the reaction. The sediment and water in each core tube were carefully mixed and a sample of the slurry was removed with a syringe inserted into a head-space anaerobic bottle containing 100 μl of ZnCl2. Then, the masses wherein N2 was produced with the m/z values of 28, 29, and 30 in the slurry samples were extracted and measured using a gas chromatographer coupled with an isotope ratio mass spectrometer (Gasbench-MAT253, Thermo Fisher Scientific, USA). The N production rates of denitrification and anammox were calculated using the formula proposed by Risgaard-Petersen et al. (2003).
2.4 Measurement of Denitrification and Anammox Bacterial Abundance and Biodiversity
DNA was extracted from the 0.50 g wet weight of the triplicate sediment samples using the PowerSoil DNA Isolation Kit (Mo Bio Laboratories, USA). All qPCRs were carried out using a 7500 Fast Real-Time PCR System (Applied Biosystems, USA). The PCR reaction mixtures contained 12.5 μl of 2 × SYBR Premix Ex Taq (TaKaRa Biotechnology, China), 200 nmol of forward primer and 200 nmol of reverse primer, and 1 μl of DNA template in a final volume of 25 μl. The primer sets and conditions used for the qPCRs are summarized in Table S1. Attempts were made to detect the hzo gene which encodes the hydrazine oxidoreductase that dehydrogenates hydrazine to dinitrogen gas. However, these attempts were not successful for any samples using the primer set of hzocl1F1-hzocl1R2 (Li et al. 2010).
Standard DNA plasmids were prepared by cloning the target gene fragment, which was amplified by the sediment samples using the same primer as that used for qPCR. The gene fragments in the standard plasmid DNA were confirmed by their sequences, which are presented in Table S2. The copy number in the standard solution was calculated using a previously proposed method (Li et al. 2015). Standard samples and no-template controls were analyzed in triplicate. For every qPCR run, the standard target DNAs were included in the 96-well plate. The data were analyzed against the standard curve using the 7500 software ver. 2.0.6 (Applied Biosystems), and then converted into copies per gram of sediment.
The clone library approach was employed to characterize the functional microbial community and determine the participants to the N removal processes in the two rivers. Based on the denaturant gradient gel electrophoresis (DGGE) gel images (for detailed methods and results, see supplementary material S1), several samples were selected to construct the nirK, nirS, and hzsA clone libraries for phylogenetic analysis. The primer sets and conditions used for amplification are summarized in Table S1. The PCR products were cloned and then sequenced after confirming that the inserted DNA fragments had appropriate sizes. The sequences were clustered into operational taxonomic units (OTUs) using the Mothur program, which employs employing the furthest neighbor algorithm (Schloss et al. 2009). The partial sequences were aligned within the same region of the closest strains available in the GenBank database using the BLAST facility. The sequence alignment was achieved using ClustalW. The phylogenetic trees were constructed in Mega 5.0 using the maximum-likelihood (ML) method (Tamura et al. 2011).
The unique functional nucleotide sequences generated in this study were deposited in the GenBank database with the following accession numbers: MK497295 to MK497556 for nirK, MK497557 to MK497726 for nirS, and MK497258 to MK497294 for hzsA.
2.5 Statistical Analysis
Before these statistical analyses, normal distribution of all dataset was checked by carrying out the Shapiro-Wilk test. The non-normally distributed data were subjected to natural log or square root transformation.
One-way analysis of variance (ANOVA) tests were conducted to evaluate the significant differences among the environmental parameters, functional bacterial abundances, and N removal rates in sediments that were seasonally sampled along the two rivers considered in this study. A P value < 0.05 was considered a difference, and < 0.01 as a significant difference. Two-way ANOVA was conducted to test the effects of the environmental variables and the functional gene abundances in the N removal process rates; their relationships are represented by the Pearson correlation coefficient. Variation partitioning analysis was used to identify which environmental variables explained most of the variation in the denitrification and anammox rates. A series of partial multiple regression analyses were carried out to decompose the variation in the N removal rates explained by the four variable groups, namely, the physicochemical properties, water nutrients, sediment characteristics, and functional bacterial abundances, which are explained jointly by these factor categories, and not explained by any of these factors (Heikkinen et al. 2004). The abovementioned statistical analyses were carried out using R (The R Foundation for Statistical Computing) with the “vegan” package (Dixon, 2003).
3 Results
3.1 Sediment Environmental Parameters
The environmental parameter arrays, including the physicochemical properties, nutrient concentrations, and sediment characteristics, which likely influence the microbial growth and activities, or reaction processes, were measured in the riparian sediment of each site (Table S3). The summer peak flows were reflected on the salinity in August, which decreased by the increase of freshwater runoff. During the investigation period, high nitrogen contents were observed in the sediments of the two rivers. The interstitial water NH4+ concentration varied between 0.004 and 12.558 mg L−1. Downstream of JL and close to the river mouth, the NH4+ content increased in the range of 9.436–12.558 μmol L−1 in April and October. The NO2− concentration in the sediment varied from a minimum of 0.008 to a maximum of 2.827 mg L−1, while the NO3− concentration ranged between 0.023 and 3.414 mg L−1. Relatively higher NO2− and NO3− contents were detected in April at all XQ sites.
The fluctuation between the measured parameters of the two rivers was observed in the seasonally collected samples (Table 1). The NO3− and OC concentration and mean diameter in the XQ sediments were significantly higher than those in the JL sediments (P < 0.01, and F = 11.561, 39.624, and 32.789, respectively). Similarly, the JL sediments contained significantly higher NH4+ compared with the XQ sediments (with P < 0.01). However, these fluctuations did not exhibit differences among the considered sites, except for the mean diameter (P < 0.001, F = 8.782), which exhibited a significantly larger sediment size along the two rivers.
3.2 N Removal Rates
The microbial denitrification and anammox rates were determined in laboratory incubations using the seasonal sediments collected from the two rivers (Fig. 1). The sediment denitrification rates exhibited large temporal and spatial variations, with the lowest values observed for JL2 in August (39.38 ± 12.13 nmol N2 m−2 h−1) and the highest values observed for XQ2 also in August (1433.01 ± 57.58 nmol N2 m−2 h−1). The occurrence of anammox in both sediments was confirmed by the substantial 29N2 production in the incubations. In the sediment, 15NO3− labelling revealed anammox rates ranging from 15.91 ± 7.50 nmol N2 m−2 h−1 for XQ3 to 1206.97 ± 178.02 nmol N2 m−2 h−1 for XQ2, both of which were observed in August. The rates of the two processes always had the same order of magnitude in the same sample. Denitrification and anammox appeared to be equally important processes for the removal of N in both rivers, with anammox contributing between 14 and 74%. The XQ sediments had an N removal rate (average of 592.19 nmol N2 m−2 h−1) that was slightly, but not significantly, greater than that of the JL sediments (average of 521.89 nmol N2 m−2 h−1). Apart from the anammox between April and October (P = 0.026), there was no other difference in the denitrification and anammox rates across sites and seasons. The correlation of the denitrification and anammox rates with the environmental variables was not observed. Notably, anammox and denitrification exhibited a significantly positive relationship (n = 24, r = 0.898, P < 0.01).
3.3 Functional Gene Abundances
The copy numbers of the corresponding genes using absolute quantitative PCR estimated the abundance of the selected genes involved in the N removal processes. The denitrifier was counted using the nirK and nirS gene copy numbers, which encode the copper and cytochrome cd1-containing nitrite reductases, respectively. In riparian sediments, the abundance of the nirK gene ranged from 1.60 × 104 to 3.03 × 106 copies g−1, while the abundance of the nirS gene varied from 8.21 × 102 to 1.79 × 105 copies g−1 (Table 2). In all samples, the nirK copies outnumbered the nirS copies, and exhibited a positive correlation (n = 24, r = 0.455, P = 0.025). Although a significant difference was not observed in the denitrification gene copy numbers across sites and seasons, JL4 exhibited higher fluctuations, while the maximal/minimal values of the nirK and nirS copy numbers appeared in different seasons (Table 2). The denitrifier abundance encoded by nirK was positively correlated with the pH change (P = 0.012).
Gene hzsA was used to estimate the number of anammox bacteria. This gene encodes one of the subunits of hydrazine synthase, which is unique and irreplaceable for the anammox metabolism catalyzing the hydrazine synthesis from nitric oxide and ammonium (Harhangi et al. 2012). In all samples, the copy numbers of this gene were detected within the range of 1.72 × 103 to 1.25 × 106 copies g−1. The average gene hzsA copies in the XQ and JL sediments were 2.49 × 105 and 2.80 × 104 copies g−1, respectively, which indicates their difference with a P value = 0.042. The abundance of anammox bacteria revealed by hzsA exhibited a significantly positive correlation with the water and OC content of the sediment (P = 0.008 and 0.007, respectively), and positive correlation with the mean diameter and TN content of the sediment (P = 0.010 and 0.041, respectively). Correlation was not observed between the gene numbers of the denitrifier or anammox bacteria and the denitrification rates (n = 24, P = 0.185 and 0.883, respectively) or anammox rates (n = 24, P = 0.863).
3.4 Presence of Denitrifier and Anammox Bacteria
The identification and distribution of denitrifier and anammox bacteria inhabiting the XQ and JL were assessed by the nirK, nirS, and hzsA sequences retrieved from the river sediments. More samples were selected to construct the denitrifier clone library encoded by nirK gene, owing to its dominance in terms of having more abundance and larger community diversity compared with the nirS gene in these samples (Table 2 and S1 result in Supplementary Material). Gene hzsA exhibited low diversity (S1 result in Supplementary Material) and only two samples were selected for clone library analysis. The fact that the anammox bacteria frequently have low diversity in lake and river ecosystems is another reason for selecting few samples to investigate anammox diversity (Wang et al. 2012; Zhang et al. 2007).
In the nirK clone library analysis, 280 sequences were obtained, among which 59, 57, 55, 56, and 53 clones were obtained from the XQ4-SP, JL3-SP, JL2-SU, XQ1-FA, and JL3-FA samples, respectively. The 110 OTUs were grouped by a sharing homology of 85% in nucleotide acid sequences and distributed within eight phylogenetic clades (enumerated from I to VIII) in the ML phylogenetic tree (Fig. 2). All clades were affiliated with nirK from Proteobacteria, including the known genera of Rhizobium, Nitrosomonas, Pseudomonas, and Rhodobacter. A total of 200 nirS sequences were retrieved from the XQ4-SP, XQ1-FA, and JL3-FA samples, and subsequently allotted into 68 OTUs at a 10% nucleotide acid divergence. The nirS phylogenetic tree was divided into seven clusters (enumerated from I to VII; Fig. 3). Phylogenetic analysis indicated the known denitrifier genera, including Paracoccus, Pseudomonas, Rhodobacter, and Hakomanas. Additionally, unknown denitrifier-like groups were observed in the XQ and JL sediments. In total, 37 clones of the hzsA gene were sequenced from the XQ4-SP and JL3-SP samples, which represented 11 OTUs at a 3% nucleotide acid divergence. All sequences shared a homology over 78% and were divided into two clusters (referred to as I and II; Fig. 4). Phylogenetic analysis revealed that 25 clone sequences were closely related to the known anammox bacteria belonging to the Candidatus Brocadia group. The remaining clones were related to Candidatus Kuenenia.
ML tree using the GTR + G + I model representing the phylogenetic position of nirK sequences obtained from the sediments of rivers XQ and JL. The values for nodes with over 50% bootstrap support are shown. The numbers in parentheses following each OTU indicate the numbers of clones recovered from the samples of XQ4-SP (a), JL3-SP (b), JL2-SU (c), XQ1-FA (d), and JL3-FA (e)
ML tree using the GTR + G + I model representing the phylogenetic position of the nirS sequences obtained from the sediments of XQ and JL. See Fig. 2 for more details
ML tree using the NT + G + I model representing the phylogenetic position of the hzsA sequences obtained from the sediments of XQ and JL. See Fig. 2 for more details
3.5 Variance Partitioning of Environmental Variables
The results obtained by multiple regression analyses indicated that all environmental factors considered in this study accounted for 73% and 82% of the variances in the sediment denitrification and anammox rates, respectively (Fig. 5). The variance partitioning analyses demonstrated that the joint effect exerted by the four categories is the primary explanation for the variances of the sediment denitrification and anammox rates, which implies that the environmental factor groups could regulate the rates by cooperating with one another. Additionally, the N removal processes were mainly influenced by the sediment characteristics, which accounted for 23% of the variance in each rate. However, the denitrifying and anammox bacterial communities had a very small and statistically insignificant effect on their corresponding rates.
4 Discussion
In XQ and JL, the river sediments exhibited higher levels of N (NH4+, NO3−, and TN) and C (OC) compared with typical eutrophic rivers such as Ishikari River in Japan (Jha and Minagawa 2013), Saint Louis River in France (Loken et al. 2016), and Cape Fear River in the USA (Lisa et al. 2015). XQ and JL have similar trophic status, but the main form of dissolved inorganic nitrogen was much different in the sense that the nitrate concentration was high in XQ while JL had high ammonia concentration. Additionally, the obvious effect of river flow on the environmental conditions was that, in the summer rainy season, the salinity and nutrient concentrations changed accordingly, particularly for JL which has smaller flow. In this study, we attempted to verify the contributions of denitrification and anammox in the XQ and JL sediments to clarify the effects of eutrophication in relatively shallow rivers with occasionally oxygen-exposed environments.
The denitrification and anammox rates measured in XQ and JL are comparable with the results obtained by previous studies in similar environments. The denitrification rates ranged from 39.38 to 1433.01 nmol N2 m−2 h−1. By scaling on the basis of weight using the wet bulk density and height of sediments, the variation range was between 0.013 and 0.48 ng N g−1 h−1, which is within the range reported for other river sediments (Jia et al. 2016; Liu et al. 2018; Ritz et al. 2018). The rates of anammox (15.91–1206.97 nmol N2 m−2 h−1, equivalent to 0.005–0.40 ng N g−1 h−1) were within the lower quartile of the values reported for other eutrophic aquatic sediments (Zheng et al. 2016). Moreover, the positive relationship between the denitrification and the anammox rates was significant. This conclusion is corroborated by earlier studies in Yangtze River which have reported that the denitrification rates are significantly coupled with the anammox rates with a linear correlation index of 0.803 (Zheng et al. 2016). These findings are also supported by the hypothesis that nitrite as the anammox substrate could be released by the denitrifier owing to the imbalance of the required nitrate and carbon condition (Brin et al. 2014). The relative contribution of anammox in the N2 production (ra%) ranged between 14 and 74%, with an average value of 43.5%. This indicates that anammox is another main process in the removal of N from river sediments. Notably, this result is contrary to that obtained by several previous studies investigating N removal processes in river sediments which have reported an anammox contribution of less than 10% (Wang et al. 2012; Zhu et al. 2013; Naeher et al. 2015). The difference in the findings may have resulted from the high concentration of inorganic nitrogen and relatively lower supply of organic matter (Lu et al. 2018).
In XQ and JL, the average abundance of the nirK gene (6.56 × 105 copies g−1) was approximately 20 times higher than that of nirS (3.43 × 104 copies g−1). The total nir-gene abundances were lower by approximately 1–3 orders of magnitude compared with previously reported findings for other rivers, such as the urban stream of Campus Greeks (Graham et al. 2010), Cape Fear River Estuary (Lisa et al. 2015), and Santa Fe River (Kim et al. 2016). The dominance of the nirK denitrifier over the nirS denitrifier is typically observed in the water-column and surface sediment of shallow rivers, where there occasionally exist oxygen-exposed environments stimulating the existence of the nirK denitrifier in overwhelming amounts (Graham et al. 2010). Conversely, the overwhelming dominance of the nirS denitrifier has been observed in the eutrophic sediments of lakes and estuaries under limited oxygen conditions (Abell et al. 2010; Yao et al. 2018). Tomasek et al. (2017) reported a similar trend of the nirS denitrifier being a majority group in flood locations, but a minority group in nonflood zones. In this study, a strong positive relationship between the abundance of the nirK gene and pH was observed. Their correlation has rarely been detected in aquatic habitats, but has been repeatedly confirmed under field and incubation conditions, and with a wider pH range in terrestrial environments (Šimek and Cooper 2010). The effect of pH is attributed to the fact that less organic carbon and inorganic nitrogen are available in relatively acidic environments. The denitrifier community composition exhibited high diversity without a sample-specific clade. Moreover, this diverse and homogeneous distribution has also been observed in other estuarine sediments, which implies broad adaptability to the environmental conditions (Jiao et al. 2018).
Gene hzsA is a more unambiguous functional marker for the anammox bacteria compared with the anammox-specific 16S rRNA gene, which is also present in other Planctomycetes (Harhangi et al. 2012). Considering the typical 3- to 6-fold ratio between the 16S rRNA and hzsA genes, the abundance of anammox detected in this study (1.72 × 103–1.25 × 106 copies g−1) is consistent with that detected in most previous studies, including those conducted at Cape Fear River (1.3 × 105–8.4 × 106 copies g−1) (Dale et al. 2009), Dongjiang River (2.66 × 105–1.07 × 107 copies g−1) (Sun et al. 2014), and Pearl River Estuary (Wang et al. 2012). Significant positive correlations were detected between the anammox bacterial abundance and the parameters of sediment characteristics including the mean diameter, water content, OC, and TN. Considering the relationships of these four variables, partial correlation analysis was further used to verify their effects. However, the correlation of each variable with anammox bacterial abundance was insignificant (P > 0.05). Hence, it was hypothesized that the significant joint influences exerted by the four variables were likely caused by the potential interactions between grain-associated anammox bacteria and heterotrophic bacteria. Various studies have revealed that grain particles can create an anaerobic microenvironment through the aerobic respiration and oxidation of anaerobic metabolites, which provide a suitable environment for anammox bacteria to oxidize ammonium (Jia et al. 2016; Xia et al. 2019). Additionally, grain particles with larger size (in the range of 0.8–8 μm) and high OC concentration are capable of sustaining more bacterial cells to form oxygen-limited conditions and facilitate the growth of anammox bacteria (Zeng et al. 2018). The extremely low diversity of the anammox bacteria was revealed by the phylogenetic analyses of the hzsA gene, which were mainly restricted to the Bracadia and Kuenenia genera that have been reported to be most common in freshwater and terrestrial ecosystems (Lisa et al. 2015). Bracadia dominated the anammox assemblages in the XQ4-SP sample and Kuenenia was only observed in the JL3-SP sample. Differential competition with substrates and differential sensitivity to salinity may explain their distribution. With regard to nitrite, Bracadia may be outcompeted by other anammox bacteria, particularly under eutrophic conditions, which determines its predominance in N-overloaded environments (Naeher et al. 2015). Additionally, Kuenenia has been reported to have a higher tolerance to salinity compared with Bracadia and has been discovered in JL, where obvious salinity fluctuations occur owing to the smaller flow and lower saltwater intrusion (Zheng et al. 2016).
The variation partitioning analysis results revealed that the sediment characteristics play a critical role in determining the sediment denitrification and anammox rates in the two investigated rivers (Fig. 5). Considering the close coupling of denitrification and anammox, and the influence of abiotic conditions on the denitrifier and anammox bacteria, it could be attempted to clarify the mechanism of N removal processes and the reason for which the single variables cannot explain the rate changes. In relatively shallow river sediments, the grain particles may provide a suitable microenvironment under suboxic conditions by the respiration of attached microorganisms for anaerobic reactions such as denitrification and anammox. In this case, sediment characteristics such as the particle size, bulk density, water content, and organic content may influence the N removal rates by promoting conditions favorable to the attached organisms. Similar results have been reported for other river and stream systems (Jha and Minagawa 2013; Xia et al. 2017). Owing to the slow growth of autotrophic organisms and low affinity for nitrate, in the substrate, the anammox bacteria are outcompeted by denitrification, and even rely on the supply of nitrite by denitrification in a stepwise manner (Naeher et al. 2015). Complicated environmental conditions such as the physiological reaction, potential connections between denitrification and anammox, and unstable environments eventually result in weak correlations between abiotic or biotic conditions and the rates of nitrification and denitrification.
The N removal capacity of denitrification and anammox for XQ and JL can be estimated by multiplying the measured rates of denitrification and anammox rates at each site and in each season by the total sediment area in the sector of the river centered on that site. The total denitrification and anammox in the investigated downstream reaches were approximately 5.84 and 5.45 Mmol N per year for XQ, respectively, and approximately 1.16 and 1.11 Mmol N per year for JL. The total annual flux of DIN into Jiaozhou Bay from XQ was 59.0 Mmol N, as calculated by the data provided by the Chinese Ocean Environmental Quality Bulletin in 2012. The DIN fluxes from JL were estimated to be approximately one-fifth of XQ, according to the nutrient concentration and flow discharge ratio data for the two rivers (Zhang and Gao 2016; Zhang et al. 2012). Thus, the denitrification and anammox both achieved 16.1% removal of the annual flux of the total nitrogen in XQ and JL. It was estimated that the denitrification and anammox account for the removal of approximately 13 to 50% of N sources in rivers and estuaries globally, as calculated based on the relationship between the N removal rates and the water travel time (Seitzinger et al. 2006). This removal percentage is comparable with that occurring in XQ and JL, which indicates that the denitrification and anammox in the two rivers play a significant role in mitigating the nitrogen inputs to the estuarine.
5 Conclusions
Sediment is an important natural system for removing ammonia and nitrate because denitrification and anammox can mitigate the biologically available nitrogen exported to coastal ecosystems. Because the availability of the N substrate increases with the N input, it was considered that the denitrification and anammox rates are higher in eutrophic lakes, rivers, or estuaries (Seitzinger et al. 2006). Denitrification and anammox were detected in the sediments along XQ and JL and identified by conducting molecular and 15N isotope experiments. Their rates in the two rivers fall in the lower range of values compared with other river ecosystems. Hence, higher substrate availability dose not promote the processes. During the study period, significant spatial and temporal variations were observed for the rates, and the variance partitioning analysis characteristics of the sediments can explain the major portion of the variability. Considering the relationships between the environmental conditions and the functional bacterial abundances, it is proposed that the anoxic conditions on the surface of grain particles are limiting factors for denitrification and anammox. This result may be universal for all relatively shallow rivers, where the sediments are aerobic with a surface thickness of a few centimeters (Naeher et al. 2015). Future work should assess the effect of particle characters and dissolved oxygen on the removal of N in river systems. Although there was no significant increase in the denitrification and anammox rates in eutrophic rivers, their contribution to the reduction of N exports from rivers to coastal ecosystems cannot be disregarded. In XQ and JL, the denitrification and anammox approximately accounted for the removal of 16.1% of N loading from rivers. Owing to the positive effects exerted by reducing the eutrophication associated with excess N input, technical approaches can be proposed to modify the geochemical conditions and increase the denitrification and anammox rates in sediments. However, this study shows that it is difficult to establish clear links between a single environmental variable and the N removal capacities, which implies that it is probably unrealistic to enhance the denitrification and anammox rates simply by managing the physicochemical conditions or launching functional bacterial inoculum. Overall, the results of this study stress the significant contribution of denitrification and anammox to the reduction of N loading in coastal zones. Future studies on the regularity of nitrogen removal processes should refer to the findings of this study to develop a targeted technique for enhancing the capacity of removing N in river systems.
References
Abell, G. C. J., Revill, A. T., Smith, C., Bissett, A. P., Volkman, J. K., & Robert, S. S. (2010). Archaeal ammonia oxidizers and nirS-type denitrifiers dominate sediment nitrifying and denitrifying populations in a subtropical macrotidal estuary. ISME Journal, 4, 286–300.
Alldred, M., & Baines, S. B. (2016). Effects of wetland plants on denitrification rates: a meta-analysis. Ecological Applications, 26(3), 676–685.
Breemen, N. V., Boyer, E. W., Goodale, C. L., Jaworski, N. A., Paustian, K., Seitzinger, S. P., Lajtha, K., Mayer, B., Dam, D. V., & Howarth, R. W. (2002). Where did all the nitrogen go? Fate of nitrogen inputs to large watersheds in the northeastern U.S.A. Biogeochemistry, 57-58(1), 267–293.
Brin, L. D., Giblin, A. E., & Rich, J. J. (2014). Environmental controls of anammox and denitrification in Southern New England estuarine and shelf sediments. Limnology and Oceanography, 59(3), 851–860.
Crowe, S. A., Treusch, A. H., Forth, M., Li, J., Magen, C., Canfield, D. E., Bo, T., & Katsev, S. (2017). Novel anammox bacteria and nitrogen loss from Lake Superior. Scientific Reports, 7(1), 13757.
Dale, O. R., Tobias, C. R., & Song, B. (2009). Biogeographical distribution of diverse anaerobic ammonium oxidizing (anammox) bacteria in Cape Fear River estuary. Environmental Microbiology, 11(5), 1194–1207.
Diaz, R. J., & Rosenberg, R. (2008). Spreading dead zones and consequences for marine ecosystems. Science, 321(5891), 926–929.
Dixon, P. (2003). VEGAN, a package of R functions for community ecology. Journal of Vegetation Science, 14, 927–930.
Dong, L. F., Nedwell, D. B., Underwood, G. J. C., Thornton, D. C. O., & Rusman, I. (2002). Nitrous oxide formation in the Colne estuary in England: the central role of nitrite. Applied and Environmental Microbiology, 68(3), 1240–1249.
Donner, S. D., & Kucharik, C. J. (2008). Corn-based ethanol production compromises goal of reducing nitrogen export by the Mississippi River. Proceedings of the National Academy of Sciences of the United States of America, 105(11), 4513–4518.
Fozia, Zheng, Y., Hou, L., Zhang, Z., Gao, D., Yin, P., Han, P., Dong, H., Liang, X., Yang, Y., & Liu, M. (2020). Community dynamics and activity of nirS-harboring denitrifiers in sediments of the Indus River estuary. Marine Pollution Bulletin, 153, 110971.
Graham, D. W., Trippett, C., Dodds, W. K., O'Brien, J. M., Banner, E. B. K., Head, I. M., Smith, M. S., Yang, R. K., & Knapp, C. W. (2010). Correlations between in situ denitrification activity and nir-gene abundances in pristine and impacted prairie streams. Environmental Pollution, 158(10), 3225–3229.
Guentzel, K. S., Hondzo, M., Badgley, B. D., Finlay, J. C., Sadowsky, M. J., & Kozarek, J. L. (2014). Measurement and modeling of denitrification in sand-bed streams under various land uses. Journal of Environmental Quality, 43(3), 1013–1023.
Harhangi, H. R., Le Roy, M., van Alen, T., Hu, B.-L., Groen, J., Kartal, B., Tringe, S. G., Quan, Z.-X., Jetten, M. S., & den Camp, H. J. O. (2012). Hydrazine synthase, a unique phylomarker with which to study the presence and biodiversity of anammox bacteria. Applied and Environmental Microbiology, 78(3), 752–758.
Heikkinen, R. K., Luoto, M., Virkkala, R., & Rainio, K. (2004). Effects of habitat cover, landscape structure and spatial variables on the abundance of birds in an agricultural-forest mosaic. Journal of Applied Ecology, 41(5), 824–835.
Jha, P. K., & Minagawa, M. (2013). Assessment of denitrification process in lower Ishikari river system, Japan. Chemosphere, 93(9), 1726–1733.
Jia, Z., Liu, T., Xia, X., & Xia, N. (2016). Effect of particle size and composition of suspended sediment on denitrification in river water. Science of the Total Environment, 541, 934–940.
Jiao, L., Wu, J., He, X., Wen, X., Li, Y., & Hong, Y. (2018). Significant microbial nitrogen loss from denitrification and anammox in the land-sea interface of low permeable sediments. International Biodeterioration & Biodegradation, 135, 80–89.
Kim, H., Bae, H.-S., Reddy, K. R., & Ogram, A. (2016). Distributions, abundances and activities of microbes associated with the nitrogen cycle in riparian and stream sediments of a river tributary. Water Research, 106, 51–61.
Lansdown, K., McKew, B. A., Whitby, C., Heppell, C. M., Dumbrell, A. J., Binley, A., Olde, L., & Trimmer, M. (2016). Importance and controls of anaerobic ammonium oxidation influenced by riverbed geology. Nature Geoscience, 9, 357.
Li, J., Nedwell, D. B., Beddow, J., Dumbrell, A. J., McKew, B. A., Thorpe, E. L., & Whitby, C. (2015). amoA gene abundances and nitrification potential rates suggest that benthic ammonia-oxidizing bacteria and not archaea dominate N cycling in the Colne estuary, United Kingdom. Applied and Environmental Microbiology, 81(1), 159–165.
Li, M., Hong, Y., Klotz, M. G., & Gu, J. D. (2010). A comparison of primer sets for detecting 16S rRNA and hydrazine oxidoreductase genes of anaerobic ammonium-oxidizing bacteria in marine sediments. Applied Microbiology and Biotechnology, 86(2), 781–790.
Lin, J., Chen, N., Wang, F., Huang, Z., Zhang, X., & Liu, L. (2020). Urbanization increased river nitrogen export to western Taiwan Strait despite increased retention by nitrification and denitrification. Ecological Indicators, 109, 105756.
Lindemann, S., Zarnoch, C. B., Castignetti, D., & Hoellein, T. J. (2016). Effect of eastern oysters (Crassostrea virginica) and seasonality on nitrite reductase gene abundance (nirS, nirK, nrfA) in an urban estuary. Estuaries and Coasts, 39(1), 218–232.
Lisa, J. A., Song, B., Tobias, C. R., & Hines, D. E. (2015). Genetic and biogeochemical investigation of sedimentary nitrogen cycling communities responding to tidal and seasonal dynamics in Cape Fear River estuary. Estuarine Coastal and Shelf Science, 167, A313–A323.
Liu, G. D., Wu, W. L., & Zhang, J. (2005). Regional differentiation of non-point source pollution of agriculture-derived nitrate nitrogen in groundwater in northern China. Agriculture, Ecosystems and Environment, 107, 211–220.
Liu, G. Q., Wang, S. Y., Zhu, X. J., Liu, S. M., & Zhang, J. (2007). Groundwater and nutrient discharge into Jiaozhou Bay, North China. Water Air and Soil Pollution, 7(6), 593–605.
Liu, W., Wang, Z., Zhang, Q., Cheng, X., Lu, J., & Liu, G. (2015). Sediment denitrification and nitrous oxide production in Chinese plateau lakes with varying watershed land uses. Biogeochemistry, 123(3), 379–390.
Liu, W., Yao, L., Jiang, X., Guo, L., Cheng, X., & Liu, G. (2018). Sediment denitrification in Yangtze lakes is mainly influenced by environmental conditions but not biological communities. Science of the Total Environment, 616-617, 978–987.
Loken, L. C., Small, G. E., Finlay, J. C., Sterner, R. W., & Stanley, E. H. (2016). Nitrogen cycling in a freshwater estuary. Biogeochemistry, 127(2–3), 199–216.
Lu, X., Bade, D. L., Leff, L. G., & Mou, X. (2018). The relative importance of anammox and denitrification to total N2 production in Lake Erie. Journal of Great Lakes Research, 44(3), 428–435.
Masclaux-Daubresse, C., Daniel-Vedele, F., Dechorgnat, J., Chardon, F., Gaufichon, L., & Suzuki, A. (2010). Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Annals of Botany, 105(7), 1141–1157.
Naeher, S., Huguet, A., Roose-Amsaleg, C. L., Laverman, A. M., Fosse, C., Lehmann, M. F., Derenne, S., & Zopfi, J. (2015). Molecular and geochemical constraints on anaerobic ammonium oxidation (anammox) in a riparian zone of the Seine estuary (France). Biogeochemistry, 123(1), 237–250.
Painting, S. J., Devlin, M. J., Malcolm, S. J., Parker, E. R., Mills, D. K., Mills, C., Tett, P., Wither, A., Burt, J., Jones, R., & Winpenny, K. (2007). Assessing the impact of nutrient enrichment in estuaries: susceptibility to eutrophication. Marine Pollution Bulletin, 55(1), 74–90.
Petersen, D. G., Blazewicz, S. J., Firestone, M., Herman, D. J., Turetsky, M., & Waldrop, M. (2012). Abundance of microbial genes associated with nitrogen cycling as indices of biogeochemical process rates across a vegetation gradient in Alaska. Environmental Microbiology, 14(4), 993–1008.
Qu, H. J., & Kroeze, C. (2010). Past and future trends in nutrients export by rivers to the coastal waters of China. Science of the Total Environment, 408(9), 2075–2086.
Risgaard-Petersen, N., Nielsen, L. P., Rysgaard, S., Dalsgaard, T., & Meyer, R. L. (2003). Application of the isotope pairing technique in sediments where anammox and denitrification coexist. Limnology and Oceanography: Methods, 1, 63–73.
Ritz, S., Dähnke, K., & Fischer, H. (2018). Open-channel measurement of denitrification in a large lowland river. Aquatic Sciences, 80(1), 11.
Rocca, J. D., Hall, E. K., Lennon, J. T., Evans, S. E., Waldrop, M. P., Cotner, J. B., Nemergut, D. R., Graham, E. B., & Wallenstein, M. D. (2015). Relationships between protein-encoding gene abundance and corresponding process are commonly assumed yet rarely observed. ISME Journal, 9(8), 1693.
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., Lesniewski, R. A., Oakley, B. B., Parks, D. H., & Robinson, C. J. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied and Environmental Microbiology, 75(23), 7537–7541.
Seitzinger, S., Harrison, J. A., Böhlke, J. K., Bouwman, A. F., Lowrance, R., Peterson, B., Tobias, C., & Drecht, G. V. (2006). Denitrification across landscapes and waterscapes: a synthesis. Ecological Applications, 16(6), 2064–2090.
Šimek, M., & Cooper, J. E. (2010). The influence of soil pH on denitrification: progress towards the understanding of this interaction over the last 50 years. European Journal of Soil Science, 53(3), 345–354.
Small, G. E., Finlay, J. C., Mckay, R. M. L., Rozmarynowycz, M. J., Brovold, S., Bullerjahn, G. S., Spokas, K., & Sterner, R. W. (2016). Large differences in potential denitrification and sediment microbial communities across the Laurentian great lakes. Biogeochemistry, 128(3), 353–368.
Speir, S. L., Taylor, J. M., & Scott, J. T. (2017). Seasonal differences in relationships between nitrate concentration and denitrification rates in ditch sediments vegetated with rice cutgrass. Journal of Environmental Quality, 46(6), 1500–1509.
Strokal, M., Yang, H., Zhang, Y., Kroeze, C., Li, L., Luan, S., Wang, H., Yang, S., & Zhang, Y. (2014). Increasing eutrophication in the coastal seas of China from 1970 to 2050. Marine Pollution Bulletin, 85(1), 123–140.
Sun, W., Xu, M.-Y., Wu, W.-M., Guo, J., Xia, C.-Y., Sun, G.-P., & Wang, A.-J. (2014). Molecular diversity and distribution of anammox community in sediments of the Dongjiang River, a drinking water source of Hong Kong. Journal of Applied Microbiology, 116(2), 464–476.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., & Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28(10), 2731–2739.
Tang, Y., Li, M., Xu, D., Huang, J., & Sun, J. (2018). Application potential of aerobic denitrifiers coupled with a biostimulant for nitrogen removal from urban river sediment. Environmental Science and Pollution Research, 25(6), 5980–5993.
Tomasek, A., Kozarek, J. L., Hondzo, M., Lurndahl, N., Sadowsky, M. J., Wang, P., & Staley, C. (2017). Environmental drivers of denitrification rates and denitrifying gene abundances in channels and riparian areas. Water Resources Research, 53(8), 6523–6538.
van Wijnen, J., Ivens, W. P. M. F., Kroeze, C., & Löhr, A. J. (2015). Coastal eutrophication in Europe caused by production of energy crops. Science of the Total Environment, 511, 101–111.
Wang, S., Zhu, G., Peng, Y., Jetten, M. S. M., & Yin, C. (2012). Anammox bacterial abundance, activity, and contribution in riparian sediments of the Pearl River estuary. Environmental Science & Technology, 46(16), 8834–8842.
Xia, X., Liu, T., Yang, Z., Michalski, G., Liu, S., Jia, Z., & Zhang, S. (2017). Enhanced nitrogen loss from rivers through coupled nitrification-denitrification caused by suspended sediment. Science of the Total Environment, 579, 47–59.
Xia, X., Li, Z., Zhang, S., Zhang, L., Zhang, L., & Wang, G. (2019). Occurrence of anammox on suspended sediment (SPS) in oxic river water: effect of the SPS particle size. Chemosphere, 235, 40–48.
Yao, L., Chen, C., Liu, G., & Liu, W. (2018). Sediment nitrogen cycling rates and microbial abundance along a submerged vegetation gradient in a eutrophic lake. Science of the Total Environment, 616-617, 899.
Zeng, J., Chen, M., Zheng, M., Qiu, Y., He, W., He, Y., & Liu, X. (2018). Effects of particles on potential denitrification in the coastal waters of the Beibu Gulf in China. Science of the Total Environment, 624, 1274–1286.
Zhang, J., & Gao, X. (2016). Nutrient distribution and structure affect the acidification of eutrophic ocean margins: a case study in southwestern coast of the Laizhou Bay, China. Marine Pollution Bulletin, 111(1), 295–304.
Zhang, R., Zhang, G., Zheng, Q., Tang, J., Chen, Y., Xu, W., Zou, Y., & Chen, X. (2012). Occurrence and risks of antibiotics in the Laizhou Bay, China: impacts of river discharge. Ecotoxicology and Environmental Safety, 80, 208–215.
Zhang, Y., Ruan, X.-H., Op den Camp, H. J. M., Smits, T. J. M., Jetten, M. S. M., & Schmid, M. C. (2007). Diversity and abundance of aerobic and anaerobic ammonium-oxidizing bacteria in freshwater sediments of the Xinyi River (China). Environmental Microbiology, 9(9), 2375–2382.
Zhang, Z., Zhu, M., Wang, Z., & Wang, J. (2006). Monitoring and managing pollution load in Bohai Sea, PR China. Ocean & Coastal Management, 49(9), 706–716.
Zhang, X., Zhang, Q., Yang, A., Hou, L., Zheng, Y., Zhai, W., & Gong, J. (2018). Incorporation of microbial functional traits in biogeochemistry models provides better estimations of benthic denitrification and anammox rates in coastal oceans. Journal of Geophysical Research: Biogeosciences, 123, 3331–3352.
Zheng, Y., Jiang, X., Hou, L., Min, L., & Rong, W. (2016). Shifts in the community structure and activity of anaerobic ammonium oxidation bacteria along an estuarine salinity gradient: shift in anammox along salinity gradient. Journal of Geophysical Research: Biogeosciences, 121(6), 1632–1645.
Zhu, G., Wang, S., Wang, W., Wang, Y., Zhou, L., Jiang, B., Op den Camp, H. J. M., Risgaard-Petersen, N., Schwark, L., Peng, Y., Hefting, M. M., Jetten, M. S. M., & Yin, C. (2013). Hotspots of anaerobic ammonium oxidation at land-freshwater interfaces. Nature Geoscience, 6, 103.
Acknowledgments
We would like to thank Jia Yue, Zhengyi Liu, and Zhonghua Wang for their help with field sampling. We also thank the Center for Analysis and Test of the Yantai Institute of Coastal Zone Research, Chinese Academy of Sciences, for measuring the environmental parameters.
Funding
This study was financially supported by the National Natural Science Foundation of China (No. 41106100) and the Science and Technology program of Yantai (No. 2016YTZD0007).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that there they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
ESM 1
(DOCX 703 kb)
Rights and permissions
About this article
Cite this article
Li, J., Yu, S. & Qin, S. Removal Capacities and Environmental Constrains of Denitrification and Anammox Processes in Eutrophic Riverine Sediments. Water Air Soil Pollut 231, 274 (2020). https://doi.org/10.1007/s11270-020-04593-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11270-020-04593-z