Abstract
Due to the limited host range of HBV, research progress has been hindered by the absence of a suitable animal model. The natural history of woodchuck hepatitis virus (WHV) infection in woodchuck closely mirrors that of HBV infection in human, making this species a promising candidate for establishing both in vivo and in vitro HBV infection models. Therefore, this animal may be a valuable species to evaluate HBV vaccines and anti-HBV drugs. A significant milestone in HBV and hepatitis D virus (HDV) infection is the discovery of sodium taurocholate cotransporting polypeptide (NTCP) as the functional receptor. In an effort to enhance susceptibility to HBV infection, we introduced hNTCP into the woodchuck hepatocytes by multiple approaches including transduction of vLentivirus-hNTCP in woodchuck hepatocytes, transfection of p-lentivirus-hNTCP-eGFP plasmids into these cells, as well as transduction of vAdenovirus-hNTCP-eGFP. Encouragingly, our findings demonstrated the successful introduction of hNTCP into woodchuck hepatocytes. However, it was observed that these hNTCP-expressing hepatocytes were only susceptible to HDV infection but not HBV. This suggests the presence of additional crucial factors mediating early-stage HBV infection that are subject to stringent species-specific restrictions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hepatitis B virus (HBV) is a major global health concern, with approximately 250 million individuals worldwide living with chronic HBV infection and approximately 15–40% of those cases progress to serious liver disease, manifesting as cirrhosis, fibrosis, and hepatocellular carcinoma [1]. Despite the presence of drugs like pegylated interferon and nucleoside analogues (NUCs) that can inhibit HBV replication and mitigate the advancement of liver disease, a definitive cure for HBV remains elusive [1, 2]. Moreover, around 5% of HBV patients are co-infected with Hepatitis D virus (HDV), a single-stranded RNA virus, and subviral satellite of HBV. This co-infection heightens the risk of hepatocellular carcinoma and mortality compared to HBV mono-infection [3, 4].
HBV is a highly species and organ restricted virus. It mainly infects liver parenchymal cells. Few higher primates other than humans, and under certain experimental conditions the tree shrew Tupaia belangeri are susceptible to HBV [5]. By the reason of ethical constraints or lack of corresponding antibodies, in vivo research has been stopped in these animal models [6]. Better-defined and easier-to-operate laboratory animals, such as mice and rats, are required urgently.
Emerging evidences point to the species-specific nature of HBV being closely tied to receptor-dependent mechanisms [7]. A pivotal discovery is the identification of the HBV functional receptor, sodium taurocholate co-transporting polypeptide (NTCP) [8], a multiple transmembrane transporter mainly expressed on the membrane of hepatocytes responsible for the vast majority of sodium-dependent uptake of bile salts from the enterohepatic circulation. Studies reveal that HBV enters into the hepatocytes through the interaction between the myristoylated N-terminus of the pre-S1 domain of the large HBV surface antigen (HBsAg) and the host receptor, NTCP [7, 9]. Introducing human NTCP (hNTCP) to human hepatocellular carcinoma cell lines, HepG2, results in their susceptibility to HBV infection [10].
NTCP protein sequences vary across species. hNTCP encoded by the solute carrier family 10 member 1 (SLC10A1), supports successful infection and serves as the sole limiting host factor in the animals like pigs and macaques [11]. NTCPs from certain non-susceptible species cannot efficiently mediate virus entry due to sequence variations within amino acids 84–87 [10, 12] or 157–165 [8, 10]. In mouse [10] and crab-eating macaque [8] models, substituting these motifs of NTCPs with their human counterparts renders these NTCPs to function as receptors for HBV in HepG2 cells. Moreover, as the inner nucleocapsid of HDV hijacks the HBV surface proteins to construct infectious viral particles, several mouse liver cell lines containing hNTCP support HDV infection [11, 13]. Consequently, hNTCP appears to be the crucial homologue facilitating the HBV entry process, underscoring that the host range constraint of HBV predominantly hinges on hNTCP.
The woodchuck hepatitis virus (WHV) exhibits a high degree of homology with HBV and can induce both acute and chronic infections [6]. WHV and the woodchuck have been a longstanding surrogate model for studying HBV-related pathogenesis and antiviral research [6, 14]. Importantly, WHV-infected woodchuck can be superinfected with HDV [15]. Given the significant role of the woodchuck model in HBV studies, it may offer a valuable platform for establishing an in vivo and in vitro HBV infection model. Woodchuck NTCP (wNTCP) is capable of supporting HBV/HDV infection in HepG2 cells, but 90% less efficiently than hNTCP [16]. Therefore, we inferred that, this discrepancy between hNTCP and wNTCP may explain why the woodchuck is less susceptible to HBV/HDV infection, and in the presence of hNTCP on the cell surface, woodchuck hepatocytes may be a surrogate model used for studying HBV.
In this study, hNTCP was introduced into the woodchuck hepatocyte in an attempt to make the woodchuck hepatocyte susceptible to HBV, but we found that woodchuck hepatocytes containing hNTCP were only susceptible to HDV infection but not to HBV. The results indicate that there exist some other key factors in the HBV infection process and hNTCP is not the only host-specific determinant that is needed for HBV infection.
Materials and methods
Sequences alignment of wNTCP and hNTCP
Sequences of wNTCP and hNTCP were obtained in FASTA format from the NCBI website, and the data were analyzed using Protein BLAST to compare the similarity of two NTCPs. The tertiary structure prediction of wNTCP protein was carried out using the AWISS-Model in the ExPASy online software.
Cell lines
The HepG2-NTCP-K7 cell line, generated from HepG2 cells expressing hNTCP, that could express hNTCP stably and support long-term HBV infection, was used [17]. Cells were cultured with Dulbecco modified Eagle medium (DMEM, Invitrogen) supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, penicillin/streptomycin, 30 μg/ml blasticidin, 1 mM sodium pyruvate and non-essential amino acids (Thermo-Fisher Scientific, Waltham, MA, USA). Woodchuck primary hepatocytes prepared [18] and cultured in Ham’s F12 Nutrient Mix, Gluta Max™ medium containing 10% FBS and penicillin/streptomycin. 293FT cell line were maintained in DMEM medium containing 10% FBS, 4 mM L-Glutamine, 1 mM MEM Sodium Pyruvate, 0.1 mM MEM Non-Essential Amino Acids, penicillin/streptomycin, and 500 μg/ml Geneticin.
Transduction of woodchuck hepatocytes with vLentivirus-hNTCP
An expression cassette containing the hNTCP (hNTCP, GenBank Accession #JQ814895.1) under control of the liver-specific transthyretin (TTR) promoter, along with the blasticidin resistance gene and human beta-globin polyadenylation sequence, was cloned into a p-Lenti6.3 expression vector (Invitrogen). ViraPower™ Lentiviral Expression System (Invitrogen) was used to generate vLentivirus-hNTCP which was then used to transduce woodchuck hepatocyte. Following the transduction procedure, blasticidin selection was applied and cellular morphology was observed.
Expression of hNTCP was confirmed by immunofluorescence analysis
Rabbit anti-human hNTCP was kindly provided by Prof. Bruno Stieger (Clinical pharmacology and Toxicology, University of Zurich). Immunofluorescence analysis was performed to confirm the expression of hNTCP in transduced woodchuck hepatocytes. Rabbit anti-human hNTCP antibody was utilized for staining. Cells were fixed, permeabilized, and blocked before being incubated with primary and secondary antibodies. Expression of hNTCP was observed under a fluorescence microscope.
Transfection of Woodchuck hepatocytes with p-lentivirus-hNTCP-eGFP plasmids.
P-lentivirus-hNTCP-eGFP plasmids were kindly provided by Dr. Martin (post-doctor in Prof. Ulrike Protzer’s Lab). P-lentivirus-hNTCP-eGFP plasmids were transfected into woodchuck hepatocytes, and transfection efficiency was assessed by the expression of enhanced green fluorescent protein (eGFP) under a fluorescence microscope.
Transduction of woodchuck hepatocytes with vAdenovirus-hNTCP-eGFP
Adenoviruses expressing both hNTCP and eGFP constructed by Dr. Martin (post-doctor in Prof. Ulrike Protzer’s Lab.) were used to transduce woodchuck hepatocytes with multiplicity of infection (MOI) of 4 or 20. Transduction efficiency was evaluated by eGFP expression under a fluorescence microscope.
Infection of HBV and HDV in hNTCP-expressing woodchuck hepatocytes
After differentiation with 2% DMSO, the three kinds of hNTCP-expressing woodchuck hepatocytes were infected with concentrated HBV stock solution (purified from HepAD38) diluted in Ham’s F12 Nutrient Mix. HBV infection was performed with an MOI of 100 or 500. HepG2-hNTCP-K7 and woodchuck hepatocytes were used as the positive and negative control, respectively. The supernatants were collected for analysis of viral markers every 3 days. For the HDV infection experiment, the vAdenovirus-hNTCP-eGFP transduced cells were infected by HDV stock solution (purified from serum of the patient) with an MOI of 200 at 3 day post transduction. The cell morphology was observed at day 3 and day 7 after infection.
Detection of HBV e antigen (HBeAg) and quantification of HDV viral genome
Supernatants of HBV-infected cells were assayed for HBeAg at day 7 and 10 p.i. by using an electro-chemiluminescence immunoassay (ECLIA) on a modular analytics E170 analyzer (Roche, Indianapolis, IN, USA) according to the manufacturer’s instructions.
HDV viral genome quantification was performed using real-time PCR with a HDV-specific TaqMan probe. The probe was mixed with the HDV-specific primers listed on Table 1 and then they were added to the master mix for the PCR reaction. The performed PCR program is listed on Table 2. HDV viral genome equivalent copies were calculated based on a standard curve generated with known copy numbers of HDV genome containing plasmids.
Statistical analysis
The results are presented as the mean ± SD. Statistical analysis was carried out using GraphPad Prism 6 software (GraphPad Software, Inc.) Student’s t test or one-way ANOVA followed by Least Significant Difference post hoc test. P < 0.05 was considered to indicate a statistically significant difference.
Results
Sequence comparison and tertiary structure prediction of wNTCP and hNTCP
We predicted the tertiary structure of wNTCP by bioinformatics analysis (Fig. 1A), and compared the sequence with hNTCP. The results revealed an 82% identity between the two proteins. Critical regions in hNTCP, such as aa 84 to 87 and aa 157 to 165, were reported to play crucial roles in HBV binding and entry process. Aa 84 to 87 is more likely involved in the binding process, but aa 157 to 165 may be essential for maintaining a spatial conformation of NTCP which is important for the entry process. Additionally, it was reported that V263 in hNTCP and I263 in wNTCP, with similar chemical properties, played dramatically different roles in supporting viral infection.
Notably, there are five different amino acids at positions 84–87, 157–165, and 263 between wNTCP and hNTCP (red box), suggesting a basis for the disparity in HBV infectivity.
Expression of hNTCP in vLentivirus-hNTCP transduced woodchuck hepatocytes
We inferred the difference between hNTCP and wNTCP may be an important reason why the woodchuck is not susceptible to HBV infection. To test our hypothesis, we first constructed an hNTCP expressing woodchuck hepatocyte by transducing woodchuck hepatocytes with the vLentivirus encoding hNTCP. Under the microscope, the cells that failed to be transduced with vLentivirus-hNTCP rapidly deteriorated and died after blasticidin selection (Fig. 2A). Immunofluorescence confirmed successful expression, with hNTCP localized to both the cytoplasm and cell membrane of the successfully transduced cells (Fig. 2B). This expression was specific to hNTCP, as woodchuck hepatocytes lacked such immunoreactivity.
Transduction of woodchuck hepatocytes with vLentivirus-hNTCP. vLentivirus-hNTCP containing expression cassette of the human NTCP (hNTCP, GenBank Accession #JQ814895.1) under control of the liver-specific transthyretin (TTR) promoter was constructed by using ViraPower™ Lentiviral Expression System (Invitrogen) A The morphology of vLentivirus-hNTCP transduced cells after blasticidin selection was observed by a bright-field microscope. The black arrow indicates the living cells, the white arrow indicates the dead cells; B Woodchuck hepatocytes were seeded and transduced with vlentivirus encoding hNTCP (vLentivirus-hNTCP). After vLentivirus-hNTCP transduction, the expression, and the location of hNTCP were analyzed by immunofluorescence. Green, hNTCP staining; blue, nucleus; (C) vLentivirus-hNTCP transduced woodchuck hepatocytes were infected with HBV stocks with an MOI of 100 or 500, and supernatants were collected at day 7 and 10 p.i. for detection of HBeAg. HepG2-hNTCP-k7 cells served as the positive control and woodchuck hepatocytes served as the negative control
HBV failed to infect vLentivirus-hNTCP transduced woodchuck hepatocytes
To further assess the susceptibility of HBV infection in hNTCP containing woodchuck hepatocytes, we infected these cells with HBV at an MOI of 100 or 500. HepG2-hNTCP-K7 cells used as a positive control and woodchuck hepatocytes used as a negative control. The viral replication marker HBeAg secreted into the supernatant was quantified at day 7 and 10 p.i.. The results showed that HBeAg could only be detected in HBV infected HepG2-hNTCP-K7 cells and increased over time. Unfortunately, woodchuck hepatocytes were not susceptible to HBV infection whether they expressed the receptor, hNTCP or not (Fig. 2C).
HBV failed to infect p-lentivirus-hNTCP-eGFP plasmids transfected woodchuck hepatocytes
As an integrating vector, the main disadvantage for lentiviral vector is insertional mutagenesis at the integration site, which is caused by either disrupting or inappropriately acting transcription of a nearby host gene. So, we hypothesized that vlentivirus might affect the expression of some key genes in woodchuck hepatocytes to some extent, leading to HBV infection failure. Therefore, we changed the strategy and transfected woodchuck hepatocytes with an hNTCP-expressing plasmid, hoping to enable the cells to acquire the ability to express hNTCP without affecting the expression of other genes. Transfection efficiency was confirmed by the expression of eGFP. The results showed that the transfection efficiency did not reach a very high level (Fig. 3A). To further evaluate whether these cells are susceptible to HBV, we challenged parallel p-lentivirus-hNTCP-eGFP plasmids transfected woodchuck hepatocytes with HBV at an MOI of 100 or 500. HepG2-hNTCP-K7 were cells used as a positive control and woodchuck hepatocytes used as a negative control. Both of them were inoculated with HBV at an MOI of 100. Nevertheless, the expression of HBeAg could only be detected in the HBV infected HepG2-hNTCP-K7 cells, indicating that expression of hNTCP is not sufficient for HBV to establish successful infection in woodchuck hepatocytes (Fig. 3B).
Transfection of woodchuck hepatocytes with p-lentivirus-hNTCP-eGFP plasmids. Transfection was performed by incubating woodchuck hepatocytes with the mixture of p-lentivirus-hNTCP-eGFP plasmids and lipofectamine (Sigma Aldrich). The nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) at 72 h after transfection and the expression of eGFP was observed under the fluorescence microscope to check the transfection efficiency at the same time. A Tranfection efficiency was monitored by the expression level of eGFP. Green, hNTCP staining cells; blue, nucleus; B P-lentivirus-hNTCP-eGFP transfected woodchuck hepatocytes were infected with HBV stocks with an MOI of 100 or 500 at 4 days post transfection, and supernatants were collected at day 7 and 10 p.i. for detection of HBeAg. HepG2-hNTCP-K7 cells served as the positive control and woodchuck cells served as the negative control
HBV failed to infect vAdenovirus-hNTCP-eGFP transduced woodchuck hepatocytes
Different from lentivirus, the adenovirus vector genome is maintained as an episome in the nucleus and the integration of the viral genomic DNA into the host’s genome is minimal, reducing the risk of insertional mutagenesis [19]. For the low expression level of hNTCP in plasmid-transfected cells, which may be the possible reason leading to the failure of HBV infection, we tried to establish hNTCP-expressing woodchuck hepatocytes by transducing woodchuck hepatocytes with vAdenovirus-hNTCP-eGFP with an MOI of 4 or 20, and transduction efficiency was checked by the expression of eGFP. The results showed that the estimated percentage of hNTCP-expressing woodchuck hepatocytes was 60–70% of the transduced cells with an MOI of 4, and 80–90% of the transduced cells with an MOI of 20 (Fig. 4A). Subsequently, vAdenovirus-hNTCP-eGFP transduced woodchuck hepatocytes were inoculated with HBV with an MOI of 100 or 500 on day 3 after transduction, and the supernatants were collected at day 7 and 10 p.i. for detection of HBeAg (Fig. 4B). Consistent with the previous results, HBV also failed to infect vAdenovirus-hNTCP-eGFP transduced woodchuck hepatocytes.
Transduction of woodchuck hepatocytes with vAdenovirus-hNTCP-eGFP. A vAdenovirus-hNTCP-eGFP was transduced into woodchuck hepatocytes with an MOI of 4 or 20. The expression of eGFP was observed under the fluorescence microscope to check the transduction efficiency. Green, hNTCP staining cells. Bars, 200 µm. B vAdenovirus-hNTCP-eGFP transduced woodchuck hepatocytes were infected with with HBV stocks with an MOI of 100 or 500 at 3 days post transduction, and supernatants were collected at day 7 and 10 p.i. for detection of HBeAg. HepG2-hNTCP-K7 cells used as the positive control and woodchuck hepatocytes used as the negative control
vAdenovirus-hNTCP transduced woodchuck hepatocytes gain susceptibility to HDV
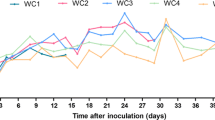
Following confirmation of hNTCP expression, the established vAdenovirus-hNTCP-eGFP (MOI 20) transduced woodchuck hepatocytes were subsequently subjected to HDV infection. The human liver cell line HepG2-hNTCP-K7 and woodchuck hepatocytes were used as positive and negative control respectively. Three days after vAdenovirus-hNTCP-eGFP transduction, HDV infection was performed with an MOI of 200. Morphological changes of cells were observed on the 3 day after infection, and became more significant at day 7 p.i. (Fig. 5A). HDV viral RNA was detected by using the real-time PCR at day 3, 7, 10, 13 after virus infection. The results showed that the peak of HDV genome copy number was at day 7 p.i., indicating that hNTCP-expressing woodchuck hepatocytes could be infected by HDV successfully, whereas woodchuck hepatocytes without transduction of vAdenovirus-hNTCP-eGFP could not be infected by HDV (Fig. 5B). We can speculate that the expression of hNTCP is sufficient to support HDV infection in woodchuck hepatocytes.
vAdenovirus-hNTCP transduced woodchuck hepatocytes gain susceptibility to HDV. On the third day after transduction with vAdenovirus-hNTCP, the transduced woodchuck hepatocytes were then infected with HDV. A Morphological changes of cells were observed on the 3 day after infection, and became more significant at day 7 p.i. Bars, 200 µm. B Total RNA was extracted and reverse transcribed, and intracellular HDV genomes were quantified by qPCR with HDV-specific primers and probe at day 3, 7, 10, 13 p.i.
Taken together, hNTCP made woodchuck hepatocytes susceptible to HDV infection suggesting its function as a receptor for HBV-preS1 binding. However, successful infection of HBV observed in HepG2-hNTCP-K7 cells but not in hNTCP expressing woodchuck hepatocytes, indicating that hNTCP is not sufficient for HBV to establish successful infection in woodchuck hepatocytes.
Discussion
HBV is characterized by strict tissue and species specificity. However, the lack of robust HBV infection cell culture models and corresponding animal models impeded further investigation of the interaction between virus and host factors [12]. Accumulating evidence indicated that the distinct species specificity of HBV and HDV is hNTCP-dependent. The discovery of the HBV receptor, hNTCP, led to the successful establishment of a cell line that was susceptible to both HBV and HDV. For instance, Zhou et al. demonstrated that primary pig hepatocytes infected with hNTCP recombinant lentivirus became susceptible to HBV [20]. Similarly, Burwitz et al. showed that macaque hepatocytes could support HBV replication in vitro following transduction with viral vectors encoding hNTCP and detectable levels of HBV DNA were observed in serum over time in vivo [21]. Lempp et al. expanded on this showing that transduced primary hepatocytes originated from various species, including mouse, rat, dog, pig, rhesus and cynomolgus macaque, became fully susceptible to both HBV and HDV with adeno-associated virus (AAV) carrying hNTCP gene [11].
Recent research has shown that woodchuck hepatic cell lines support the entire HBV life cycle, including replication, cccDNA formation, and virion secretion after transfection with the HBV replicon, except for the entry steps [22]. In this study, we introduced hNTCP into woodchuck hepatocytes using various methods: by transducing with vLentivirus-hNTCP, transfecting with p-lentivirus-hNTCP-eGFP plasmids, or transducing with vAdenovirus-hNTCP-eGFP. We confirmed the expression of hNTCP in both cytoplasm and membrane of woodchuck hepatocyte and then evaluated their susceptibility to HBV infection. Surprisingly, hNTCP expressing woodchuck hepatocytes were unable to be successful infected by HBV. However, vAdenovirus-hNTCP-eGFP transduced woodchuck hepatocytes could be infected by HDV. Our findings were in line with recent reports [11, 13]. In the hNTCP-expressing mouse cell lines [13] and primary hepatocytes from mouse, rat, and dog [11], susceptibility to HDV was conferred, but HBV was still restricted. It is confusing that the two viruses, HBV and HDV, with the same surface antigen, showed completely different infection characteristics with the hepatocytes expressing the specific receptor hNTCP.
Notably, HDV does not use the HBV replication machinery, and relies on host factors for its lifecycle. Our presumption is that the species restriction of NTCPs is just related to the entry process, and primarily limits HDV infection. Prior mutagenesis studies on NTCP orthologs had highlighted the role of at least by two critical regions of NTCP, aa 84 to 87 [10, 12] and aa 157 to 165 [8, 10], in receptor recognition by pre-S1 of HBV and HDV. Among these aa 84 to 87 appears to be involved in the binding process, while aa 157 to 165 is crucial for maintaining the spatial conformation of NTCP [10]. Several cell lines, regardless of their species, supported HDV infection when expressing hNTCP or other-NTCP variants with human counterparts of aa 84 to 87 or 157 to 165 supplemented by human counterparts [8, 12, 23]. Interestingly, mouse NTCP (mNTCP) or rat NTCP (rNTCP) can bind to the MyrB, a synthetic L-protein derived lipopeptide, but dose not support HBV and HDV infection [12]. However, replacing mNTCP’s aa 84 to 87 with the human counterpart and transfer into HepG2 cells rescued receptor activity [12, 23]. Crab eating monkey NTCP (mkNTCP) neither bound to pre-S1 peptide nor supported HBV and HDV infection, however, alteration of aa 157 to 165 of mkNTCP by the human homologous sequence converted mkNTCP to a functional receptor for HBV infection [8].
Interestingly, our study revealed that even with hNTCP expression, HBV remained restricted in woodchuck hepatocytes. Previous research showed that the hepatocytes that expressing hNTCP could successfully be infected by HBV if they came from species closely related to humans, such as pigs [11, 19] and macaques [11, 21]. Based on the evolutionary distance, pigs and macaque were chosen. Macaque hepatocytes and primary pig hepatocytes expressing hNTCP exhibited increased expression levels of HBsAg and HBeAg, as well as increased cccDNA formation after HBV infection [13]. Therefore, we speculate that, the restriction of HBV infection is not only related to hNTCP-dependent steps but is also influenced by some additional species-specific factors. Our next step will try to look for the key factors which restrict HBV infection in woodchuck hepatocytes, and then try to establish a more robust in vivo and in vitro HBV infection model for investigating HBV related basic research as well as evaluating the effect of clinical antiviral research.
Conclusion
In conclusion, woodchucks play a pivotal role in HBV infection research, making them valuable candidates for in vivo and in vitro models. However, the inability of HBV to infect woodchuck hepatocytes limits their applicability. Our hypothesis suggests that the disparity in HBV receptor expression between human hepatocytes and woodchuck hepatocytes is a significant factor restricting HBV infection. In this study, hNTCP was introduced into the woodchuck hepatocytes as an attempt to make the woodchuck hepatocytes susceptible to HBV. Our results indicated that hNTCP-expressing woodchuck hepatocytes were only sensitive to HDV infection but not to HBV. We propose that the species restriction of HBV infection involves factors beyond hNTCP. Our further work will delve into identifying the additional key factors limiting HBV infection in woodchuck hepatocytes, ultimately aiming to establish a more comprehensive in vivo and in vitro model for in-depth HBV research.
Data availability
The datasets generated during the current study are available from the corresponding author on reasonable request.
References
Nicolini LA, Orsi A, Tatarelli P, Viscoli C, Icardi G, Sticchi L (2019) A global view to HBV chronic infection: evolving strategies for diagnosis, treatment and prevention in immunocompetent individuals. Int J Environ Res Public Health. https://doi.org/10.3390/ijerph16183307
Eller C, Heydmann L, Colpitts CC, Verrier ER, Schuster C, Baumert TF (2018) The functional role of sodium taurocholate cotransporting polypeptide NTCP in the life cycle of hepatitis B, C and D viruses. Cell Mol Life Sci 75(21):3895–3905
Gilman C, Heller T, Koh C (2019) Chronic hepatitis delta: a state-of-the-art review and new therapies. World J Gastroenterol 25(32):4580–4597
Koh C, Da BL, Glenn JS (2019) HBV/HDV coinfection: a challenge for therapeutics. Clin Liver Dis 23(3):557–572
Hwang JR, Park SG (2018) Mouse models for hepatitis B virus research. Lab Anim Res 34(3):85–91
Ortega-Prieto AM, Cherry C, Gunn H, Dorner M (2019) In vivo model systems for hepatitis B virus research. ACS Infect Dis 5(5):688–702
Yu Y, Li S, Liang W (2018) Bona fide receptor for hepatitis B and D viral infections: mechanism, research models and molecular drug targets. Emerg Microbes Infect 7(1):134
Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, Huang Y, Qi Y, Peng B, Wang H, Fu L, Song M, Chen P, Gao W, Ren B, Sun Y, Cai T, Feng X, Sui J, Li W (2012) Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife. https://doi.org/10.7554/eLife.00049
Jacquet S, Pons JB, De Bernardo A, Ngoubangoye B, Cosset FL, Regis C, Etienne L, Pontier D (2019) Evolution of hepatitis B virus receptor NTCP reveals differential pathogenicities and species specificities of hepadnaviruses in primates, rodents, and bats. J Virol. https://doi.org/10.1128/JVI.01738-18
Yan H, Peng B, He W, Zhong G, Qi Y, Ren B, Gao Z, Jing Z, Song M, Xu G, Sui J, Li W (2013) Molecular determinants of hepatitis B and D virus entry restriction in mouse sodium taurocholate cotransporting polypeptide. J Virol 87(14):7977–7991
Lempp FA, Wiedtke E, Qu B, Roques P, Chemin I, Vondran FWR, Le Grand R, Grimm D, Urban S (2017) Sodium taurocholate cotransporting polypeptide is the limiting host factor of hepatitis B virus infection in macaque and pig hepatocytes. Hepatology 66(3):703–716
Ni Y, Lempp FA, Mehrle S, Nkongolo S, Kaufman C, Falth M, Stindt J, Koniger C, Nassal M, Kubitz R, Sultmann H, Urban S (2014) Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology 146(4):1070–1083
Lempp FA, Mutz P, Lipps C, Wirth D, Bartenschlager R, Urban S (2016) Evidence that hepatitis B virus replication in mouse cells is limited by the lack of a host cell dependency factor. J Hepatol 64(3):556–564
Balsitis S, Gali V, Mason PJ, Chaniewski S, Levine SM, Wichroski MJ, Feulner M, Song Y, Granaldi K, Loy JK, Thompson CM, Lesniak JA, Brockus C, Kishnani N, Menne S, Cockett MI, Iyer R, Mason SW, Tenney DJ (2018) Safety and efficacy of anti-PD-L1 therapy in the woodchuck model of HBV infection. PLoS One 13(2):e0190058
Fiedler M, Roggendorf M (2001) Vaccination against hepatitis delta virus infection: studies in the woodchuck (Marmota monax) model. Intervirology 44:154–161
Fu L, Hu H, Liu Y, Jing Z, Li W (2017) Woodchuck sodium taurocholate cotransporting polypeptide supports low-level hepatitis B and D virus entry. Virology 505:1–11
Ko C, Chakraborty A, Chou WM, Hasreiter J, Wettengel JM, Stadler D, Bester R, Asen T, Zhang K, Wisskirchen K, McKeating JA, Ryu WS, Protzer U (2018) Hepatitis B virus genome recycling and de novo secondary infection events maintain stable cccDNA levels. J Hepatol 69(6):1231–1241
Lu M, Hilken G, Yang DL, Kemper T, Roggendorf M (2001) Replication of naturally occurring woodchuck hepatitis virus deletion mutants in primary hepatocytes cultures and after transmission to naive woodchucks. J Virol 75:3811–3818
Chen YH, Keiser MS, Davidson BL (2018) Viral vectors for gene transfer. Curr Protoc Mouse Biol 8(4):e58
Zhou M, Qin B, Deng XS, Zeng XL, Lu Y, Huang ZG, Wu CC, Mou LS (2019) hNTCPexpressing primary pig hepatocytes are a valuable tool for investigating hepatitis B virus infection and antiviral drugs. Mol Med Rep 20(4):3820–3828
Burwitz BJ, Wettengel JM, Muck-Hausl MA, Ringelhan M, Ko C, Festag MM, Hammond KB, Northrup M, Bimber BN, Jacob T, Reed JS, Norris R, Park B, Moller-Tank S, Esser K, Greene JM, Wu HL, Abdulhaqq S, Webb G, Sutton WF, Klug A, Swanson T, Legasse AW, Vu TQ, Asokan A, Haigwood NL, Protzer U, Sacha JB (2017) Hepatocytic expression of human sodium-taurocholate cotransporting polypeptide enables hepatitis B virus infection of macaques. Nat Commun 8(1):2146
Hong XP, Kawasawa YI, Menne S, Hu JM (2022) Host cell-dependent late entry step as determinant of hepatitis B virus infection. PLoS Pathog 18(6):e1010633
He W, Cao Z, Mao F, Ren B, Li Y, Li D, Li H, Peng B, Yan H, Qi Y, Sun Y, Wang F, Sui J, Li W (2016) Modification of three amino acids in sodium taurocholate cotransporting polypeptide renders mice susceptible to infection with hepatitis D virus in vivo. J Virol 90(19):8866–8874
Acknowledgements
We thank the Prof. Bruno Stieger who gave us the hNTCP antibody.
Funding
This work was supported by the National Natural Science Foundation of China (81860113) and Shenzhen Science and Technology Program Project (JCYJ20190812172005670).
Author information
Authors and Affiliations
Contributions
JS designed the experiments, performed the experiments, MC and LY analyzed the data, DZ and JW did the statistical analysis. DZ, LY and JS carried out experiments. JS and LY wrote the paper. ML, DY, UP and R.M. conceived the experiments and revised the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Edited by Juergen Richt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, L., Zhou, D., Martin, K. et al. Aborted infection of human sodium taurocholate cotransporting polypeptide (hNTCP) expressing woodchuck hepatocytes with hepatitis B virus (HBV). Virus Genes 59, 823–830 (2023). https://doi.org/10.1007/s11262-023-02031-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-023-02031-w