Abstract
Respiratory syncytial virus (RSV) causes lower respiratory tract infections and bronchiolitis, mainly affecting children under 2 years of age and immunocompromised patients. Currently, there are no available vaccines or efficient pharmacological treatments against RSV. In recent years, tremendous efforts have been directed to understand the pathological mechanisms of the disease and generate a vaccine against RSV. Although RSV is highly infectious, not all the patients who get infected develop bronchiolitis and severe disease. Through various sequencing studies, single nucleotide polymorphisms (SNPs) have been discovered in diverse receptors, cytokines, and transcriptional regulators with crucial role in the activation of the innate immune response, which is implicated in the susceptibility to develop or protect from severe forms of the infection. In this review, we highlighted how variations in the key genes affect the development of innate immune response against RSV. This data would provide crucial information about the mechanisms of viral infection, and in the future, could help in generation of new strategies for vaccine development or generation of the pharmacological treatments.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Respiratory syncytial virus (RSV) infection is one of the main causes worldwide of acute respiratory infections (ARIs) among children under 5 years. 22% of ARIs are associated with this virus, being clinically manifested as bronchiolitis from 40 to 90% [1, 2]. In 2015, 3.2 million of hospitalizations were globally associated to RSV, of which 59,600 cases resulted in deaths [2, 3]. RSV has a seasonal behavior with outbreaks mainly from early autumn to late spring in the North Hemisphere [4]. Viral spreading shows an important reduction during the summer, although rare outbreaks have been reported [5]. Importantly, co-infection of RSV and SARS-CoV-2 might have an important effect on the treatment and prognosis of the disease, since this viral coinfection may be associated to a higher level of care, increased hospital stay, and progression to acute respiratory distress syndrome [6]. Currently, no vaccine has been licensed against RSV [7], and the search for an effective and safe preventive method for RSV infection continues to be one of the greatest challenges for the scientific community. Most treatment options are inaccessible, such as palivizumab which is used in severe disease, and in some cases, infected children are mainly treated with palliative treatments, such as oxygen and bronchodilators [8]. This suggests that there is need for an effective and safe treatment strategy against RSV infection. In this review, we have discussed general biological characteristics of RSV and further focused on the genetics of main components of the innate immune response activated during RSV infection. We have revised the polymorphism in various genes of innate immune response associated with disease severity, particularly those that impact the probabilities of hospital care and co-infections among the infected individuals.
General characteristics of respiratory syncytial virus (RSV)
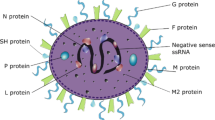
RSV was isolated in 1955 from a colony of 20 chimpanzees suffering from infectious coryza in the Walter Reed Army Institute of Research, Maryland, USA [9]. Two years later, similar viruses were isolated from children with respiratory illness (bronchopneumonia and laryngotracheobronchitis) [10]. RSV is a representative member of the order Mononegavirales, family Pneumoviridae, and subfamily Orthopneumovirus [11]. RSV is typically rounded (diameter 150–250 nm) or filamentous (diameter 90–100 nm × length 10 µm) [12]. The genome is 15.2 kb single stranded, non-segmented, negative-sense RNA and contains ten genes that encode eleven different proteins, which are classified as structural, regulatory, and non-structural [13]. The F (fusion) and G (attachment) envelope glycoproteins, the M, M2-1, M2-2 matrix proteins, SH, N and P proteins, give structure to the virion. The large polymerase protein (L) is responsible for the viral RNA synthesis. Finally, the NS (non-structural)1 and NS2 proteins are important disruptors and regulators of the expression of cellular responses against the RSV [14, 15] (Fig. 1).
Schematic representation of respiratory syncytial virus (RSV) genome and structure. The length of the genome is more than 15 kb, encoding 11 proteins. The negative-sense ssRNA is attached to N, P, and L proteins forming a nucleocapsid, which is covered by M proteins and enveloped with a membrane consisting of G, F, and SH proteins. Created with BioRender.com (access date: March 25th, 2022)
The G and F proteins are the major glycoproteins on the surface of the virion and have important roles in the virus entry to the host cell [12]. The G glycoprotein works as an attachment protein that binds virions to the surface molecules in the target cells. The F glycoprotein also facilitates attachment and mediates fusion of the viral and host cell membranes [16,17,18,19]. Diverse molecules have been proposed as receptor for RSV in the host cells, such as the intercellular adhesion molecule-1 which binds the F protein [20], heparin which interacts with the G and F proteins [21, 22], annexin II which binds to G protein [23], and recently, and still controversial, nucleolin which binds to the viral F protein [24,25,26]. Although Toll-like receptor (TLR)4 and fractalkine receptor can recognize viral proteins and have been associated with the innate antiviral response against RSV, their possible involvement in viral attachment and entry is still elusive [27, 28]. Once the viral RNA is located into the host cell, transcription and translation of the viral genome occurs in a well-coordinated mechanism [29]. The three viral glycoproteins are anchored to the cell membrane to facilitate the release of viral particles, while the assembly of nucleocapsid is achieved in the cytoplasmic inclusions [30]. Finally, viral assembly occurs in plasma membrane, where the viral particle acquires its viral envelope [30, 31] (Fig. 2).
Life cycle of RSV. For attachment and recognition of the virion mediated by glycoprotein (G) and fusion (F), proteins in the virion interact with the receptors in the host cell surface (1). The fusion (F) protein exposes the fusion peptide and change to a pre-harping conformation which facilitates the fusion of the virus to the host cell membrane and leads to nucleocapsid delivery (2). Viral genome is thereafter, contained in the cytoplasmic inclusions where primary transcription occurs (3). This is followed by translation of genome in the cytoplasmic ribosomes (4) and replication (5). The SH, F, and G proteins are sorted via endoplasmic reticulum (6) and Golgi apparatus (7). Viral proteins are assembled into viral filaments for viral budding process (8). Created with BioRender.com (Access date: March 25th, 2022)
Innate immune response in respiratory syncytial virus (RSV) infection and associated polymorphisms
Polymorphisms in the genes of immune system is considered as an important aspect behind the resistance or susceptibility of the host to an infectious disease. Over the years, researchers have explored many genetic factors having role in immune surveillance against infectious diseases [32]. Among them, single genetic mutations (such as single nucleotide polymorphisms -SNPs-) has been associated to the predisposition and development of the severe forms of the infections [33].
Different components of the innate immune response participate in the control of RSV infection, including various pattern recognition receptors (PRRs), diverse cell types, and a large array of cytokines and chemokines. An appropriate innate immune response has an essential role in the resolution of RSV infection, as it promotes virus clearance, avoids virus replication and spreading to the lower respiratory tract, and promotes the development of an adequate adaptive immune response [34].
The panoply of molecules giving structure to the RSV and those implied in its replicative cycle are classified as pathogen-associated molecular patterns (PAMPs) and share common general characteristics with those expressed by other viral pathogens. They are recognized by PRRs that promote the innate immune response leading to the activation of an antiviral state. These PRRs are anchored to the cell membrane or located in the intracellular vesicles, such as TLRs, but could also be dispersed in the cytoplasm, as in the case of RIG-I-like receptors, and NOD-like receptors. Thus, different viral molecules at each step of their infection cycle can be recognized by the host cells for inducing an immune response.
Once host cells recognize the presence of RSV components, such as the cytoplasmic viral RNA by the RIG-I receptor, the production of type I interferons (IFNs) is triggered. These innate anti-viral cytokines are represented by various isoforms such as IFN-α, IFN-β, and recently described IFN-ε, -κ, -ω [35,36,37,38]. Besides, upon RSV infection, diverse innate immune cells are activated to produce inflammatory mediators that orchestrate a type 1 antiviral response, characterized by production of IFN-γ (type II IFN) by natural killer (NK) and NKT cells. This response is combined with a type 2 response, which is accomplished by the group 2 innate lymphoid cells, mast cells, and airway epithelial. These cells release interleukin (IL)-5, IL-13, IL-25, IL-33, thymic stromal lymphopoietin (TSLP), CXCL8 (IL-8), CXCL10, CCL4, regulated on activation normal T cell expressed and presumably secreted (RANTES, CCL5), and the high mobility group box I alarmin [39,40,41,42]. Among the primary cytokines produced by airway epithelial cells after RSV infection, tumor necrosis factor (TNF)-α, IL-1α, and IL-1β induce the secretion of IL-6, IL-8, and CCL5 in an autocrine manner [43]. Altogether, these mediators attract eosinophils, neutrophils, monocytes, NK, NKT, dendritic cells (DC), and T cells to the airways, which are involved in the immune response against RSV and viral clearance [39, 41].
Nevertheless, many of these cells are also involved in the severity of the RSV-associated respiratory disease. Bronchoalveolar lavage fluid (BALF) or nasopharyngeal aspirates of infants with severe RSV bronchiolitis are characterized by a predominance of neutrophils [44], a significant increase in the activated conventional DC [45,46,47], and accumulation of granzyme B-expressing NK cells [48]. Pro-inflammatory cytokines IL-1β, IL-6, IL-8, and TNF-α are also significantly higher in the BALF of RSV cases [47]. Additionally, a lower number of plasmacytoid DC (pDC) producing antiviral IFN-α in BALF has been found in preterm infant with RSV bronchiolitis [47]. Various studies have suggested the crucial role of pDCs and type I IFN responses in limiting the viral load and pulmonary inflammation, and in promoting viral clearance as an early response to RSV [49, 50].
It is also known that genetic polymorphism in the immune system genes influence the ability to respond to the RSV and also influences the severity of the infection. In this section, we reviewed the polymorphism in important genes of innate immune system which have been associated with disease severity. For all the discussed genes in this study, we explored the role of protein encoded by those genes in host response to RSV infection and the signaling pathway involved. We have further described the genetic studies in pediatric population which have correlated genetic polymorphism with RSV disease, emphasizing their protective or predisposing participation.
Pattern recognition receptors (PRRs)
One of the initial contacts between RSV and the host cell is mediated by the recognition of the F protein through TLR4. This interaction is proposed as an initiator of the innate immune response, that probably facilitates the virus entry and has been considered as one of the pathogenic triggers, which exacerbates airway inflammation by the release of cytokines and chemokines during RSV infection [43, 51, 52]. Besides, when human lung epithelial cells are infected with RSV, the expression of TLR4 mRNA is increased, suggesting that RSV plays a role in the inflammatory sensitization of the airway epithelium [53]. Innate inflammatory cytokines are expressed once cell activation is initiated by TLR4 through MyD88-dependent or -independent signaling pathways [54]. It has been observed that in splenocytes of TLR4-/- or MyD88-/- mice, the production of IFN-β or TNF-α is highly diminished, which negatively impacts the RSV-specific antibody levels [55]. Moreover, cytokine and chemokine production is also dependent on the nuclear transcription factor kappa-light-chain-enhancer of activated B cells (NF-κB), once the TLR4/CD14 complex has been activated [45, 56]. Genetic polymorphisms alter the function of TLR4, and these alterations have been associated with the severity of RSV infection. Two of the most studied polymorphisms in TLR4 gene are 299Gly and 399Ile, due to their importance in the establishment of an effective immune response. The change of A to G at the position 896 generates a modification of Asp to Gly at the position 299, and the change of C to T at the position 196 leads to substitution of Thr to Ile, although the molecular effect of these changes is still elusive [57]. Both genetic polymorphisms are present in high frequency (in a heterozygous genotype) in ethnically diverse premature infants with symptomatic RSV infection [58]. In 1–12 months old Israeli infants, severe RSV bronchiolitis is significantly associated with polymorphisms 299Gly and/or 399Ile in TLR4, with increased odd ratios (OR) of 4.9 (299Gly/399Ile), 5.1 (299Gly), and 4.0 (399Ile) of hospital admission [59]. On the contrary, the peripheral blood mononuclear cells from Canadian pediatrics subjects (7–9 years old) heterozygous for 299Gly and 399Ile, and acutely exposed to RSV, showed no difference in the production of IFN-γ, CXCL10, IL-10, and CCL5, when compared to that obtained from normal homozygous infants [60]. The role of these alleles has been evidenced in human bronchial epithelial cells which express TLR4 gene with 299Gly or 399Ile polymorphisms. These cells showed reduced production of IL-8, IL-10, IL-12p35, IL-8, and CCL8, indicating that impaired TLR4-response may affect the establishment of an effective immune responses against RSV [57].
TLR2 receptor recognizes common viral motif in RNA viruses, such as dengue virus, human immunodeficiency virus, hepatitis C virus, and rhinovirus, through the dimerization with either TLR1 or TLR6 [61,62,63,64]. Heterodimers TLR2/TLR1 and TLR2/TLR6 recruit MyD88 to the Toll/IL-1 receptor (TIR) domain, which is located in the cytosolic C-terminal region [65, 66]. It has been shown in a study that peritoneal macrophages from C57BL/6 mice stimulated with RSV elicited TNF-α production, which was found to be significantly reduced in TLR2 knock-out and TLR6 knock-out mice. This indicated that only TLR2/TLR6 heterodimers are responsible for RSV recognition and activation of the innate immune responses [67]. Human primary small airway epithelial cells exposed to viral G protein have been found to activate the TLR2/TLR6 signaling and further expression of TNF-α [68]. In the human macrophage cell line U937 infected with RSV, TLR2/MyD88/NF-κB signaling is required for pro-IL-1β and NLRP3 gene expression. This has been found to later on trigger the inflammasome assembly and the subsequent caspase-1 activation and mature IL-1β secretion. After this, a coordinated participation of different pathways is required to orchestrate the innate immune response [69]. Although some TLR2 polymorphisms have been associated with the severity of viral infections, the contribution or mechanism of some of them have not been clearly described yet. The polymorphism rs18998830, known as -15,607 A/G and laid on the first intron of the TLR2 gene, has been significantly associated to severe bronchiolitis induced by RSV with fatalities in the Brazilian infants [70, 71]. Another polymorphism in TLR2 gene, named as rs3804099, which is a synonymous SNPs in the single exon that means a C/T change coding Asn, was associated to a reduced proinflammatory cytokine secretion in hepatitis B virus chronic infection in a Chinese population [72]. In a study conducted in Germany with 156 infants suffering from severe RSV infection, no association of disease severity with rs3804099 polymorphism was reported [73].
During its life cycle, the RSV synthetizes a positive-sense RNA antigenome and various subgenomic mRNAs, which generate an intermediate double-stranded (ds)RNAs within cytosolic inclusions in the host cell [74]. Endosomal TLR3 recognizes dsRNA produced during viral replication [75]. The sensing of dsRNA is crucial to achieve an antiviral state during viral infection, which is characterized by the expression of IFN-α and IFN-β, and other proinflammatory cytokines, such as TNF-α, IL-6, IL-8, and IL-12 [76,77,78,79]. The induction of antiviral cytokine genes is triggered via the TIR domain-containing adaptor-inducing interferon-β (TRIF) signaling, which activates the interferon regulatory factor 3 (IRF3), NF-κB and the activator protein (AP)-1 [80, 81]. Nevertheless, TLR3 activation has also been found to cause RSV-induced airway hyperreactivity and eosinophilia, since IL-33 production is partly TLR3-dependent in alveolar macrophages [82, 83]. There is limited evidence for the presence of polymorphic variants of the TLR3 gene in association with the severity of RSV infection. In the previously mentioned study where 156 German infants were analyzed for TLRs polymorphisms, no association was found between severity of RSV infection and polymorphisms in TLR3 gene rs3775291 (Leu412Phe G/A in exon 4), rs3775290 (F459F in exon 4 1337C/T), rs3775296 (in exon 2, untranslated region 299698T/G) [73]. Moreover, in 129 full-term Finn infants hospitalized for bronchiolitis, there were no significant association between rs3775291 SNP and RSV infection [84]. Although the association between TLR3 polymorphisms and severity of RNA virus infection has been suggested [85,86,87], more data are needed to clarify it.
Finally, it is worthy to consider that RSV RNA can also be recognized by the retinoic acid-inducible gene-I (RIG-I), that facilitates the oligomerization of the mitochondrial activator of signaling (MAVS) on the mitochondrial surface. Thereafter, diverse adaptors, like IκB kinase γ (IKKγ) and TNF receptor-associated factors (TRAFs), are activated with the subsequent activation of NF-κB [88,89,90]. It has been shown that after RSV infection of A549 cells, RIG-I activates the dimerization of the IRF3 and its translocation to the nucleus, leading to the expression of type I IFNs [91, 92]. Based on the biochemical and structural modeling approaches, two variants of human RIGI gene have been identified: the P229fs, a frameshift mutation that generates a truncated constitutively active receptor; and the S183I (a Ser to Ile mutation), which drastically inhibits antiviral signaling due to unintended stable complexes of RIG-I with itself and with MAVS [93]. However, these genetic alterations have not been detected in RSV-infected patients till date. Moreover, the information about SNPs in the RIG-I receptor in these patients is scarce. A study conducted in Canada detected rs10813831 (C/T Arg7Cys) and rs17217280 (T/A Asp580Glu) SNPs, however the differences in particular genotype or allele frequency between the children hospitalized with severe RSV bronchiolitis (n = 140) and children who tested positive for RSV but without hospitalization (n = 100) were not significant [94]. Thus, the possibility of detecting changes in this receptor in diverse childhood populations with severe RSV infection remains open and important to be studied as genetic variants of the RIGI gene have been reported to favor severe infections with other RNA viruses, like hepatitis C virus [95].
Cytokines and chemokines
Type I interferon (IFN)
IFN-α has 13 different subtypes in humans, while there is only one subtype of IFN-β [96]. Once, type I IFNs, are released from the initially infected cells, they induce an antiviral state in the neighboring uninfected cells. To accomplish this, IFNs bind to the IFN-α receptors consisting of IFNAR1 and IFNAR2 chains which further activates JAK-STAT signaling [97, 98]. Type I IFN binding drives the assembly of the two IFNAR chains and the consequent phosphorylation of IFNAR1-associated Tyk2 and IFNAR2-associated Jak1 tyrosine kinases, which phosphorylate IFNAR1 and IFNAR2 [99]. Phosphorylation of the IFNAR1 chain results in phosphorylation of STAT1 and STAT2, which translocate into the nucleus and together with IRF9 form the interferon-stimulated gene factor 3 (ISGF3) transcriptional complex. ISGF3 recognizes the type I IFN-stimulated response elements in promoters of interferon-stimulated genes (ISGs). This initiates transcription and translation of various genes having antiviral activity, antiproliferative activity and have potential to induce robust adaptive immune response [99,100,101,102,103]. RSV has the potential to evade IFN type I-mediated immune response. NS1 binds to RIG-I, thereby inhibiting the activation of MAVS pathway and disrupting the downstream IFN antiviral and inflammatory response [104]. It has been shown in a study that NS2 expression in airway epithelial cells via vaccinia vector decreases the STAT2 levels in human tracheobronchial epithelial cells (hTBE) [98]. The genes coding for type I IFNs are grouped in a locus at the chromosome 9 and consists of 17 different functional genes, among which main are IFNA5 and IFNA13 [104]. The effect of these genes on the severity of infection caused by RSV has been studied. Genetic resistance to severe RSV infection has been associated to the polymorphism rs10757212 of IFNA5 gene. The minor allele T was found to provide a protective effect to the hetero- and homozygous carriers (ORs C/T 0.80 and T/T 0.53, respectively) in a cohort of Dutch children [105]. Additionally, in other study conducted in The Netherlands, the change c.-603G/A (rs643070) of IFNA13 gene conferred protection against RSV bronchiolitis in preterm children (OR 0.68) [106]. In the same study, but in context of IFN-α receptor genes, it was described that the polymorphism of rs7279064 of IFNAR2 gene, which changes Phe10Val, increases the risk of severe RSV infection (OR 1.64) in the same population [106]. Despite the great importance of an adequate and timely type I IFN response to control viral replication and spreading, no other polymorphisms of type I IFN genes that impact RSV severity have been described so far.
Regulated on activation normal T cell expressed and previously secreted (RANTES)
Nasal epithelial cells, fibroblasts, and mast cells produce RANTES protein after RSV infection [39, 107]. RANTES plays a critical role during RSV infection and pathophysiology of bronchiolitis, since it induces chemotaxis of eosinophils, T cells, and monocytes [108]. RANTES levels are associated with risk of recurrent wheezing in RSV bronchiolitis [109]. In an animal model of RSV-infected mice, RANTES production was found to be dependent on the RSV infection, and neutralization of RANTES with anti-RANTES antibody reduced the airway hyperreactivity [110]. Genetic predisposition to severe bronchiolitis has been associated to the variations in the RANTES gene. The mutations -403G/A and -28C/G in the promoter were found to increase the transcription of RANTES which was evident by the serum levels of RANTES [111,112,113]. These variants have been related to increased risk of RSV bronchiolitis in two studies in China. One study analyzed 320 children with RSV bronchiolitis in Southern China, and found that the heterozygous genotype G/A in -403 G/A polymorphism was associated with increased recurrent wheezing risk after RSV bronchiolitis [113]. The second study evaluated 238 infants (under 12 months) in the Nanjing Children's Hospital, and the results showed that the presence of -28G allele increased the risk of RSV bronchiolitis to 2.09, showing an absolute eosinophil count in peripheral blood of RSV-infected children higher than that of control infants [114]. On the contrary, in a study conducted in 106 Greek infants (1–24 months old), there was no association of two SNPs (-403G/A, -28C/G,) in the promoter region of the RANTES gene to severity of RSV [115]. These results suggest that more studies are needed to determine the possible association of polymorphism in these genes in different population with a sufficient population size.
Interleukin (IL)-8
During RSV infection, neutrophil recruitment is dependent on the IL-8 production. Neutrophil influx is a remarkable characteristic among the patients with severe RSV infection as the increased concentration of plasma levels of IL-8 has been associated with severe RSV infection in the children [116,117,118]. Neutrophils have been found to show protective effect during RSV infection, such as reduction of viral dissemination [119], or production of the anti-viral cathelicidin LL-37 [120, 121]. Nevertheless, many reports have evidenced a detrimental role of this innate immune cell in the pathogenesis of severe cases of RSV infection, mainly mediated through the release of elastase [122], mucin production [123], or NETosis [124]. The IL8 gene has polymorphic variations that may contribute to the severity of RSV infection. The SNP -251A/T is ubicated in the promoter and has been associated with a higher risk of severe symptoms in RSV infection. In a cohort of 117 infants in the United Kingdom, the presence of allele -251A was increased in patients with RSV bronchiolitis. As a functional approach, the authors also reported that the -251A allele was related to the increased IL-8 secretion after stimulating whole blood cells with LPS [125]. In a cohort of 320 Chinese children with severe bronchiolitis for RSV, the 54.6% presented wheeze and had increased prevalence of the -251A allele [126]. On the contrary, the allele -251T showed increased frequency in 101 Chinese children with severe RSV pneumonia, which increased the OR to 2.08 [127].
Transcriptional factors
Nuclear transcription factor κ-light-chain-enhancer of activated B cells (NF-κB)
The NF-κB is a transcription factor with pivotal role in the regulation of the expression of hundreds of genes that participate in the immune responses, such as enzymes, receptors, chemokines, cytokines [128]. Structurally, NF-κB consist of a family of dimeric transcriptional factors formed of two class of proteins. One is represented by RelA (p65), RelB and c-Rel subunits, that contain the DNA-binding/dimerization domain called Rel homology domain (RHD), and the transcriptional activation domain. The other class comprises of p50 and p52 subunits, which are expressed as large precursors p105 and p100, respectively, and contains the RHD and, additionally, an ankyrin repeat domain. In a canonical pathway, p105 is cleaved with the participation of a phosphorylation-mediated activation of IKK complexes. This releases the inhibitory κB (IκB) proteins and C-terminal ankyrin repeats from p105, thereafter allowing the release of the heterodimers RelA:p50 (prominently), RelA:c-Rel and c-Rel:p50, which drive the expression of diverse genes of immune response [129,130,131]. RSV evades immune response by redirecting the RelA protein to the cytoplasmic inclusions, making it unavailable to translocate to the nucleus for the transcriptional transactivation [132]. In this context, IL-8 expression after RSV infection has been found to be dysregulated. It has been shown by an in vitro study that this dysregulation is dependent on the translocation of RelA into the nucleus and binding to the IL-8 promoter [133]. It has also been found that host genetics might influence the activity of NF-κB during RSV infection. The promoter of the NFKBIA gene (coding IκB protein) possesses variants that influence the grade of response after stimuli. The polymorphism rs2233406 (-839 C/T) alters the binding regions for CCAAT/enhancer binding protein α (C/EBPα). In 352 Canadian children the minor allele was more prevalent in the group with severe RSV infection and significantly increased the OR to 1.83 [134].
Activator protein (AP)-1
The activator protein-1 (AP-1) is a dimeric transcriptional factor consisting of the members of the family Jun (c-Jun, JunB, JunD) and Fos (c-Fos, FosB, Fra-1, Fra-2) proteins, that after their interaction bind to AP-1 regulatory elements located in the promoters and enhancers of diverse genes related to the immune response [135, 136]. The increased expression of AP-1 in A549 cells (human type II pulmonary epithelial cells) infected with RSV demonstrated the importance of AP-1 in RSV infection. Binding of AP-1 to a region located from − 132 to − 99 in the IL8 promoter was found to induce the expression of IL-8 [45, 137]. The silent polymorphism rs11688 (c.750 G/A) in JUN gene, which encodes the c-Jun protein, has been recognized as a gene marker for the predisposition of severe forms of RSV bronchiolitis. This was evident in a cohort of 480 children in The Netherlands, in whom this allele increased the OR to 1.48 and 3.45 in hetero- and homozygous patients, respectively [105].
Vitamin D receptor
Vitamin D is a steroid hormone obtained from dietary constituents such as oily fish, and endogenous sources including photochemical transformation of precursor 7-dehydrocholesterol [138, 139]. The active form of vitamin D is produced when CYP27B1 hydroxylates 25-hydroxyvitamin D3 into 1,25-dihydroxyvitamin D3 (1,25D), which has been found to induce the production of antimicrobial molecules cathelicidin and LL-37 in macrophages and monocytes [140, 141]. Vitamin D might play a regulatory role in RSV-induced inflammation. The hTBE cells have reduced activation of NF-κB in presence of 1,25D due to the increased expression of NFKBIA mRNA. This effect was found to alter the response in RSV-infected hTBE, since the treatment with 1,25D reduced the levels of IFNB, CXCL10, and ISG15 mRNA. However, the reduction of IFN-β did not alter the viral replication [142]. The effects of vitamin D are facilitated by the binding of vitamin D to the vitamin D receptor (VDR), which once complexed act as transcriptional factor triggering the expression of vitamin D responsive genes [143, 144]. Some genetic variations in the VDR gene have been described to alter the response to vitamin D in diverse pathologies [145]. In terms of RSV infection, the polymorphism rs10735810 has been found to be significantly involved. It causes a change C/T (Thr1Met), which can be determined with the restriction enzyme FokI, and generates a new start codon located three codons upstream from the wild-type start site (ATG). Then, the polymorphic version of VDR contains three amino acids extra in the N-terminal side [146,147,148]. The role of the polymorphism rs10735810 in RSV severity was indicated in 470 children hospitalized in The Netherlands, by its significant association with bronchiolitis as evident by OR of 1.30 [105]. Similar results have been found in a study conducted among 296 South African children with an average age of 3 months, reporting that patients with allele T were more susceptible to severe RSV infection [149].
Conclusion
As most cases of severe RSV infection occur in otherwise healthy infants who have no identifiable risk factors, it is suggested that additional subclinical factors, such as population genetic variations, should also been studied as these might also influence the course of RSV infection. As we highlighted in this review, various studies have shown that different polymorphisms associated with innate immune genes play crucial roles in the physiopathology, susceptibility, or protection to RSV infection in children (summarized in Fig. 3).
Overall, literature suggests that the identification of more SNPs associated with RSV infection would help to decipher the mechanisms involved in the severity of RSV infection. This mechanistic elucidation could further lead to development of novel therapeutic strategies against RSV infection. Since until now there are no vaccine for protection or specific treatment for helping to patient during infection, an accurate and properly modulation of the children immune response against the virus might be key in the prompt clearance of RSV from the host.
References
Miller S (2010) A community health concern: respiratory syncytial virus and children. J Pediatr Nurs 25:551–554. https://doi.org/10.1016/j.pedn.2010.06.011
Shi T, McAllister DA, O’Brien KL, Simoes EAF, Madhi SA, Gessner BD, Polack FP, Balsells E, Acacio S, Aguayo C, Alassani I, Ali A et al (2017) Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. Lancet 390:946–958. https://doi.org/10.1016/S0140-6736(17)30938-8
Mazur NI, Löwensteyn YN, Willemsen JE, Gill CJ, Forman L, Mwananyanda LM, Blau DM, Breiman RF, Madhi SA, Mahtab S, Gurley ES, El Arifeen S, Assefa N, Scott JAG, Onyango D, Tippet Barr BA, Kotloff KL, Sow SO, Mandomando I, Ogbuanu I, Jambai A, Bassat Q, CHAMPS Network the RSV GOLD Study Group, Caballero MT, Polack FP, Omer S, Kazi AM, Simões EAF, Satav A, Bont LJ (2021) Global respiratory syncytial virus-related infant community deaths. Clin Infect Dis 73:S229–S237. https://doi.org/10.1093/cid/ciab528
Ruzin A, Pastula ST, Levin-Sparenberg E, Jiang X, Fryzek J, Tovchigrechko A, Lu B, Qi Y, Liu H, Jin H, Yu L, Hackett J, Villafana T, Esser MT (2018) Characterization of circulating RSV strains among subjects in the OUTSMART-RSV surveillance program during the 2016–17 winter viral season in the United States. PLoS ONE. https://doi.org/10.1371/journal.pone.0200319
Thielen BK, Bye E, Wang X, Maroushek S, Friedlander H, Bistodeau S, Christensen J, Reisdorf E, Shilts MH, Martin K, Como-Sabetti K, Strain AK, Ferrieri P, Lynfield, (2020) Summer outbreak of severe RSV-B disease, Minnesota, 2017 associated with emergence of a genetically distinct viral lineage. J Infect Dis 222:288–297. https://doi.org/10.1093/infdis/jiaa075
Zandi M, Soltani S, Fani M, Abbasi S, Ebrahimi S, Ramezani A (2021) Severe acute respiratory syndrome coronavirus 2 and respiratory syncytial virus coinfection in children. Osong Public Health Res Perspect 12:286–292. https://doi.org/10.24171/j.phrp.2021.0140
Modjarrad K, Giersing B, Kaslow DC, Smith PG, Moorthy VS, WHO RSV Vaccine Consultation Expert Group (2016) WHO consultation on respiratory syncytial virus vaccine development report from a world health organization meeting held on 23–24 March 2015. Vaccine 34:190–197. https://doi.org/10.1016/j.vaccine.2015.05.093
Committee on Infectious Diseases (2009) From the American academy of pediatrics: policy statements—modified recommendations for use of palivizumab for prevention of respiratory syncytial virus infections. Pediatrics 124:1694–1701. https://doi.org/10.1542/peds.2009-2345
Blount Jr RE, Morris JA, Savage RE (1956) Recovery of cytopathogenic agent from chimpanzees with coryza. Proc Soc Exp Biol Med 92:544–549. https://doi.org/10.3181/00379727-92-22538
Chanock R, Roizman B, Myers R (1957) Recovery from infants with respiratory illness of a virus related to chimpanzee coryza agent (CCA). I. Isolation, properties and characterization. Am J Hyg 66:281–290. https://doi.org/10.1093/oxfordjournals.aje.a119901
Amarasinghe GK, Ayllón MA, Bào Y, Basler CF, Bavari S, Blasdell KR, Briese T, Brown PA, Bukreyev A, Balkema-Buschmann A, Buchholz UJ (2019) Taxonomy of the order Mononegavirales: update 2019. Arch Virol 164:1967–1980. https://doi.org/10.1007/s00705-019-04247-4
Bächi T, Howe C (1973) Morphogenesis and ultrastructure of respiratory syncytial virus. J Virol 12:1173–1180. https://doi.org/10.1128/JVI.12.5.1173-1180.1973
Yun MR, Kim AR, Lee HS, Kim DW, Lee WJ, Kim K, Kim SS, Kim YJ (2015) Complete genome sequences of human respiratory syncytial virus genotype a and B isolates from South Korea. Genome Announc 3:e00332-e415. https://doi.org/10.1128/genomeA.00332-15
Ban J, Lee NR, Lee NJ, Lee JK, Quan FS, Inn KS (2018) Human respiratory syncytial virus NS 1 targets TRIM25 to suppress RIG-I ubiquitination and subsequent RIG-I-mediated antiviral signaling. Viruses 10:716. https://doi.org/10.3390/v10120716
Ramaswamy M, Shi L, Varga SM, Barik S, Behlke MA, Look DC (2006) Respiratory syncytial virus nonstructural protein 2 specifically inhibits type I interferon signal transduction. Virology 344:328–339. https://doi.org/10.1016/j.virol.2005.09.009
Sugrue RJ, Brown C, Brown G, Aitken J, McL Rixon HW (2001) Furin cleavage of the respiratory syncytial virus fusion protein is not a requirement for its transport to the surface of virus-infected cells. J Gen Virol 82:1375–1386. https://doi.org/10.1099/0022-1317-82-6-1375
Zimmer G, Budz L (2001) Herrler G (2001) Proteolytic activation of respiratory syncytial virus fusion protein. Cleavage at two furin consensus sequences. J Biol Chem. https://doi.org/10.1074/jbc.M102633200
Melero JA, Mas V, McLellan JS (2017) Structural, antigenic and immunogenic features of respiratory syncytial virus glycoproteins relevant for vaccine development. Vaccine 35:461–468. https://doi.org/10.1016/j.vaccine.2016.09.045
Wertz GWPL, Collins PL, Huang Y, Gruber C, Levine S, Ball LA (1985) Nucleotide sequence of the G protein gene of human respiratory syncytial virus reveals an unusual type of viral membrane protein. Proc Natl Acad Sci U S A 82:4075–4079. https://doi.org/10.1371/journal.ppat.1005318
Behera AK, Matsuse H, Kumar M, Kong X, Lockey RF (2001) Mohapatra SS (2001) Blocking intercellular adhesion molecule-1 on human epithelial cells decreases respiratory syncytial virus infection. Biochem Biophys Res Commun 280:88–95. https://doi.org/10.1006/bbrc.2000.4093
Crim RL, Audet SA, Feldman SA, Mostowski HS, Beeler JA (2007) Identification of linear heparin-binding peptides derived from human respiratory syncytial virus fusion glycoprotein that inhibit infectivity. J Virol 81:261–271. https://doi.org/10.1128/JVI.01226-06
Krusat T, Streckert HJ (1997) Heparin-dependent attachment of respiratory syncytial virus (RSV) to host cells. Arch Virol 142:1247–1254. https://doi.org/10.1007/s007050050156
Malhotra R, Ward M, Bright H, Priest R, Foster MR, Hurle M, Blair E, Bird M (2003) Isolation and characterisation of potential respiratory syncytial virus receptor(s) on epithelial cells. Microbes Infect 5:123–133. https://doi.org/10.1016/s1286-4579(02)00079-5
Mastrangelo P, Hegele RG (2013) RSV fusion: time for a new model. Viruses 5:873–885. https://doi.org/10.3390/v5030873
Tayyari F, Marchant D, Moraes TJ, Duan W, Mastrangelo P, Hegele RG (2011) Identification of nucleolin as a cellular receptor for human respiratory syncytial virus. Nat Med 17:1132–1135. https://doi.org/10.1038/nm.2444
Bilawchuk LM, Griffiths CD, Jensen LD, Elawar F, Marchant DJ (2017) The susceptibilities of respiratory syncytial virus to nucleolin receptor blocking and antibody neutralization are dependent upon the method of virus purification. Viruses 9:207. https://doi.org/10.3390/v9080207
Harcourt J, Alvarez R, Jones LP, Henderson C, Anderson LJ, Tripp RA (2006) Respiratory syncytial virus G protein and G protein CX3C motif adversely affect CX3CR1+ T cell responses. J Immunol 176:1600–1608. https://doi.org/10.4049/jimmunol.176.3.1600
Wang D, Wang J (2020) Antiviral immune mechanism of Toll-like receptor 4-mediated human alveolar epithelial cells type II. Exp Ther Med 20:2561–2568. https://doi.org/10.3892/etm.2020.8963
Cowton VM, McGivern DR, Fearns R (2006) Unravelling the complexities of respiratory syncytial virus RNA synthesis. J Gen Virol 87:1805–1821. https://doi.org/10.1099/vir.0.81786-0
Feldman SA, Crim RL, Audet SA, Beeler JA (2001) Human respiratory syncytial virus surface glycoproteins F, G and SH form an oligomeric complex. Arch Virol 146:2369–2383. https://doi.org/10.1007/s007050170009
Shaikh FY, Crowe JE Jr (2013) Molecular mechanisms driving respiratory syncytial virus assembly. Future Microbiol 8:123–131. https://doi.org/10.2217/fmb.12.132
Mukherjee S, Huda S, Sinha Babu SP (2019) Toll-like receptor polymorphism in host immune response to infectious diseases: a review. Scand J Immunol. https://doi.org/10.1111/sji.12771
Tangye SG, Al-Herz W, Bousfiha A, Chatila T, Cunningham-Rundles C, Etzioni A, Franco JL, Holland SM, Klein C, Morio T, Ochs HD, Oksenhendler E, Picard C, Puck J, Torgerson TR, Casanova JL, Sullivan KE (2020) Human inborn errors of immunity: 2019 update on the classification from the international union of immunological societies expert committee. J Clin Immunol 40:24–64. https://doi.org/10.1007/s10875-019-00737-x
Sun Y, López CB (2017) The innate immune response to RSV: Advances in our understanding of critical viral and host factors. Vaccine 35:481–488. https://doi.org/10.1016/j.vaccine.2016.09.030
DeCarlo CA, Severini A, Edler L, Escott NG, Lambert PF, Ulanova M, Zehbe I (2010) IFN-kappa, a novel type I IFN, is undetectable in HPV-positive human cervical keratinocytes. Lab Invest 90:1482–1491. https://doi.org/10.1038/labinvest.2010.95
Flores I, Mariano TM, Pestka S (1991) Human interferon omega (omega) binds to the alpha/beta receptor. J Biol Chem 266:19875–19877
Hijano DR, Vu LD, Kauvar LM, Tripp RA, Polack FP, Cormier SA (2019) Role of type I interferon (IFN) in the respiratory syncytial virus (RSV) immune response and disease severity. Front Immunol 10:566. https://doi.org/10.3389/fimmu.2019.00566
Marks ZRC, Campbell N, deWeerd NA, Lim SS, Gearing LJ, Bourke NM, Hertzog PJ (2019) Properties and functions of the novel type i interferon epsilon. Semin Immunol. https://doi.org/10.1016/j.smim.2019.101328
Al-Afif A, Alyazidi R, Oldford SA, Huang YY, King CA, Marr N, Haidl ID, Anderson R, Marshall JS (2015) Respiratory syncytial virus infection of primary human mast cells induces the selective production of type I interferons, CXCL10, and CCL4. J Allergy Clin Immunol 136:1346–54.e1. https://doi.org/10.1016/j.jaci.2015.01.042
Becker S, Reed W, Henderson FW, Noah TL (1997) RSV infection of human airway epithelial cells causes production of the beta-chemokine RANTES. Am J Physiol 272:L512–L520. https://doi.org/10.1152/ajplung.1997.272.3.L512
Glaser L, Coulter PJ, Shields M, Touzelet O, Power UF, Broadbent L (2019) Airway epithelial derived cytokines and chemokines and their role in the immune response to respiratory syncytial virus infection. Pathogens 8:106. https://doi.org/10.3390/pathogens8030106
Norlander AE, Peebles RSJr (2020) Innate type 2 responses to respiratory syncytial virus infection. Viruses 12:521. https://doi.org/10.3390/v12050521
Marchant D, Singhera GK, Utokaparch S, Hackett TL, Boyd JH, Luo Z, Si X, Dorscheid DR, McManus BM, Hegele RG (2010) Toll-like receptor 4-mediated activation of p38 mitogen-activated protein kinase is a determinant of respiratory virus entry and tropism. J Virol 84:11359–11373. https://doi.org/10.1128/JVI.00804-10
Everard ML, Swarbrick A, Wrightham M, McIntyre J, Dunkley C, James PD, Sewell HF, Milner AD (1994) Analysis of cells obtained by bronchial lavage of infants with respiratory syncytial virus infection. Arch Dis Child 71:428–432. https://doi.org/10.1136/adc.71.5.428
Gill MA, Palucka AK, Barton T, Ghaffar F, Jafri H, Banchereau J, Ramilo O (2005) Mobilization of plasmacytoid and myeloid dendritic cells to mucosal sites in children with respiratory syncytial virus and other viral respiratory infections. J Infect Dis 191:1105–1115. https://doi.org/10.1086/428589
Gill MA, Long K, Kwon T, Muniz L, Mejias A, Connolly J, Roy L, Banchereau J, Ramilo O (2008) Differential recruitment of dendritic cells and monocytes to respiratory mucosal sites in children with influenza virus or respiratory syncytial virus infection. J Infect Dis 198:1667–1676. https://doi.org/10.1086/593018
Kerrin A, Fitch P, Errington C, Kerr D, Waxman L, Riding K, McCormack J, Mehendele F, McSorley H, MacKenzie K, Wronski S, Braun A, Levin R, Theilen U, Schwarze J (2017) Differential lower airway dendritic cell patterns may reveal distinct endotypes of RSV bronchiolitis. Thorax 72:620–627. https://doi.org/10.1136/thoraxjnl-2015-207358
Bem RA, Bos AP, Bots M, Wolbink AM, van Ham SM, Medema JP, Lutter R, van Woensel JB (2008) Activation of the granzyme pathway in children with severe respiratory syncytial virus infection. Pediatr Res 63:650–655. https://doi.org/10.1203/PDR.0b013e31816fdc32
Wang H, Peters N, Schwarze J (2006) Plasmacytoid dendritic cells limit viral replication, pulmonary inflammation, and airway hyperresponsiveness in respiratory syncytial virus infection. J Immunol 177:6263–6270. https://doi.org/10.4049/jimmunol.177.9.6263
Tsuchida T, Matsuse H, Fukahori S, Kawano T, Tomari S, Fukushima C, Kohno S (2012) Effect of respiratory syncytial virus infection on plasmacytoid dendritic cell regulation of allergic airway inflammation. Int Arch Allergy Immunol 157:21–30. https://doi.org/10.1159/000324676
Funchal GA, Jaeger N, Czepielewski RS, Machado MS, Muraro SP, Stein RT, Bonorino CB, Porto BN (2015) Respiratory syncytial virus fusion protein promotes TLR-4-dependent neutrophil extracellular trap formation by human neutrophils. PLoS ONE. https://doi.org/10.1371/journal.pone.0124082
Marzec J, Cho HY, High M, McCaw ZR, Polack F, Kleeberger SR (2019) Toll-like receptor 4-mediated respiratory syncytial virus disease and lung transcriptomics in differentially susceptible inbred mouse strains. Physiol Genomics 51:630–643. https://doi.org/10.1152/physiolgenomics.00101.2019
Monick MM, Yarovinsky TO, Powers LS, Butler NS, Carter AB, Gudmundsson G, Hunninghake GW (2003) Respiratory syncytial virus up-regulates TLR4 and sensitizes airway epithelial cells to endotoxin. J Biol Chem 278:53035–53044. https://doi.org/10.1074/jbc.M308093200
Zhou Y, Yang J, Deng H, Xu H, Zhang J, Jin W, Gao H, Liu F, Zhao D (2014) Respiratory syncytial virus infection modulates interleukin-8 production in respiratory epithelial cells through a transcription factor-activator protein-1 signaling pathway. Mol Med Rep 10:1443–1447. https://doi.org/10.3892/mmr.2014.2357
Cyr SL, Angers I, Guillot L, Stoica-Popescu I, Lussier M, Qureshi S, Burt DS, Ward BJ (2009) TLR4 and MyD88 control protection and pulmonary granulocytic recruitment in a murine intranasal RSV immunization and challenge model. Vaccine 27:421–430. https://doi.org/10.1016/j.vaccine.2008.10.073
Bao X, Indukuri H, LiuT LSL, Tian B, Brasier AR, Garofalo RP, Casola A (2010) IKKε Modulates RSV-induced NF- κB-dependent gene transcription. Virology 408:224–231. https://doi.org/10.1016/j.virol.2010.09.016
Tulic MK, Hurrelbrink RJ, Prêle CM, Laing IA, Upham JW, Le Souef P, Sly PD, Holt PG (2007) TLR4 polymorphisms mediate impaired responses to respiratory syncytial virus and lipopolysaccharide. J Immunol 179:132–140. https://doi.org/10.4049/jimmunol.179.1.132
Awomoyi AA, Rallabhandi P, Pollin TI, Lorenz E, Sztein MB, Boukhvalova MS, Hemming VG, Blanco JC, Vogel SN (2007) Association of TLR4 polymorphisms with symptomatic respiratory syncytial virus infection in high-risk infants and young children. J Immunol 179:3171–3177. https://doi.org/10.4049/jimmunol.179.5.3171
Tal G, Mandelberg A, Dalal I, Cesar K, Somekh E, Tal A, Oron A, Itskovich S, Ballin A, Houri S, Beigelman A, Lider O, Rechavi G, Amariglio N (2004) Association between common Toll-like receptor 4 mutations and severe respiratory syncytial virus disease. J Infect Dis 189:2057–2063. https://doi.org/10.1086/420830
Douville RN, Lissitsyn Y, Hirschfeld AF, Becker AB, Kozyrskyj AL, Liem J, Bastien N, Li Y, Victor RE, Sekhon M, Turvey SE, HayGlass KT (2010) TLR4 Asp299Gly and Thr399Ile polymorphisms: no impact on human immune responsiveness to LPS or respiratory syncytial virus. PLoS ONE. https://doi.org/10.1371/journal.pone.0012087
Aguilar-Briseño JA, Upasani V, Ellen BMT, Moser J, Pauzuolis M, Ruiz-Silva M, Heng S, Laurent D, Choeung R, Dussart P, Cantaert T, Smit JM, Rodenhuis-Zybert IA (2020) TLR2 on blood monocytes senses dengue virus infection and its expression correlates with disease pathogenesis. Nat Commun 11:3177. https://doi.org/10.1038/s41467-020-16849-7
Henrick BM, Yao XD, Rosenthal KL, INFANT study team (2015) HIV-1 structural proteins serve as PAMPs for TLR2 heterodimers significantly increasing infection and innate immune activation. Front Immunol 6:426. https://doi.org/10.3389/fimmu.2015.00426
Neamatallah M, El-Bendary M, Elalfy H, Besheer T, El-Maksoud MA, Elhammady D, Abed S, Elegezy M, Kandeel L, Eldeib D, Mousa N, Abd El-Hafeez M, El-Gilany AH, Esmat G (2020) Impact of toll-like receptors 2(TLR2) and TLR 4 gene variations on HCV susceptibility, response to treatment and development of hepatocellular carcinoma in cirrhotic HCV patients. Immunol Invest 49:462–476. https://doi.org/10.1080/08820139.2019.1673772
Triantafilou K, Vakakis E, Richer EA, Evans GL, Villiers JP, Triantafilou M (2011) Human rhinovirus recognition in non-immune cells is mediated by Toll-like receptors and MDA-5, which trigger a synergetic pro-inflammatory immune response. Virulence 2:22–29. https://doi.org/10.4161/viru.2.1.13807
Botos I, Segal DM, Daviesa DR (2011) The structural biology of toll-like receptors. Structure 19:447–459. https://doi.org/10.1016/j.str.2011.02.004
O’Neill LA, Bowie AG (2007) The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol 7:353–364. https://doi.org/10.1038/nri2079
Murawski MR, Bowen GN, Cerny AM, Anderson LJ, Haynes LM, Tripp RA, Kurt-Jones EA, Finberg RW (2009) Respiratory syncytial virus activates innate immunity through Toll-like receptor 2. J Virol 83:1492–1500. https://doi.org/10.1128/JVI.00671-08
Alshaghdali K, Saeed M, Kamal MA, Saeed A (2021) Interaction of ectodomain of respiratory syncytial virus G protein with TLR2/TLR6 heterodimer: an in vitro and in silico approach to decipher the role of RSV G protein in pro-inflammatory response against the virus. Curr Pharm Des 27:4464–4476. https://doi.org/10.2174/1381612827666210716160030
Segovia J, Sabbah A, Mgbemena V, Tsai SY, Chang TH, Berton MT, Morris IR, Allen IC, Ting JP, Bose S (2012) TLR2/MyD88/NF-kappaB pathway, reactive oxygen species, potassium efflux activates NLRP3/ASC inflammasome during respiratory syncytial virus infection. PLoS ONE. https://doi.org/10.1371/journal.pone.0029695
Alvarez AE, Marson FAL, Bertuzzo CS, Bastos JCS, Baracat ECE, Brandão MB, Tresoldi AT, das Neves Romaneli, MT, Almeida CCB, de Oliveira T, Schlodtmann PG, Corrêa E de Miranda MLF, Dos Reis MC, De Pieri JV, Arns CW, Ribeiro JD (2018) Association between single nucleotide polymorphisms in TLR4, TLR2, TLR9, VDR, NOS2 and CCL5 genes with acute viral bronchiolitis. Gene 645:7–17. https://doi.org/10.1016/j.gene.2017.12.022
Chedid P, Salami A, Shamieh SE (2020) The association of rs1898830 in toll-like receptor 2 with lipids and blood pressure. J Cardiovasc Dev Dis 7:24. https://doi.org/10.3390/jcdd7030024
Lin Y, Gao ZX, Shen X, Chen MJ, Li YT, Li SL, Lin HL, Zhao QF, Liu F, Niu JJ (2018) Correlation between polymorphisms in toll-like receptor genes and the activity of hepatitis B virus among treatment-naïve patients: a case-control study in a Han Chinese population. BMC Infect Dis 18:28. https://doi.org/10.1186/s12879-018-2943-x
Mailaparambil B, Krueger M, Heinze J, Forster J, Heinzmann A (2008) Polymorphisms of toll like receptors in the genetics of severe RSV associated diseases. Dis Markers 25:59–65. https://doi.org/10.1155/2008/619595
Groskreutz DJ, Monick MM, Powers LS, Yarovinsky TO, Look DC, Hunninghake GW (2006) Respiratory syncytial virus induces TLR3 protein and protein kinase R, leading to increased double-stranded RNA responsiveness in airway epithelial cells. J Immunol 176:1733–1740. https://doi.org/10.4049/jimmunol.176.3.1733
Wong JP, Christopher ME, Viswanathan S, Dai X, Salazar AM, Sun LQ, Wang M (2009) Antiviral role of toll-like receptor-3 agonists against seasonal and avian influenza viruses. Curr Pharm Des 15:1269–1274. https://doi.org/10.2174/138161209787846775
Alexopoulou L, Holt AC, Medzhitov R, Flavell RA (2001) Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413:732–738. https://doi.org/10.1038/35099560
Cheung MB, Sampayo-Escobar V, Green R, Moore ML, Mohapatra S, Mohapatra SS (2016) Respiratory syncytial virus-infected mesenchymal stem cells regulate immunity via interferon beta and indoleamine-2,3-dioxygenase. PLoS ONE. https://doi.org/10.1371/journal.pone.0163709
Dou Y, Zhao Y, Zhang ZY, Mao HW, Tu WW, Zhao XD (2013) Respiratory syncytial virus infection induces higher Toll-like receptor-3 expression and TNF-alpha production than human metapneumovirus infection. PLoS ONE. https://doi.org/10.1371/journal.pone.0073488
Liu D, Chen Q, Zhu H, Gong L, Huang Y, Li S, Yue C, Wu K, Wu Y, Zhang W, Huang G, Zhang L, Pu S, Wang D (2018) Association of respiratory syncytial virus toll-like receptor 3-mediated immune response with COPD exacerbation frequency. Inflammation 41:654–666. https://doi.org/10.1007/s10753-017-0720-4
Behzadi P, García-Perdomo HA, Karpiński TM (2021) Toll-like receptors: general molecular and structural biology. J Immunol Res 2021:9914854. https://doi.org/10.1155/2021/9914854
Klouwenberg PK, Tan L, Werkman W, van Bleek GM, Coenjaerts F (2009) The role of Toll-like receptors in regulating the immune response against respiratory syncytial virus. Crit Rev Immunol 29:531–550. https://doi.org/10.1615/critrevimmunol.v29.i6.40
Qi F, Wang D, Liu J, Zeng S, Xu L, Hu H, Liu B (2015) Respiratory macrophages and dendritic cells mediate respiratory syncytial virus-induced IL-33 production in TLR3- or TLR7-dependent manner. Int Immunopharmacol 29:408–415. https://doi.org/10.1016/j.intimp.2015.10.022
Wu YH, Lai AC, Chi PY, Thio CL, Chen WY, Tsai CH, Lee YL, Lukacs NW, Chang YJ (2020) Pulmonary IL-33 orchestrates innate immune cells to mediate respiratory syncytial virus-evoked airway hyperreactivity and eosinophilia. Allergy 75:818–830. https://doi.org/10.1111/all.14091
Nuolivirta K, He Q, Vuononvirta J, Koponen P, Helminen M, Korppi M (2012) Toll-like receptor 3 L412F polymorphisms in infants with bronchiolitis and postbronchiolitis wheezing. Pediatr Infect Dis J 31:920–923. https://doi.org/10.1097/INF.0b013e31825aff25
Alagarasu K, Bachal RV, Memane RS, Shah PS, Cecilia D (2015) Polymorphisms in RNA sensing toll like receptor genes and its association with clinical outcomes of dengue virus infection. Immunobiology 220:64–68. https://doi.org/10.1016/j.imbio.2014.09.020
Esposito S, Molteni CG, Giliani S, Mazza C, Scala A, Tagliaferri L, Pelucchi C, Fossali E, Plebani A, Principi N (2012) Toll-like receptor 3 gene polymorphisms and severity of pandemic A/H1N1/2009 influenza in otherwise healthy children. Virol J 9:270. https://doi.org/10.1186/1743-422X-9-270
Habibabadi HM, Parsania M, Pourfathollah AA, Haghighat S, Sharifi Z (2020) Association of TLR3 single nucleotide polymorphisms with susceptibility to HTLV-1 infection in Iranian asymptomatic blood donors. Rev Soc Bras Med Trop. https://doi.org/10.1590/0037-8682-0026-2020
Jamaluddin M, Tian B, Boldogh I, Garofalo RP, Brasier AR (2009) Respiratory syncytial virus infection induces a reactive oxygen species-MSK1-phospho-Ser-276 RelA pathway required for cytokine expression. J Virol 83:10605–10615. https://doi.org/10.1128/JVI.01090-09
Liu P, Li K, Garofalo RP, Brasier AR (2008) Respiratory syncytial virus induces RelA release from cytoplasmic 100-kDa NF-κB2 complexes via a novel retinoic acid-inducible gene-I·NF-κB-inducing kinase signaling pathway. J Biol Chem 283:23169–23178. https://doi.org/10.1074/jbc.M802729200
Yoboua F, Martel A, Duval A, Mukawera E, Grandvaux N (2010) Respiratory syncytial virus-mediated NF-κB p65 phosphorylation at serine 536 is dependent on RIG-I, TRAF6, and IKKβ. J Virol 84:7267–7277. https://doi.org/10.1128/JVI.00142-10
Stephens LM, Varga SM (2020) Function and modulation of type I interferons during respiratory syncytial virus infection. Vaccines (Basel) 8:177. https://doi.org/10.3390/vaccines8020177
Yamamoto K, Yamamoto S, Ogasawara N, Takano K, Shiraishi T, Sato T, Miyata R, Kakuki T, Kamekura R, Kojima T, Tsutsumi H, Himi T, Yokota SI (2016) Clarithromycin prevents human respiratory syncytial virus-induced airway epithelial responses by modulating activation of interferon regulatory factor-3. Pharmacol Res 111:804–814. https://doi.org/10.1016/j.phrs.2016.07.033
Pothlichet J, Burtey A, Kubarenko AV, Caignard G, Solhonne B, Tangy F, Ben-Ali M, Quintana-Murci L, Heinzmann A, Chiche JD, Vidalain PO, Weber AN, Chignard M, Si-Tahar M (2009) Study of human RIG-I polymorphisms identifies two variants with an opposite impact on the antiviral immune response. PLoS ONE. https://doi.org/10.1371/journal.pone.0007582
Marr N, Hirschfeld AF, Lam A, Wang S, Lavoie PM, Turvey SE (2014) Assessment of genetic associations between common single nucleotide polymorphisms in RIG-I-like receptor and IL-4 signaling genes and severe respiratory syncytial virus infection in children: a candidate gene case-control study. PLoS ONE. https://doi.org/10.1371/journal.pone.0100269
Wu X, Zang F, Liu M, Zhuo L, Wu J, Xia X, Feng Y, Yu R, Huang P, Yang S (2019) Genetic variants in RIG-I-like receptor influences HCV clearance in Chinese Han population. Epidemiol Infect. https://doi.org/10.1017/S0950268819000827
Gilbert K, Schlaak JF, Yang D, Dittmer U (2013) IFN-α subtypes: distinct biological activities in anti-viral therapy. Br J Pharmacol 168:1048–1058. https://doi.org/10.1111/bph.12010
Kong X, San Juan H, Kumar M, Behera AK, Mohapatra A, Hellermann GR, Mane S, Lockey RF, Mohapatra SS (2003) Respiratory syncytial virus infection activates STAT signaling in human epithelial cells. Biochem Biophys Res Commun 306:616–622. https://doi.org/10.1016/s0006-291x(03)01008-8
Ramaswamy M, Shi L, Monick MM, Hunninghake GW, Look DC (2004) Specific inhibition of type I interferon signal transduction by respiratory syncytial virus. Am J Respir Cell Mol Biol 30:893–900. https://doi.org/10.1165/rcmb.2003-0410OC
Zanin N, Viaris de Lesegno C, Lamaze C, Blouin CM (2021) Interferon receptor trafficking and signaling: journey to the cross roads. Front Immunol. https://doi.org/10.3389/fimmu.2020.615603
Goritzka M, Durant LR, Pereira C, Salek-Ardakani S, Openshaw PJ, Johansson C (2014) Alpha/beta interferon receptor signaling amplifies early proinflammatory cytokine production in the lung during respiratory syncytial virus infection. J Virol 88:6128–6136. https://doi.org/10.1128/JVI.00333-14
Payelle-Brogard B, Pellegrini S (2010) Biochemical monitoring of the early endocytic traffic of the type I interferon receptor. J Interferon Cytokine Res 30:89–98. https://doi.org/10.1089/jir.2009.0044
Platanitis E, Demiroz D, Schneller A, Fischer K, Capelle C, Hartl M, Gossenreiter T, Müller M, Novatchkova M, Decker T (2019) A molecular switch from STAT2-IRF9 to ISGF3 underlies interferon-induced gene transcription. Nat Commun 10:2921. https://doi.org/10.1038/s41467-019-10970-y
Schoggins JW (2019) Interferon-stimulated genes: what do they all do? Annu Rev Virol 6:567–584. https://doi.org/10.1146/annurev-virology-092818-015756
Wittling MC, Cahalan SR, Levenson EA, Rabin RL (2021) Shared and unique features of human interferon-beta and interferon-alpha subtypes. Front Immunol 11:605673. https://doi.org/10.3389/fimmu.2020.605673
Janssen R, Bont L, Siezen CLE, Hodemaekers HM, Ermers MJ, Doornbos G, van ’t Slot R, Wijmenga C, Goeman JJ, Kimpen JLL, van Houwelingen HC, Kimman TG, Hoebee B (2007) Genetic susceptibility to respiratory syncytial virus bronchiolitis is predominantly associated with innate immune genes. J Infect Dis 196:826–834. https://doi.org/10.1086/520886
Siezen CLE, Bont L, Hodemaekers HM, Ermers MJ, Doornbos G, Van’t Slot R, Wijmenga C, van Houwelingen HC, Kimpen JLL, Kimman TG, Hoebee B, Janssen R (2009) Genetic susceptibility to respiratory syncytial virus bronchiolitis in preterm children is associated with airway remodeling genes and innate immune genes. Pediatr Infect Dis J 28:333–335. https://doi.org/10.1097/INF.0b013e31818e2aa9
Jung SY, Shin SY, Eun YG, Kim SW, Cho JS (2013) Changes of histamine receptors and CC chemokines in nasal epithelial cells and fibroblasts after respiratory syncytial virus infection. Am J Rhinol Allergy 27:e17-21. https://doi.org/10.2500/ajra.2013.27.3863
Appay V, Rowland-Jones SL (2001) RANTES: a versatile and controversial chemokine. Trends Immunol 22:83–87. https://doi.org/10.1016/s1471-4906(00)01812-3
Hasegawa K, Piedra PA, Bauer CS, Celedón JC, Mansbach JM, Spergel JM, Espinola JA, Camargo CA Jr, MARC-35 Investigators (2018) Nasopharyngeal CCL5 in infants with severe bronchiolitis and risk of recurrent wheezing: a multi-center prospective cohort study. Clin Exp Allergy 48:1063–1067. https://doi.org/10.1111/cea.13166
Tekkanat KK, Maassab H, Miller A, Berlin AA, Kunkel SL, Lukacs NW (2002) RANTES (CCL5) production during primary respiratory syncytial virus infection exacerbates airway disease. Eur J Immunol 32:3276–3284. https://doi.org/10.1002/1521-4141(200211)32:11%3c3276::AID-IMMU3276%3e3.0.CO;2-5
al Sharif F, Ollier WE, Hajeer AH (1999) A rare polymorphism at position -28 in the human RANTES promoter. Eur J Immunogenet 26:373–374. https://doi.org/10.1046/j.1365-2370.1999.00162.x
Drysdale SB, Milner AD, Greenough A (2012) Respiratory syncytial virus infection and chronic respiratory morbidity - is there a functional or genetic predisposition? Acta Paediatr 101:1114–1120. https://doi.org/10.1111/j.1651-2227.2012.02825.x
Tian M, Liu F, Wen G, Shi S, Chen R, Zhao D (2009) Effect of variation in RANTES promoter on serum RANTES levels and risk of recurrent wheezing after RSV bronchiolitis in children from Han, Southern China. Eur J Pediatr 168:963–977. https://doi.org/10.1007/s00431-008-0870-3
Zhao D, Wen G, Tian M, Shi S, Chen R (2008) Association of RANTES gene promoter -28C/G polymorphism with respiratory syncytial virus bronchiolitis. Zhonghua Er Ke Za Zhi 46:89–93
Amanatidou V, Sourvinos G, Apostolakis S, Neonaki P, Tsilimigaki A, Krambovitis E, Spandidos DA (2008) RANTES promoter gene polymorphisms and susceptibility to severe respiratory syncytial virus-induced bronchiolitis. Pediatr Infect Dis J 27:38–42. https://doi.org/10.1097/INF.0b013e31814d4e42
Brand HK, Ferwerda G, Preijers F, de Groot R, Neeleman C, Staal FJ, Warris A, Hermans PW (2013) CD4+ T-cell counts and interleukin-8 and CCL-5 plasma concentrations discriminate disease severity in children with RSV infection. Pediatr Res 73:187–193. https://doi.org/10.1097/INF.0b013e31814d4e42
Brown PM, Schneeberger DL, Piedimonte G (2015) Biomarkers of respiratory syncytial virus (RSV) infection: specific neutrophil and cytokine levels provide increased accuracy in predicting disease severity. Paediatr Respir Rev 16:232–240. https://doi.org/10.1016/j.prrv.2015.05.005
Lukens MV, van de Pol AC, Coenjaerts FE, Jansen NJ, Kamp VM, Kimpen JL, Rossen JW, Ulfman LH, Tacke CE, Viveen MC, Koenderman L, Wolfs TF, van Bleek GM (2010) A systemic neutrophil response precedes robust CD8(+) T-cell activation during natural respiratory syncytial virus infection in infants. J Virol 84:2374–2383. https://doi.org/10.1128/JVI.01807-09
Deng Y, Herbert JA, Robinson E, Ren L, Smyth RL, Smith CM (2020) Neutrophil-airway epithelial interactions result in increased epithelial damage and viral clearance during respiratory syncytial virus infection. J Virol 94:e02161-e2219. https://doi.org/10.1128/JVI.02161-19
Currie SM, Gwyer Findlay E, McFarlane AJ, Fitch PM, Böttcher B, Colegrave N, Paras A, Jozwik A, Chiu C, Schwarze J, Davidson DJ (2016) Cathelicidins have direct antiviral activity against respiratory syncytial virus in vitro and protective function in vivo in mice and humans. J Immunol 196:2699–2710. https://doi.org/10.4049/jimmunol.1502478
Mansbach JM, Hasegawa K, Ajami NJ, Petrosino JF, Piedra PA, Tierney CN, Espinola JA, Camargo CA (2017) Serum LL-37 levels associated with severity of bronchiolitis and viral etiology. Clin Infect Dis 65:967–975. https://doi.org/10.1093/cid/cix483
Yasui K, Baba A, Iwasaki Y, Kubo T, Aoyama K, Mori T, Yamazaki T, Kobayashi N, Ishiguro A (2005) Neutrophil-mediated inflammation in respiratory syncytial viral bronchiolitis. Pediatr Int 47:190–195. https://doi.org/10.1111/j.1442-200x.2005.02039.x
Stokes KL, Currier MG, Sakamoto K, Lee S, Collins PL, Plemper RK, Moore ML (2013) The respiratory syncytial virus fusion protein and neutrophils mediate the airway mucin response to pathogenic respiratory syncytial virus infection. J Virol 87:10070–10082. https://doi.org/10.1128/JVI.01347-13
Muraro SP, De Souza GF, Gallo SW, Da Silva BK, De Oliveira SD, Vinolo MAR, Saraiva EM, Porto BN (2018) Respiratory syncytial virus induces the classical ROS-dependent NETosis through PAD-4 and necroptosis pathways activation. Sci Rep 8:14166. https://doi.org/10.1038/s41598-018-32576-y
Hull J, Thomson A, Kwiatkowski D (2000) Association of respiratory syncytial virus bronchiolitis with the interleukin 8 gene region in UK families. Thorax 55:1023–1027. https://doi.org/10.1136/thorax.55.12.1023
Tian M, Zhao DY, Wen GY, Shi SY, Chen RH (2007) Association between interleukin-8 gene-251 locus polymorphism and respiratory syncytial virus bronchiolitis and post-bronchiolitis wheezing in infants. Zhonghua Er Ke Za Zhi 45:856–859
Lu A, Wang L, Zhang X (2010) Haplotype of IL-8 -251T and 781C is associated with the susceptibility to respiratory syncytial virus. J Trop Pediatr 56:242–246. https://doi.org/10.1093/tropej/fmp101
Mitchell JP, Carmody RJ (2018) NF-kappaB and the transcriptional control of inflammation. Int Rev Cell Mol Biol 335:41–84. https://doi.org/10.1016/bs.ircmb.2017.07.007
Gilmore TD (2006) Introduction to NF-kappaB: players, pathways, perspectives. Oncogene 25:6680–6684. https://doi.org/10.1038/sj.onc.1209954
Kabacaoglu D, Ruess DA, Ai J, Algül H (2019) NF-kappaB/Rel transcription factors in pancreatic cancer: focusing on RelA, c-Rel, and RelB. Cancers (Basel) 11:937. https://doi.org/10.3390/cancers11070937
Santoro MG, Rossi A, Amici C (2003) NF-kappaB and virus infection: who controls whom. EMBO J 22:2552–2560. https://doi.org/10.1093/emboj/cdg267
Jobe F, Simpson J, Hawes P, Guzman E, Bailey D (2020) Respiratory syncytial virus sequesters NF-kappaB subunit p65 to cytoplasmic inclusion bodies to inhibit innate immune signaling. J Virol 94:e01380-e1420. https://doi.org/10.1128/JVI.01380-20
Garofalo R, Sabry M, Jamaluddin M, Yu RK, Casola A, Ogra PL, Brasier AR (1996) Transcriptional activation of the interleukin-8 gene by respiratory syncytial virus infection in alveolar epithelial cells: nuclear translocation of the RelA transcription factor as a mechanism producing airway mucosal inflammation. J Virol 70:8773–8781. https://doi.org/10.1128/JVI.70.12.8773-8781.1996
Ali S, Hirschfeld AF, Mayer ML, Fortuno ES III, Corbett N, Kaplan M, Wang S, Schneiderman J, Fjell CD, Yan J, Akhabir L, Aminuddin, F, Marr N, Lacaze-Masmonteil T, Hegele RG, Becker A, Chan-Yeung M, Hancock RE, Kollmann TR, Daley D, Sandford AJ, Lavoie PM, Turvey SE (2013) Functional genetic variation in NFKBIA and susceptibility to childhood asthma, bronchiolitis, and bronchopulmonary dysplasia. J Immunol 190:3949–3958. https://doi.org/10.4049/jimmunol.1201015
Bejjani F, Evanno E, Zibara K, Piechaczyk M, Jariel-Encontre I (2019) The AP-1 transcriptional complex: local switch or remote command? Biochim Biophys Acta Rev Cancer 1872:11–23. https://doi.org/10.1016/j.bbcan.2019.04.003
Schonthaler HB, Guinea-Viniegra J, Wagner EF (2011) Targeting inflammation by modulating the Jun/AP-1 pathway. Ann Rheum Dis 70:109–112. https://doi.org/10.1136/ard.2010.140533
Casola A, Garofalo RP, Jamaluddin M, Vlahopoulos S, Brasier AR (2000) Requirement of a novel upstream response element in respiratory syncytial virus-induced IL-8 gene expression. J Immunol 164:5944–5951. https://doi.org/10.4049/jimmunol.164.11.5944
Ismailova A, White JH (2021) Vitamin D, infections and immunity. Rev Endocr Metab Disord 29:1–13. https://doi.org/10.1007/s11154-021-09679-5
Hetta HF, Muhammad K, El-Masry EA, Taha AE, Ahmed EA, Phares C, Kader HA, Waheed Y, Zahran AM, Yahia R, Meshaal AK, El-Saber Batiha G (2021) The interplay between vitamin D and COVID-19: protective or bystander? Eur Rev Med Pharmacol Sci 25:2131–2145. https://doi.org/10.26355/eurrev_202102_25119
Bacchetta J, Chun RF, Gales B, Zaritsky JJ, Leroy S, Wesseling-Perry K, Boregaard N, Rastogi A, Salusky IB, Hewison M (2014) Antibacterial responses by peritoneal macrophages are enhanced following vitamin D supplementation. PLoS One. https://doi.org/10.1371/journal.pone.0116530
Liu PT, Stenger S, Li H, Wenzel L, Tan BH, Krutzik SR, Ochoa MT, Schauber J, Wu K, Meinken C, Kamen DL, Wagner M, Bals R, Steinmeyer A, Zügel U, Gallo RL, Eisenberg D, Hewison M, Hollis BW, Adams JS, Bloom BR, Modlin RL (2006) Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response. Science 311:1770–1773. https://doi.org/10.1126/science.1123933
Hansdottir S, Monick MM, Lovan N, Powers L, Gerke A, Hunninghake GW (2010) Vitamin D decreases respiratory syncytial virus induction of NF-kappaB-linked chemokines and cytokines in airway epithelium while maintaining the antiviral state. J Immunol 184:965–974. https://doi.org/10.4049/jimmunol.0902840
Carlberg C, Seuter S (2009) A genomic perspective on vitamin D signaling. Anticancer Res 29:3485–3493
Laplana M, Royo JL, Fibla J (2018) Vitamin D receptor polymorphisms and risk of enveloped virus infection: a meta-analysis. Gene 678:384–394. https://doi.org/10.1016/j.gene.2018.08.017
Valdivielso JM, Fernandez E (2006) Vitamin D receptor polymorphisms and diseases. Clin Chim Acta 371:1–12. https://doi.org/10.1016/j.cca.2006.02.016
Gross C, Krishnan AV, Malloy PJ, Eccleshall TR, Zhao XY, Feldman D (1998) The vitamin D receptor gene start codon polymorphism: a functional analysis of FokI variants. J Bone Miner Res 13:1691–1699. https://doi.org/10.1359/jbmr.1998.13.11.1691
Jurutka PW, Remus LS, Whitfield GK, Thompson PD, Hsieh JC, Zitzer H, Tavakkoli P, Galligan MA, Dang HT, Haussler CA, Haussler MR (2000) The polymorphic N terminus in human vitamin D receptor isoforms influences transcriptional activity by modulating interaction with transcription factor IIB. Mol Endocrinol 14:401–420. https://doi.org/10.1210/mend.14.3.0435
Saijo T, Ito M, Takeda E, Huq AH, Naito E, Yokota I, Sone T, Pike JW, Kuroda Y (1991) A unique mutation in the vitamin D receptor gene in three Japanese patients with vitamin D-dependent rickets type II: utility of single-strand conformation polymorphism analysis for heterozygous carrier detection. Am J Hum Genet 49:668–673
Kresfelder TL, Janssen R, Bont L, Pretorius M, Venter M (2011) Confirmation of an association between single nucleotide polymorphisms in the VDR gene with respiratory syncytial virus related disease in South African children. J Med Virol 83:1834–1840. https://doi.org/10.1002/jmv.22179
Acknowledgements
B.A. H-M. and C.B. B-G. received a master fellowship from CONACYT (#803202 and #756640).
Funding
This work was supported by National Council of Science and Technology (Grant CONACYT APN-2017-5901 to D. C-G.) and Autonomous University of Aguascalientes (Grant PIBB 19-9N to D. C-G.).
Author information
Authors and Affiliations
Contributions
All the authors contributed equally to this work. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Edited by Joachim J. Bugert.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Córdova-Dávalos, L.E., Hernández-Mercado, A., Barrón-García, C.B. et al. Impact of genetic polymorphisms related to innate immune response on respiratory syncytial virus infection in children. Virus Genes 58, 501–514 (2022). https://doi.org/10.1007/s11262-022-01932-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-022-01932-6