Abstract
Argentina exhibits low serological prevalence for Hepatitis B virus (HBV); however, occult hepatitis B infection (OBI) has been reported in blood donors, Amerindians and individuals coinfected with hepatitis C virus (HCV), and/or human immunodeficiency virus (HIV). The aim of this study was to analyze the genetic diversity of HBV and to evaluate serological marker associations and coinfections with HCV and HIV in patients attending and treated in a public hospital in the province of Buenos Aires, Argentina. A total of 189 HBV reactive samples (HBsAg and/or anti-HBc) were analyzed for HBV DNA characterization. All reactive samples were tested for anti-HCV and HIV-antigen/antibody using CMIA assays. Thirty-six samples exhibited detectable HBV DNA, 7 of which were OBI. HBV sequences were classified as subgenotypes A1, A2, B2, D3, F1b, F3 and F4. Mutations related to the ability to escape the host’s immune response, resistance to antiviral therapy and progression to disease were found in patients, partly due to the variable sensitivity of HBsAg, the reverse transcriptase, the basal core promoter and the preCore. HCV and HIV prevalence was 10% and most of the genotypes found in the sequences were genotype 1 and B/F recombinant subtype, respectively. Of the total samples analyzed, 7 exhibited coinfections. This study shows the frequency of OBI, subgenotype distribution, HBV mutations and coinfections, which may have important clinical implications in public hospital patients. Planned prevention, detection and treatment adherence are needed to reduce transmission and morbidity in vulnerable populations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hepatitis B virus (HBV) infection is a worldwide public health concern. It is estimated that 257 million individuals are chronic carriers [1]. HBV promotes liver inflammation, frequently associated with cirrhosis and/or hepatocellular carcinoma (HCC) [1]. HBV has a partially double-stranded DNA genome with four overlapping open reading frames (ORFs; PreS/S, PreCore/Core [preC/C], X and polymerase [P]) [2].
HBV infection is globally distributed, with high (≥ 8%), intermediate (2–8%) and low (≤ 2%) endemic areas [1, 2]. Most South American countries are currently low endemic areas except for Ecuador, Venezuela and southern Brazil that exhibit intermediate endemicity. There are also scattered highly endemic regions, such as Peru, southern Colombia, northern Bolivia and northern Brazil [3]. HBV is presently divided into nine confirmed genotypes (A–I) and one putative genotype (J); some of these genotypes are subdivided into numerous subgenotypes with different geographical distributions, disease progression and response to therapy [1, 2]. Occult HBV infection (OBI) is defined as the presence of HBV viral DNA in the liver (with or without detectable HBV DNA in serum) in HBsAg-non reactive individuals tested with currently available serum assays. A cutoff value of < 200 IU/ml was also introduced for HBV DNA in serum [4]. The prevalence of OBI varies widely throughout the world, and among the study population of patients, due to the sensitivity of HBsAg and HBV DNA detection assays [4].
Argentina exhibits low serological prevalence for HBV in blood donors [5, 6]. However, the HBV infection may vary according to the geographic region and/or the vulnerable population under study [2]. HBV different genotypes and subgenotypes have been reported in dissimilar populations in this country [5,6,7,8,9,10,11,12,13,14]. In addition, OBI has been detected in blood donors, Amerindians and patients coinfected with HCV and /or HIV from Argentina [6, 12, 15].
The objective of this study was to analyze the genetic diversity of HBV and evaluate serological marker associations and coinfections with HCV and HIV in patients attending and treated in a public hospital in the province of Buenos Aires, Argentina.
Materials and methods
Study population
A retrospective study was conducted on 189 HBV-reactive serum samples detected by ELISA (HBsAg and anti-HBc antibodies; Architect i2000; Abbott Diagnosis); 65 were reactive for HBsAg and anti-HBc and 124 for anti-HBc. The Architect HBsAg Qualitative II assay has a sensitivity of ≤ 0.13 IU/ml. The samples were collected from the Virology Laboratory at Hospital Interzonal General de Agudos "Dr. Pedro Fiorito”, Buenos Aires, Argentina, between 2015 and 2019. The samples sent to the laboratory were taken from patients with HBV serological markers of acute (n = 57), chronic (n = 8) and past (n = 124) infection admitted to the different hospital departments (Emergency, Internal Medicine, Infectology, Surgery, Gastroenterology and Hepatology). Antiviral therapy (entecavir or tenofovir) is available for HBV chronically infected patients with or without coinfections (HCV, HIV). Treatment indications in Argentina are based on the combination of three criteria: transaminases (> normal level), viral load (HBV DNA > 2000 IU/ml) and liver histology (inflammation and/or fibrosis), also taking into account the general condition of the patient and the availability of antiviral drugs. In Argentina, the individuals attending these public hospitals have very limited economic resources and receive free medical care. None of the individuals who participated in this study had been vaccinated for HBV.
Serological markers and load viral
Other HBV serological markers, HBeAg, IgM anti-HBc, anti-HBe and anti-HBs (Architect i2000; Abbott Diagnosis), were determined in all HBV reactive samples by ELISA. Moreover, HCV (total anti-HCV Ab; Abbott Architect i2000) and HIV (HIV-antigen/antibody; Abbott Architect i2000) serological markers were tested by CMIA tests, respectively. In HBV-positive samples, DNA quantification was performed by the COBAS Taq man HBV test (Real time; Roche Molecular Systems).
DNA and RNA extraction
DNA and RNA were extracted from all serum samples (n = 189) using the QIAmp DNA Mini Kit and the QIAamp Viral RNA Mini Kit (QIAGEN AG) respectively, according to the manufacturer's protocol.
HBV DNA amplification
PCR for the S/P overlapping genomic region (nucleotides [nt] 256–796) and a nested-PCR (n-PCR) for the preC/C regions (nt 1736–2471) were performed as previously described for HBV [5, 12, 16].
HCV RNA and HIV RNA amplifications
HCV and/or HIV reactive samples were amplified by n-PCR as previously described [9, 15].
Sequencing and phylogenetic analysis
PCR products HBV, HCV and HIV were sequenced directly in both directions with the corresponding amplification primers from Macrogen Inc. (Seoul, South Korea). The sequences obtained were aligned using the CLUSTALX v1.83 software. Phylogenetic trees were obtained by MEGA X using the neighbor-joining algorithm with the Kimura two-parameter model of molecular evolution.
Analysis of the regulatory regions and ORFs of HBV
The nucleotide sequences representing the basal core promoter (BCP; nt 1742–1814), the preC (nt 1814–1901) and partial S (amino acids [aa] 55–210)/P (aa 64–220) regions for HBV were translated into aa sequences according to their corresponding ORFs and compared with the consensus sequences from the corresponding genotypes and subgenotypes using BioEdit software.
Statistical analysis
Statistical tests were performed using the Student’s t-test and the Chi-square test (Epidat v3.1 software), as appropriate.
Results
Demographic features
Of the total samples analyzed, 72.5% (137/189) were from males and 27.5% (52/189) from females. The mean age ± SD of the subjects was 50 ± 14 years (range 18–70 years). Most of the individuals were Argentinians living in areas close to the hospital located in Avellaneda, Buenos Aires province. In addition, the subjects of other nationalities (Bolivians, Peruvians, Brazilians, Venezuelans and Paraguayans) were also treated. Furthermore, in the surveys most of the patients reported having engaged in risk behaviors such as the use of inhaled drugs, sexual contact without prevention measures or dangerous tattoos, among others.
Detection of HBV DNA and occult HBV infection (OBI)
One-hundred- and eighty-nine (189) HBV-reactive samples with acute (n = 57), chronic (n = 8), and past (n = 124) were analyzed (Supplemental Figure). Thirty-six exhibited detectable HBV DNA, 23 for the S/P region and 20 for the preC/C region (Table 1). Of the total number of positive samples for HBV DNA, 7 were classified as OBI (non-reactive HBsAg and viral load < 200 IU/ml; Table 1). All OBI samples only amplified in the preC / C region.
Phylogenetic analysis and HBsAg subtype
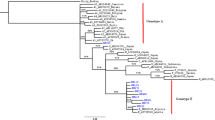
Most of the S/P region sequences from individuals belonged to genotype F (15/23), 9 to subgenotype F1b, 1 to subgenotype F3 and 5 to subgenotype F4. Of the 6 sequences containing genotype A, 2 were assigned to subgenotype A1 and 4 to subgenotype A2. Subgenotypes B2 and D3 were described in one sequence, respectively (Fig. 1). Moreover, HBsAg subtype of the S gene sequence were described in Table 1 [17].
Phylogenetic relationships of 23 HBV sequences (black frame) shown in this study were compared to representative sequences belonging to all reportedly known genotypes and subgenotypes by using the neighbor-joining method. Bootstrap statistical analysis was performed using 1000 data sets and the numbers of the nodes indicate the percentage of the number of substitutions per site (bootstrap). All sequences deposited in the GenBank are named with their corresponding accession number. This tree represents the partial S region (nt 320–775) of HBV sequences. The GenBank accession numbers of those sequences reported in this study are MT439874–MT439896
Subgenotypes in the preC/C region could not be validated in 13 sequences that only amplified for this region.
Analysis of the S and P proteins
The S sequences (n = 23) of the patients showed specific aa polymorphisms that are characteristic of genotype A, B, D and F, respectively. Of the total genotype A sequences (n = 6), 5 exhibited mutations inside and outside the main hydrophilic region, Y100C, M103I, T131N, T143M and W182Stop among them. No mutations were observed in sequence B2, whereas sequence D3 showed mutations I110L, T125M and T189I. Twelve of the total genotype F sequences did not detect aa mutations, whereas 2 subgenotype F4 sequences showed P62L, S167L and V177A mutations and one sequence exhibited L98V and S140T. In the P protein, resistance-related mutations rtS106C, rtW153R, rtL180M, rtM204V and rtV191I were observed. The remaining S and P protein variants are shown in Table 1.
Analysis of the BCP and PreC
Of the total preC/C sequences (n = 20), 11 exhibited mutation 1764, two mutation 1896 and two mutation 1899. Furthermore, C1766T, T1768A, C1773T, C1799G, A1846T, T1850A, and T1858C mutations were observed in some sequences (Table 1, MT448618-MT44863).
HCV and HIV analysis
Prevalence, molecular detection and phylogenetic analysis
Of the total samples analyzed (n = 189), 20 were anti-HCV reactive with a prevalence rate of 10.58% (20/189; 95% CI 5.9–15.2). Of the 20 reactive samples, HCV RNA from 9 samples was amplified. HCV sequences were classified as genotype 1 (103P, 116P, 128P, 141P and 175P), 2 (145P) and 3 (38P, 72P and 168P; data not shown; MT902919-MT902927; Table 2a).
A total of 19 samples were HIV Antigen/Ab reactive with an overall prevalence of 10.05% (19/189; 95% CI 5.5–16.4) and HIV RNA were detected in 11 reactive samples. All HIV sequences were classified: 2 as subtype B (165P and 222P), 1 as subtype F (204P) and BF recombinant subtype (1P, 14P, 27P, 38P, 71P, 82P, 127P, 195P; data not shown; MT902909–MT902918; Table 2a).
Serological marker associations and coinfections
Of the total samples analyzed, 20.6% (39/189; 95% CI 14.6–26.6) showed more than one serological marker of viral infection (HCV and/or HIV): two (HBV-HCV and HBV-HIV) were observed in 18.0% (34/189; 95% CI 12.2–23.7) and three (HBV-HCV-HIV) were detected in 2.7% (5/189; 95% CI 0.8–6.0; Table 2a).
Of the total amplified samples (n = 189), 56 showed some of the three viruses analyzed (HBV, HCV and HIV) and 7 exhibited coinfections. HBV–HIV and HIV–HCV coinfections were found in 6 samples (1P, 14P, 27P,71P, 204P and 222P) and 1 (38P), respectively. Sample 222P with HBV–HIV coinfection was classified as OBI. Detailed data of the genotypes, subgenotypes and subtypes associations markers are shown in Table 2b.
Discussion
The aim of this study was to analyze the genetic diversity of HBV and evaluate serological marker associations and coinfections in patients with acute, chronic and past infection attending and treated in a public hospital in the province of Buenos Aires, Argentina. These results show the frequency of OBI, HBV subgenotype distribution, HCV and HIV serological marker associations and coinfections. Mutations related to the ability to escape the host’s immune response, resistance to antiviral therapy and progression to disease were found in these patients, which may have important clinical implications.
OBI is a common condition among infected individuals belonging to endemic regions, to high-risk groups or patients with chronic liver disease [4, 18]. The causes of HBV suppression are not yet well clarified, although different factors might be associated: elapsed time and recovery from infection as a result of host immune response, low viral load, difference in sensitivity of immunoassays, formation of immune-complexes, epigenetic factors, coinfection with other infectious agents and/or variability of the HBV genome or S gene mutations [4, 18]. In this study, all OBI samples were negative in the PCR assay targeting the S-gene probably due to mutations in the S region and could not be genotyped. Moreover, OBI may impact several different clinical contexts, including the possible transmission of the infection, the risk of reactivation, the contribution to liver disease progression and the development of HCC [4]. In this regard, 3 out of 7 OBI case studies showed HCV (215P and 216P) and HIV (222P) infection. Prospective studies and meta-analyses have reported a higher incidence of HCC in patients with HCV and OBI compared with patients without OBI [19]. This status may play a synergistic role in the occurrence of HCC in HCV coinfected patients, especially in patients with advanced fibrosis and cirrhosis [19]. In HIV-positive subjects, the OBI condition has been associated with a low number of CD4-positive lymphocytes, elevation of alanine aminotransferase and more frequent AIDS-defining illnesses [20]. However, there is a lack of studies to better evaluate HBV infection progression and liver disease evolution in HIV-positive patients with OBI [20]. On the other hand, OBI status was unknown in all the patients because the diagnosis and follow-up of OBI in Argentina are not routinely performed in “anti-HBc only” individuals.
HBV genotype F is the most frequent genotype in South America. Nevertheless, global human migrations affect the pattern of genotype distribution, introducing genotypes and subgenotypes differing from those circulating in the original inhabitants [21]. In this work, A, B, D and F genotypes were documented, while subgenotypes F1b and F4 were the most common in this population (Table 3). Other studies have demonstrated the previous circulation in Argentina and border countries of the subgenotypes found in hospital patients, with the exception of subgenotype F3 that was detected for the first time in our country [Table 3; 5–14, 21–27]. The presence of this subgenotype is due to the recent migrations of individuals from Venezuela and Colombia looking for better job, health and educational opportunities. or undergraduate, graduate and postgraduate university programs [21]. Furthermore, the observed HBsAg subtype was consistent with the phylogenetic analysis [17].
The selection pressure of HBV by host immunity and/or antiviral therapies can generate strains with amino acid variants in or around the determinant a (det a) of HBsAg, the main target of neutralizing Ab. Amino acid mutations have been observed in the deduced sequences of HBsAg, among which Y100C, M103I, T131N, T143M and W182Stop stand out for genotype A, I110L and T125M for genotype D and L98V for genotype F. The Y100C mutation (44P and 84P) has been associated with OBI individuals, although in vitro studies demonstrate that it alone does not reduce the amounts of HBsAg or the affinity of HBsAg for ELISA assays, as well as both samples analyzed in this study [28]. M103I (49P) and T131N (84P) mutations have been detected together with other variants of the S region, hindering the recognition of HBsAg by the humoral immune response and favoring its escape [29, 30]. Mutation T131N was found even in children with HBV prophylaxis [30]. The T143M variant (84P) was documented to decrease the sensitivity of ELISA assays; in turn it was observed by protein modeling that it alters the antigenicity of HBsAg [31, 32]. The W182Stop found in two chronic patients (71P and 185P) generates the defective secretion of HBsAg. This mutation has carcinogenic properties and was found to induce apoptosis in vitro, exacerbating the progression of the disease [33, 34]. With regard to genotype D, mutation T125M has been previously described in this genotype in patients monoinfected with HBV and coinfected with HBV/HIV [35]. This mutation is found in sample 204P that has co-circulation of HBsAg/anti-HBs, showing evasion of the humoral immune response. Furthermore, the L98V variant (36P) was observed in genotype F in patients with chronic HBV infection or HBV reactivation [36, 37].
In the P protein, several mutations were related to antiviral therapy resistance. Mutation rtS106C (36P; acute infection) was found to be associated with resistance to tenofovir, although it has been observed in naïve patients with necroinflammation and an increased development of cirrhosis [38]. The rtW153R mutation (84P; chronic infection without therapy) is frequent in genotype A and was described in association with adefovir resistance, while rtV191I - W182Stop in ORF S-occurs more frequently in individuals with chronic HBV (71P and 158P; chronic infection) [33, 34, 39]. Mutation rtV191I/sW182* is resistant to lamivudine, and remains sensitive to adefovir and tenofovir [33]. The 71P (HCV and HIV coinfection) and 185P patients received tenofovir and entecavir antiviral therapy, respectively. However, the detectable viral load in them was related to low adherence to therapy, probably due to their socioeconomic status and addiction to inhaled drugs. Mutations rtL180M and rtM204V—in the YMDD motif of viral polymerase—are responsible for resistance to lamivudine, telbivudine, entecavir and clevudine [33, 40]. Resistance to entecavir was observed in the 49P patient and the addition of adefovir or tenofovir to the therapy was recommended (Ministry of Health of Argentina recommendations).
On the other hand, BCP and preC/C mutations have been associated with significant virological or clinical events, such as the failure to form a nucleocapsid, liver disease progression or HBeAg seroconversion [41]. In particular, mutations G1896A, A1762T, G1764A and the A1762T/G1764A may prevent the production of HBeAg by introducing a premature stop codon into the ORF or may increase the transcription of pregenomic ribonucleic acid by removing of the nuclear receptor-binding motif, contributing to an inefficient immune response that ultimately leads to hepatocarcinogenesis. [42]. A meta-analysis study revealed that the G1896A (n = 2) and G1764A (n = 11) mutations observed in this work, correlate with a statistically significant increase in the risk of HCC, even this latter mutation alone plays a significant role similar to A1762T and A1762T/G1764 [43]. This may indicate that any one site of mutation of A1762T or G1764A constitutes a danger signal [43]. Moreover, other mutations such as C1766T, A1846T and G1899A are correlated with an increased risk of acute chronic liver failure [44].
Of the total samples analyzed, 20% showed associations with HCV and/or HIV serological markers and 7 exhibited coinfections, highlighting the importance of coinfection detection in HBV-reactive samples. With regard to the genomic characterization of HCV and HIV in HBV-reactive samples, our study allowed to determine the presence of different genotypes and OBI in coinfections in this population [9, 15].
In conclusion, this study shows the presence of OBI in patients with and without HCV and HIV coinfection. This is important because OBI screening or “anti-HBc only” patient follow-up is not routinely performed in the hospital. Furthermore, HCV and / or HIV serological markers and genotype-subtype associations were found in subjects with past and active HBV infection. Drug-resistant in naïve and treated patients together with immune-escape HBV mutants have important clinical implications in the vulnerable population. Finally, planned prevention, detection and treatment adherence are needed to reduce transmission and morbidity in this vulnerable population.
References
Yuen MF, Chen DS, Dusheiko GM, Janssen HLA, Lau DTY, Locarnini SA, Peters MG, Lai CL (2008) Hepatitis B virus infection. Nat Rev Dis Primers 4:18035. https://doi.org/10.1038/nrdp.2018.35
Mathet V, Cuestas ML, Trinks J, Mathet V, Cuestas ML, Trinks J, Minassian ML, Ruiz V, Rivero CW, Andreetta AM, Weissenbacher MC, Oubiña JR (2007) Chapter X: genetic diversity and variability of Hepatitis B virus (HBV) in Latin America and the Caribbean region: implications in epidemiological, clinical, diagnostic, prophylactic and therapeutic approaches. In: Denyer DV (ed) Progress in hepatitis B. Nova Science Publishers Inc, New York, pp 277–351
Roman S, Jose-Abrego A, Fierro NA, Escobedo-Melendez G, Ojeda-Granados C, Martinez-Lopez E, Panduro A (2014) Hepatitis B virus infection in Latin America: a genomic medicine approach. World J Gastroenterol 20(23):7181–7196. https://doi.org/10.3748/wjg.v20.i23.7181
Raimondo G, Locarnini S, Pollicino T, Levrero M, Zoulim F, Lok AS; Taormina Workshop on Occult HBV Infection Faculty Members (2019) Update of the statements on biology and clinical impact of occult hepatitis B virus infection. J Hepatol 71(2):397–408. https://doi.org/10.1016/j.jhep.2019.03.034
Delfino CM, Gentile EA, Castillo AI, Cuestas ML, Pataccini G, Cánepa C, Malan R, Blejer J, Berini C, Eirin ME, Pedrozo W, Oubiña JR, Biglione MM, Mathet VL (2014) Hepatitis B virus and hepatitis D virus in blood donors from Argentina: circulation of HBsAg and reverse transcriptase mutants. Arch Virol 159(5):1109–1117. https://doi.org/10.1007/s00705-013-1917-y
Pisano MB, Blanco S, Carrizo H, Ré VE, Gallego S (2016) Hepatitis B virus infection in blood donors in Argentina: prevalence of infection, genotype distribution and frequency of occult HBV infection. Arch Virol 161(10):2813–2817. https://doi.org/10.1007/s00705-016-2960-2
Cassino L, Laufer N, Salomon H, Campos R, Quarleri J (2009) Hepatitis B precore/core promoter mutations in isolates from HBV-monoinfected and HBV-HIV coinfected patients: a 3-yr prospective study. J Clin Virol 46(4):354–359. https://doi.org/10.1016/j.jcv.2009.09.015
González López Ledesma MM, Mojsiejczuk LN, Rodrigo B, Sevic I, Mammana L, Galdame O, Gadano A, Fainboim H, Campos R, Flichman D (2015) Hepatitis B virus genotype distribution and genotype-specific BCP/preCore substitutions in acute and chronic infections in Argentina. PLoS ONE 10(3):e0121436. https://doi.org/10.1371/journal.pone.0121436
Carobene M, Bolcic F, Farías MS, Quarleri J, Avila MM (2014) HIV, HBV, and HCV molecular epidemiology among trans (transvestites, transsexuals, and transgender) sex workers in Argentina. J Med Virol 86(1):64–70. https://doi.org/10.1002/jmv.23805
Piñeiro Y, Leone FG, Pezzano SC, Torres C, Rodríguez CE, Eugenia Garay M, Fainboim HA, Remondegui C, Sorrentino AP, Mbayed VA, Campos RH (2008) Hepatitis B virus genetic diversity in Argentina: dissimilar genotype distribution in two different geographical regions; description of hepatitis B surface antigen variants. J Clin Virol 42(4):381–388. https://doi.org/10.1016/j.jcv.2008.01.018
Villar LM, de Paula VS, do Lago BV, Miguel JC, Cruz HM, Portilho MM, Marques VA, Ravier RP, Lo Castro I, Cuello H, Espul C (2020) Epidemiology of hepatitis B and C virus infection in Central West Argentina. Arch Virol 165(4):913–922. https://doi.org/10.1007/s00705-020-04540-7
Delfino CM, Berini C, Eirin ME, Malan R, Pedrozo W, Krupp R, Blejer J, Espejo R, Fierro L, Puca A, Oubiña JR, Mathet VL, Biglione MM (2012) New natural variants of hepatitis B virus among Amerindians from Argentina with mainly occult infections. J Clin Virol 54(2):174–179. https://doi.org/10.1016/j.jcv.2012.02.023
Mojsiejczuk LN, Torres C, Sevic I, Badano I, Malan R, Flichman DM, Liotta DJ, Campos RH (2016) Molecular epidemiology of hepatitis B virus in Misiones. Argentina Infect Genet Evol 44:34–42. https://doi.org/10.1016/j.meegid.2016.06.032
Gallego F, Pisano MB, Torres C, Caeiro L, Martínez Wassaf M, Balangero M, Campos R, Ré V (2014) Molecular epidemiology of hepatitis B virus in Córdoba. Argentina J Clin Virol 61(2):204–210. https://doi.org/10.1016/j.jcv.2014.06.030
Quarleri J, Moretti F, Bouzas MB, Laufer N, Carrillo MG, Giuliano SF, Pérez H, Cahn P, Salomon H (2007) Hepatitis B virus genotype distribution and its lamivudine-resistant mutants in HIV-coinfected patients with chronic and occult hepatitis B. AIDS Res Hum Retroviruses 23(4):525–531. https://doi.org/10.1089/aid.2006.0172
Birkenmeyer L, Mushahwar I (1994) Detection of hepatitis A, B and D virus by the polymerase chain reaction. J Virol Methods 49:101–112. https://doi.org/10.1016/0166-0934(94)90035-3
Gerlich WH, Glebe D, Kramvis A, Magnius LO (2020) Peculiarities in the designations of hepatitis B virus genes, their products, and their antigenic specificities: a potential source of misunderstandings. Virus Genes 56(2):109–119. https://doi.org/10.1007/s11262-020-01733-9
Makvandi M (2016) Update on occult hepatitis B virus infection. World J Gastroenterol 22(39):8720–8734. https://doi.org/10.3748/wjg.v22.i39.8720
Mak LY, Wong DK, Pollicino T, Raimondo G, Hollinger FB, Yuen MF (2020) Occult hepatitis B infection and hepatocellular carcinoma: Epidemiology, virology, hepatocarcinogenesis and clinical significance. J Hepatol 73(4):952–964. https://doi.org/10.1016/j.jhep.2020.05.042
Sarmati L, Malagnino V (2019) HBV Infection in HIV-Driven Immune Suppression. Viruses 11(11):1077. https://doi.org/10.3390/v11111077
Alvarado-Mora MV, Pinho JR (2013) Distribution of HBV genotypes in Latin America. Antivir Ther 18(3 Pt B):459–465. https://doi.org/10.3851/IMP2599
Mojsiejczuk L, Elizalde MM, López G, Figueredo D, Marquez N, Campos RH, Flichman D (2019) Molecular epidemiology of hepatitis B virus in Paraguay. Infect Genet Evol 71:91–97. https://doi.org/10.1016/j.meegid.2019.03.020
Di Lello FA, Piñeiro Y, Leone FG, Muñoz G, Campos RH (2009) Diversity of hepatitis B and C viruses in Chile. J Med Virol 81(11):1887–1894. https://doi.org/10.1002/jmv.21607
Lopez L, Flichman D, Mojsiejczuk L, Gonzalez MV, Uriarte R, Campos R, Cristina J, Garcia-Aguirre L (2015) Genetic variability of hepatitis B virus in Uruguay: D/F. A/F genotype recombinants Arch Virol 160(9):2209–2217. https://doi.org/10.1007/s00705-015-2477-0
Khan A, Tanaka Y, Saito H, Ebinuma H, Sekiguchi H, Iwama H, Wakabayashi G, Kamiya T, Kurbanov F, Elkady A, Mizokami M (2008) Transmission of hepatitis B virus (HBV) genotypes among Japanese immigrants and natives in Bolivia. Virus Res 132(1–2):174–180. https://doi.org/10.1016/j.virusres.2007.12.005
Paoli J, Wortmann AC, Klein MG, Pereira VRZB, Cirolini AM, Godoy BA, Fagundes NJR, Wolf JM, Lunge VR, Simon D (2018) HBV epidemiology and genetic diversity in an area of high prevalence of hepatitis B in southern Brazil. Braz J Infect Dis 22(4):294–304. https://doi.org/10.1016/j.bjid.2018.06.006
Lago BV, de Espirito-Santo MP, Costa VD, Marques VA, Villar LM, Lewis-Ximenez LL, Lampe E, Mello FCA (2019) Genetic diversity of the Hepatitis B Virus Subgenotypes in Brazil. Viruses 11(9):860. https://doi.org/10.3390/v11090860
Mello FC, Martel N, Gomes SA, Araujo NM (2011) Expression of Hepatitis B virus surface antigen containing Y100C variant frequently detected in occult HBV infection. Hepat Res Treat. https://doi.org/10.1155/2011/695859
Salpini R, Colagrossi L, Bellocchi MC, Surdo M, Becker C, Alteri C, Aragri M, Ricciardi A, Armenia D, Pollicita M, Di Santo F, Carioti L, Louzoun Y, Mastroianni CM, Lichtner M, Paoloni M, Esposito M, D’Amore C, Marrone A, Marignani M, Sarrecchia C, Sarmati L, Andreoni M, Angelico M, Verheyen J, Perno CF, Svicher V (2015) Hepatitis B surface antigen genetic elements critical for immune escape correlate with hepatitis B virus reactivation upon immunosuppression. Hepatology 61(3):823–833. https://doi.org/10.1002/hep.27604
Lin YM, Jow GM, Mu SC, Chen BF (2013) Naturally occurring hepatitis B virus B-cell and T-cell epitope mutants in hepatitis B vaccinated children. Sci World J. https://doi.org/10.1155/2013/571875
Araujo NM, Vianna CO, Moraes MT, Gomes SA (2009) Expression of Hepatitis B virus surface antigen (HBsAg) from genotypes A, D and F and influence of amino acid variations related or not to genotypes on HBsAg detection. Braz J Infect Dis 13(4):266–271. https://doi.org/10.1590/s1413-86702009000400005
Ie SI, Thedja MD, Roni M, Muljono DH (2010) Prediction of conformational changes by single mutation in the hepatitis B virus surface antigen (HBsAg) identified in HBsAg-negative blood donors. Virol J 7:326. https://doi.org/10.1186/1743-422X-7-326
Wang ML, Tang H (2016) Nucleos(t)ide analogues causes HBV S gene mutations and carcinogenesis. Hepatobiliary Pancreat Dis Int 15(6):579–586. https://doi.org/10.1016/s1499-3872(16)60064-4
Colledge D, Soppe S, Yuen L, Selleck L, Walsh R, Locarnini S, Warner N (2007) Stop codons in the hepatitis B surface proteins are enriched during antiviral therapy and are associated with host cell apoptosis. Virology 501:70–78. https://doi.org/10.1016/j.virol.2016.11.007
Pourkarim MR, Amini-Bavil-Olyaee S, Verbeeck J, Lemey P, Zeller M, Rahman M, Maes P, Nevens F, Van Ranst M (2010) Molecular evolutionary analysis and mutational pattern of full-length genomes of hepatitis B virus isolated from Belgian patients with different clinical manifestations. J Med Virol 82(3):379–389. https://doi.org/10.1002/jmv.21726
Inuzuka T, Ueda Y, Arasawa S, Takeda H, Matsumoto T, Osaki Y, Uemoto S, Seno H, Marusawa H (2018) Expansion of viral variants associated with immune escape and impaired virion secretion in patients with HBV reactivation after resolved infection. Sci Rep 8(1):18070. https://doi.org/10.1038/s41598-018-36093-w
Xiang KH, Michailidis E, Ding H, Peng YQ, Su MZ, Li Y, Liu XE, Dao Thi VL, Wu XF, Schneider WM, Rice CM, Zhuang H, Li T (2017) Effects of amino acid substitutions in hepatitis B virus surface protein on virion secretion, antigenicity. HBsAg and viral DNA J Hepatol 66(2):288–296. https://doi.org/10.1016/j.jhep.2016.09.005
Park ES, Lee AR, Kim DH, Lee JH, Yoo JJ, Ahn SH, Sim H, Park S, Kang HS, Won J, Ha YN, Shin GC, Kwon SY, Park YK, Choi BS, Lee YB, Jeong N, An Y, Ju YS, Yu SJ, Chae HB, Yu KS, Kim YJ, Yoon JH, Zoulim F, Kim KH (2019) Identification of a quadruple mutation that confers tenofovir resistance in chronic hepatitis B patients. J Hepatol 70(6):1093–1102. https://doi.org/10.1016/j.jhep.2019.02.006
Mirandola S, Sebastiani G, Rossi C, Velo E, Erne EM, Vario A, Tempesta D, Romualdi C, Campagnolo D, Alberti A (2012) Genotype-specific mutations in the polymerase gene of hepatitis B virus potentially associated with resistance to oral antiviral therapy. Antiviral Res 96(3):422–429. https://doi.org/10.1016/j.antiviral.2012.09.014
Menéndez-Arias L, Álvarez M, Pacheco B (2014) Nucleoside/nucleotide analog inhibitors of hepatitis B virus polymerase: mechanism of action and resistance. Curr Opin Virol 8:1–9. https://doi.org/10.1016/j.coviro.2014.04.005
Kim H, Lee SA, Do SY, Kim BJ (2016) Precore/core region mutations of hepatitis B virus related to clinical severity. World J Gastroenterol 22(17):4287–4296. https://doi.org/10.3748/wjg.v22.i17.4287
Dong Q, Chan HL, Liu Z, Chan DP, Zhang B, Chen Y, Kung HF, Sung JJ, He ML (2008) A1762T/G1764A mutations of hepatitis B virus, associated with the increased risk of hepatocellular carcinoma, reduce basal core promoter activities. Biochem Biophys Res Commun 374(4):773–776. https://doi.org/10.1016/j.bbrc.2008.07.115
Wei F, Zheng Q, Li M, Wu M (2017) The association between hepatitis B mutants and hepatocellular carcinoma: a meta-analysis. Medicine (Baltimore) 96(19):e6835. https://doi.org/10.1097/MD.0000000000006835
Hu F, Bi S, Yan H, Shi Y, Sheng J (2015) Associations between hepatitis B virus basal core promoter/pre-core region mutations and the risk of acute-on-chronic liver failure: a meta-analysis. Virol J 12:87. https://doi.org/10.1186/s12985-015-0313-5
Acknowledgements
We would like to thank all the staff at the Virology Laboratory of Hospital Interzonal General de Agudos "Dr. Pedro Fiorito”, as well as Lucas Zapata for his collaboration and participation in this study.
Funding
This work was supported by Agencia Nacional de Promoción Científica y Tecnológica, grant number PICT 2016–0698.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Ethics approval
This study was reviewed by the Bioethics Committee of “Fundación Huésped” and conducted in compliance with all federal regulations governing the protection of human subjects.
Informed consent
All participants 18 years of age or older were required to sign an informed consent.
Additional information
Edited by Wolfram H. Gerlich.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Delfino, C.M., Giorgio, M., García, G. et al. Drug-resistant and immune-escape hepatitis B virus mutants, occult hepatitis B infection and coinfections in public hospital patients from Argentina. Virus Genes 57, 327–337 (2021). https://doi.org/10.1007/s11262-021-01850-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-021-01850-z