Abstract
A novel strain of H3N8 influenza virus was isolated from domestic pigeons during the avian influenza virus (AIV) surveillance in wet markets in Anhui, China, during 2013. The virus was characterized by whole-genome sequencing with subsequent genetic comparison and phylogenetic analysis. Phylogenetic analysis revealed that the NA gene of AIV mapped to the North American lineage, and the remaining seven genes belong to a Eurasian lineage. These findings indicated that this H3N8 virus is a novel nature reassortant virus. Comparison of the hemagglutinin amino acid sequences indicated 9 substitutions. One substitution caused the loss of a potential glycosylation site, and six substitutions were not previously observed in avian H3 isolates. Q226 and T228 at the receptor binding sites suggested that Anhui-08 preferentially binds to a-2,3-linked sialic acid receptors, and the cleavage site sequence showed a low pathogenic feature. Animal experiments further confirmed that A/pigeon/Anhui/08/2013 (H3N8) is low or in pigeons. The results improve our understanding of these viruses as they evolve and also provide important information to aid ongoing risk assessment analyses because these zoonotic influenza viruses continue to circulate and adapt to new hosts.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wet markets (live-animal markets) play a critical role in the epidemiology of avian influenza viruses (AIVs). Wet markets are also considered the pathways for disease transmission and offer conditions for virus amplification, reassortment, and cross-species transmission because wet markets sell live poultry, fish, reptiles, and various mammals at a high density [1]. Pigeons, also often sold in wet markets in East Asian countries, possess both human-like a-2,6-linked sialic acids receptors and avian-like a-2,3-linked sialic acids receptors in the airway epithelial cells [2].
Although pigeons serve as an unnoticed reservoir for the transmission of influenza viruses, they carry the virus over great distances and excrete it in their droppings [3]. However, pigeons display only a mild and short-lived illness themselves [4]. Pigeons carrying AIV may be a source of infection to other animals, even including humans [3]. Several IV subtypes are isolated from pigeons, including H9N2, H3N3, H7N7, H5N1, H1N1, H3N6, and H7N9. Among different strains of IV, H5N1, and H7N9, which infect humans, are the deadliest.
H3N8, which was first isolated in Miami in 1963, is the major cause of equine influenza. Equine influenza viruses are responsible for rapidly spreading outbreaks of respiratory disease in horses [5]. Since then, H3N8 has broken the species barrier and is transmitted to other species, including donkey, dog, aquatic bird, swine, and cat. A recent outbreak in New England was recorded with seal infections caused by an avian H3N8 influenza virus, resulting in 162 seal deaths [6]. Strikingly, this virus displays increased affinity for mammalian receptors. More importantly, analysis of a panel of human sera for H3N8 neutralizing antibodies indicates that there is no population-wide immunity to this H3N8 [7]. Therefore, this virus may pose a potential threat to human beings. Moreover, the Hong Kong H3N2 AIV possesses HA and PB1 gene cassettes recruited directly from an duck-origin H3N8 virus [8, 9]. Approximately 33,800 people died from the “Hong Kong influenza” in the United States alone in 1968 [10].
During the routine surveillance for avian influenza in wet markets in Anhui Province, Central China, in August 2013, an H3N8 AIV, A/Pigeon/Anhui/08/2013(H3N8) (Anhui-08), was isolated from an apparently healthy domestic pigeon. All gene segments of Anhui-08 were sequenced and compared with reference sequences available in GenBank. The results indicated that the Anhui-08 isolated in our study is a novel reassortant genotype virus in pigeons. Therefore, strengthening the epidemiological monitoring of AIVs in wet markets is beneficial for a better understanding of cross-species transmission and the continuous evolution of AIVs.
Materials and methods
Sample collection and virus isolation
In April 2013, a routine epidemiological investigation of AIV was performed in domestic poultry in Anhui wet markets. In total, 2000 oropharyngeal and cloacal swabs of ducks, chickens, and pigeons were collected and stored in PBS supplemented with penicillin (2000 U/ml) and streptomycin (2000 U/ml). Virus isolation was conducted after filtering through a 0.22-μm syringe-driven filter. The supernatants were then inoculated into the allantoic cavities of 10-day-old specific-pathogen-free embryonated chicken eggs and, cultured in a 37 °C incubator. The embryos were checked daily, and the allantoic fluid was collected after 3–4 days post-inoculation. The harvested allantoic fluid was frozen at −80 °C for RNA extraction and genome amplification.
RNA extraction and RT-PCR
Viral RNA was extracted from virus-infected allantoic fluid with the Transgen viral RNA mini kit (Transgen, China) according to the manufacturer’s instructions. cDNAs were synthesized from viral RNAs by reverse transcription with the Uni12 primer, and were amplified by polymerase chain reaction with influenza gene-specific universal primers as described previously [11].
DNA cloning
PCR products of each RT-PCR were purified using the AxyPrepTM DNA Gel Extraction Kit (AXYGEN). Purified PCR products ligated into a TA-cloning vector, pEGMT (Promega, USA), were transformed into DH5a E. coli-competent cells. Cells harboring recombinant plasmid were screened on Luria–Bertani (LB) agar plates containing ampicillin (100 μg/ml). Next, the inserts were amplified by gene-specific primers at the same condition described above. The confirmed recombinant DNA plasmids were used for sequencing.
Gene sequencing and analysis
The purified recombinant plasmids were sequenced by Sangon Biotech Life Science Production & Services (Shanghai). Translation of the nucleotide sequence and sequence alignments were performed using the DNA Star software (DNASTAR Inc.). Phylogenetic analysis was performed using the neighbor-joining method by MEGA (version 5.1) software. The bootstrap values were determined from 1000 replicates of the original data. Glycosylation sites and transmembrane segments of the HA protein were identified using Predict Protein (http://www.cbs.dtu.dk/services/NetNGlyc//).
Nucleotide sequence accession numbers
Full-genome sequence data for this virus, named A/Pigeon/Anhui/08/2013 (H3N8), were submitted to GenBank and are available under accession numbers KJ579956 to KJ579963.
Animal study
To evaluate the pathogenicity and replication of the Anhui-08 virus in pigeons, two groups (20 per group) were inoculated intranasally with high concentrations (106 EID50) of the H3N8 or mock infected with PBS with a volume of 0.1 ml. The inoculated pigeons were monitored daily for morbidity and mortality up to day 14 post-infection (pi). Tracheal and cloacal swabs were collected on days 3, 5, and 7 pi for virus titration. Virus titration was conducted in 10-day-old SPF embryonated chicken eggs, and calculated using the method of Reed and Muench. Sera were harvested from the inoculated pigeons on day 21 pi for seroconversion confirmation using hemagglutination inhibition (HI) assays according to the OIE standard.
Hemagglutinin-inhibition test
Sera obtained from pigeons in the infected and control groups were pretreated with receptor destroying enzyme (Sigma, USA) at 37 °C overnight and then inactivated for 30 min at 56 °C. They were then processed for the detection of H3-specific antibodies using the hemagglutinin-inhibition test with 1 % chicken red blood cells and the Anhui-08 virus as an antigen.
Results
Virus isolation and homology comparison
Total 94 strains influenza viruses were isolated from pigeons at the wet markets in Anhui, An H3N8 AIV [A/Pigeon/Anhui/08/2013 (H3N8)] included. The entire genome of the isolated virus was sequenced, and BLAST analysis was performed on each sequence to identify the related reference viruses available in GenBank. The genetic analysis showed that the isolated multiple gene segments of the virus shared high homology with viruses isolated from waterfowls in the Eurasian region. As shown in Table 1, BLAST analysis revealed that the HA gene of Anhui-08 had 94 % nucleotide identity with A/duck/Siberia/100/2001(H3N8). The NA gene of Anhui-08 shared 99 % nucleotide sequence homology with the isolate A/environment/Hunan/S4350/2011(H3N8). The nucleotide homologies of the internal genes were closely related to other IV subtypes isolated from Eurasian ducks, chickens, and wild birds, with similarities reaching greater than 93 % (Table 1).
Phylogenetic analysis
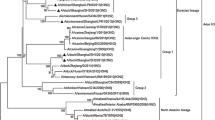
To better understand the evolutionary relationships of Anhui-08 in detail, phylogenetic analyses of each segment were conducted. In a previous study, the HA genes of H3 subtype viruses were phylogenetically divided into North American and Eurasian lineages [12, 13]. Different genes belonging to various lineages were further divided into different clades based on gene evolution distance. The Eurasian lineage was divided into four groups, namely A, B, C, and D (Fig. 2a). The HA of Anhui-08 isolate belongs to group B, which is different from the represented strains A/duck/Beijing/61/05(H3N8), A/equine/Jilin/189(H3N8), and the majority of H3 AIVs isolated from China and Japan (Fig. 1).
Phylogenetic tree of the HA, NA, PB2, PB1, PA, NP, M, and NS genes of the H3N8 influenza viruses. The trees were generated using MEGA5.1 software, and the bootstrap value was tested by 1000 replications. The Anhui-08 virus in the present study is highlighted with red. The scale bar represents the distance unit between sequence pairs
Similar to HA, the NA phylogenetic tree was also divided into North American and Eurasian lineages. The NA gene of the North American lineage was further categorized into clades A, B, and C (Figs. 1 and 2a). The NA of Anhui-08 was located in the North American lineage at clade A. All of the internal genes of Anhui-08 were mapped to various clades within the Eurasian lineage (Figs. 1 and 2b). These results demonstrate that the Anhui-08 is a novel reassortant between North American and Eurasian lineages, with a different origin for the internal gene segments.
The clades designation diagram of the 8 gene segments is based on the results from the phylogenetic analysis of H3N8 viruses. The solid rectangle is Eurasian lineage and the hollow rectangle indicated North American lineage. The different clades were marked in different colors: red rectangle is clade A, yellow one is clade B, blue one is clade C, and green one is clade D. a The eight gene segments were PB2, PB1, PA, HA, NP, NA, M, and NS, respectively. b Abbreviations Anhui-08 A/Pigeon/Anhui/08 H3N8, A/Ck/VN/G14 A/chicken/Vietnam/G14/2008 H3N8, A/dk/VN/G119 A/duck/Vietnam/G119/2006 H3N8, A/dk/NC/1681 A/duck/Nanchang/1681/1992 H3N8
Molecular analysis
In this study, the amino acid sequence motif at the cleavage site of the Anhui-08 HA gene was PEKQTR/GL, with low pathogenic AIV characteristics similar to other H3 subtypes. The amino acid sequences of HA were aligned to identify possible determinants within HA that may be related to the cross the host transmission of H3N8 to pigeons. Six amino acid mutations differentiated the HA amino acid sequences from the reference viruses (Table 2). Anhui-08 and the reference viruses are as follows: P40T, V74I, A81T, P144T, G153N, N161S, D338N, I445V, and N503D. Among these charges, P40T, V74I, A81T, D338N, I445V, and N503D have never been previously reported in avian H3N8 isolates. Five potential N-glycosylation sites (PGS) of the HA proteins are located at positions 24, 54, 70, 181, 301, and 499 for the reference isolates. The isolate Anhui-08 had an amino acid substitution of T70P that removed a PGS near the head region of the HA molecule (Fig. 3). This mutation was not observed in any Eurasian or North American isolates. Q226 and T228 at the receptor binding sites of the H3N8 were similar to all contemporary H3 AIVs, suggesting that Anhui-08 preferentially binds to a-2,3-linked sialic acid receptors, which are predominant in avian species.
Five amino acid changes differentiated the NA amino acid sequences of Anhui-08 from the contemporary H3 AIVs (Table 2). There are D215N, N337S, S464P, E467D, and one amino acid deletion at position 464 in the NA protein. Six potential PGS of NA proteins are located at positions 46, 54, 84, 144, 293, and 398, which are similar to the H3N8 reference isolates.
In addition, an amino acid analysis of the internal gene segment of the Anhui-08 isolate compared to the closest reference viruses revealed minor mutations as follows: two sites in PB2, none in PB1, two sites in PA, none in NP, none in M1, seven sites in M2, two sites in NS1, and none in NS2. No amino acid substitutions were detected in key sites that are related to host tropism and virus replication capacity, such as E627K and D701N in the PB2 protein, Y436H in the PB1 protein, and T515A in the PA protein (Table 1).
Lethality, viral shedding, and distribution in pigeons challenged with Anhui-08
Two groups of pigeon (20 animals per group) were intranasally inoculated with 106 EID50 of the H3N8 or mock infected with PBS at a volume of 0.1 mL to determine the pathogenicity and replication of the Anhui-08 virus in pigeons. Five pigeons in each group were euthanized on days 3, 5, and 7 post-challenge. Organs, including the lungs, spleen, brain, and kidney, were collected for virus titration in eggs. Viral titers in the trachea and cloacal swabs were measured, and swabs were collected from the remaining five pigeons in each group on days 3, 5, 7, and 14 pi. These pigeons were also used to observe clinical signs of disease, such as decreased activity and neurological signs for 14 days.
Throughout the 14-d observation period, all pigeons remained clinically healthy, and no gross lesions were observed in any of the two infected groups. Therefore, our results demonstrated that the virus was weak and nonpathogenic to pigeons.
On day 3 pi, Anhui-08 was recovered from multiple organs, including the spleen, kidney, and lung, from one to two pigeons with titers ranging from 100.98 to 101.75 EID50/g (Table 3). On day 5 pi, Anhui-08 replicated in the lung, brain, spleen, and kidney more efficiently than on day 3 pi, with titers ranging from 101.5 EID50/g to 102.75 EID50/g in one to three pigeons (Table 3). However, the titers quickly dropped. Anhui-08 was only recovered from the lung and spleen with titers of 100.98 EID50/g and 101.5 EID50/g, respectively, in one pigeon on day 7 pi. Additionally, the virus was not detected in the brain of pigeons at different time points. No viral shedding was detected in the tracheal and cloacal swabs among the infected pigeons on days 3 and 7 pi. On day 5 pi, two pigeons shed Anhui-08 through tracheal swabs, with titers of 101.75 EID50/ml and 101.25 EID50/ml (Table 3). However, all infections terminated in a self-limited manner, as suggested by the negative virus isolation results at 14 pi (data not shown). Notably, two of five pigeons seroconverted with titers of 23 and 24 (Table 3). Therefore, Anhui-08 can establish infections in pigeons and then spread to multiple extra-pulmonary organs but cannot efficiently replicate in these organs.
Discussion
H3N8 influenza viruses are increasingly detected in wild and domestic ducks. Available information indicates that these viruses are generated by reassortment via the poultry trade or wild bird migration [13]. It has long been considered that pigeons are resistant to influenza virus, although earlier reports described susceptibility to the hemagglutinin subtype H7 and more recently, the Asian H5N1 highly pathogenic avian influenza infection and prolonged virus shedding [3, 14]. However, in this study, H3N8 virus was isolated from domestic pigeons in wet markets in Anhui province in August 2013, and the virus was then sequenced and characterized. To the best of our knowledge, this study is the first to provide evidence that the IV subtype H3N8 can be isolated from pigeons. Genetic and phylogenetic analyses showed that the entire genome sequences of the Anhui-08 virus shared high homology with the viruses isolated from domestic ducks and wild waterfowl. Apparently, the Anhui-08 virus remains of avian origin but with a novel genotype.
Wet markets aid in the epidemiology of AIVs [4]. Since the late 1970s, abundant and diverse AIVs have been detected in wet markets. Poultries do not typically exhibit any symptoms after being infected with IV, but they can continuously shed the virus. Investigations highlighted the diversity and abundance of low pathogenic AIVs in East Asian wet markets [15, 16]. Evidently, a novel reasserting avian-origin influenza A (H7N9) virus has been isolated from wet markets [17].
HA and NA are major glycoproteins on the surface of the influenza virus and play critical roles in virus pathogenicity and host tropism [18]. For the HA protein, the amino acids of Q226 and T228 suggested that Anhui-08 receptor binding sites are still avian-like and the amino acid sequence analysis did not reveal any amino acid alterations located in cleavage region. However, the T70P substitution removed the glycosylation site at position 70. Interestingly, HA also contained five conserved amino acid substitutions, which distinguished them from contemporary H3 isolates. The NA protein had four conserved amino acid substitutions and one amino acid deletion in the tail region was not previously observed in avian species. However, whether Anhui-08 obtained from these characterizations accounts for the cross-host barrier for the virus to pigeons remains unknown.
Results from the animal study suggested that Anhui-08 showed low pathogenicity to pigeons. This finding was consistent with the molecular features of the HA gene, which lacks a multibasic HA cleavage motif. In addition, pigeons did not develop any signs of the disease. Only short-term viral shedding was detected in their tissues and swabs, which indicated that the H3N8 virus has not yet completely adapted to pigeons.
However, seroconversion of several pigeons suggested that Anhui-08 did in fact establish an infection.
A previously study showed that H5N1 virus infection occurred in a domestic cat infected after eating a pigeon carcass [19]. Therefore, we hypothesize that pigeons may assist in cross-species transmission and the evolution of IV. The capacity of the Anhui-08 to infect other animal species has not been investigated; therefore, its threat to other animal species cannot be determined. However, the frequently close contact between pigeons, humans, and other poultries in wet markets increases public health concerns regarding zoonotic potential. Therefore, ongoing surveillance of wet markets and antigenic analyses should be performed. Moreover, in-depth study is necessary to determine whether this novel reassortant virus poses a threat to other hosts.
References
R.G. Webster, Lancet 363, 234–236 (2004)
Y. Liu, C. Han, X. Wang, J. Lin, M. Ma, Y. Shu, J. Zhou, H. Yang, Q. Liang, C. Guo, J. Zhu, H. Wei, J. Zhao, Z. Ma, J. Pan, Avian Pathol. 38, 263–266 (2009)
B. Jia, J. Shi, Y. Li, K. Shinya, Y. Muramoto, X. Zeng, G. Tian, Y. Kawaoka, H. Chen, Arch. Virol. 153, 1821–1826 (2008)
G. Fournie, F.J. Guitian, P. Mangtani, A.C. Ghani, J. R. Soc. Interface 8, 1079–1089 (2011)
G.H. Waddell, M.B. Teigland, M.M. Sigel, J. Am. Vet. Med. Assoc. 143, 587–590 (1963)
S.J. Anthony, J.A. St Leger, K. Pugliares, H.S. Ip, J.M. Chan, Z.W. Carpenter, I. Navarrete-Macias, M. Sanchez-Leon, J.T. Saliki, J. Pedersen, W. Karesh, P. Daszak, R. Rabadan, T. Rowles, W.I. Lipkin, MBio 3, e00166–e00212 (2012)
E.A. Karlsson, H.S. Ip, J.S. Hall, S.W. Yoon, J. Johnson, M.A. Beck, R.J. Webby, S. Schultz-Cherry, Nat. Commun. 5, 4791 (2014)
C.W. Ward, T.A. Dopheide, Biochem. J. 195, 337–340 (1981)
W.G. Laver, R.G. Webster, Virology 51, 383–391 (1973)
G. Neumann, T. Noda, Y. Kawaoka, Nature 459, 931–939 (2009)
E. Hoffmann, J. Stech, Y. Guan, R.G. Webster, D.R. Perez, Arch. Virol. 146, 2275–2289 (2001)
J. Pu, Q.F. Liu, Y.J. Xia, Y.L. Fan, E.G. Brown, F.L. Tian, J.H. Liu, Virus Genes 38, 136–142 (2009)
B.B. Dong, C.L. Xu, L.B. Dong, H.J. Cheng, L. Yang, S.M. Zou, M. Chen, T. Bai, Y. Zhang, R.B. Gao, X.D. Li, J.H. Shi, H. Yuan, J. Yang, T. Chen, Y. Zhu, Y. Xiong, S. Yang, Y.L. Shu, Biomed. Environ. Sci. 26, 546–551 (2013)
E.F. Kaleta, A. Honicke, Dtsch. Tierarztl. Wochenschr. 111, 467–472 (2004)
A. Amonsin, C. Choatrakol, J. Lapkuntod, R. Tantilertcharoen, R. Thanawongnuwech, S. Suradhat, K. Suwannakarn, A. Theamboonlers, Y. Poovorawan, Emerg. Infect. Dis. 14, 1739–1742 (2008)
H. Chen, G.J. Smith, K.S. Li, J. Wang, X.H. Fan, J.M. Rayner, D. Vijaykrishna, J.X. Zhang, L.J. Zhang, C.T. Guo, C.L. Cheung, K.M. Xu, L. Duan, K. Huang, K. Qin, Y.H. Leung, W.L. Wu, H.R. Lu, Y. Chen, N.S. Xia, T.S. Naipospos, K.Y. Yuen, S.S. Hassan, S. Bahri, T.D. Nguyen, R.G. Webster, J.S. Peiris, Y. Guan, Proc. Natl. Acad. Sci. USA 103, 2845–2850 (2006)
Q. Zhang, J. Shi, G. Deng, J. Guo, X. Zeng, X. He, H. Kong, C. Gu, X. Li, J. Liu, G. Wang, Y. Chen, L. Liu, L. Liang, Y. Li, J. Fan, J. Wang, W. Li, L. Guan, Q. Li, H. Yang, P. Chen, L. Jiang, Y. Guan, X. Xin, Y. Jiang, G. Tian, X. Wang, C. Qiao, C. Li, Z. Bu, H. Chen, Science 341, 410–414 (2013)
W. Shi, F. Lei, C. Zhu, F. Sievers, D.G. Higgins, PLoS One 5, e14454 (2010)
S. Su, L. Wang, X. Fu, S. He, M. Hong, P. Zhou, A. Lai, G. Gray, S. Li, Emerg. Infect. Dis. 20, 2096–2099 (2014)
Acknowledgments
This research was supported by the Open Fund for the State Key Laboratory of Agricultural Microbiology (AMLKF-201210). The authors especially acknowledge Prof. Yanxiu Liu (College of Foreign Languages, Huazhong Agricultural University, Wuhan 430070, P. R China.) for editing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflicts of interest.
Additional information
Edited by Keizo Tomonaga.
Zhong Zou and Sunrui Chen have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Zou, Z., Chen, S., Liu, Z. et al. Identification and genetic analysis of H3N8 subtype influenza viruses isolated from domestic pigeons in Central China. Virus Genes 52, 38–50 (2016). https://doi.org/10.1007/s11262-015-1261-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-015-1261-2