Abstract
Two dsRNAs of estimated lengths of 5 (L1) and 3.7 (L2) kpb are commonly found in strains of the basidiomycetous yeast Xanthophyllomyces dendrorhous, and the presence of virus-like particles (VLPs) have been described in some strains. Recently, two putative totiviruses (XdV-L1A and XdV-L1B) were identified from L1 dsRNA and one (XdV-L2) from L2 dsRNA in the strain UCD 67-385. In some strains, there are smaller dsRNAs (0.9–1.4 kb) that probable are satellite elements. In this work, the VLPs from several strains of X. dendrorhous, which differ in their dsRNAs content, were separated by sucrose gradient and characterized in relation to the dsRNAs and proteins that compose them. It was found that all types of dsRNAs were encapsidated into VLPs, supporting the hypothesis that the smaller dsRNAs are satellite molecules. A main protein of approx. 76 or 37 kDa composed the virions that only have the L1-dsRNA or L2-dsRNA, respectively. In the strain UCD 67-385, these both proteins were identified as viral capsid protein (CP), allow to confirm the gag predicted ORFs in XdV-L1A, XdV-L1B, and XdV-L2, with CPs of 76.6, 76.2, and 38.8 kDa, respectively. Analysis of predicted structures of CPs of XdV-L1A and XdV-L1B, showed high similitudes with the CPs of ScV-L-A and other totiviruses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Viruses with double-stranded RNA (dsRNA) genome are widely found in fungi, in fact five of the eight families of dsRNA viruses recognized by the International Committee for the Taxonomy of Viruses (ICTV) have been reported in fungi. The Totiviridae family is composed by four genera whose members have non-segmented dsRNA genomes, encapsidated in isometric virions called Virus-Like Particles (VLPs), which have no lipids or carbohydrates [1, 2]. The virus L-A (ScV-L-A) of Saccharomyces cerevisiae is the type species of genus Totivirus and is one of the most studied mycovirus. Its dsRNA genome of 4,579 bp in length has two ORFs, the 5′ ORF encodes for the major capsid protein (CP) (Gag or CP, 76–81 kDa) and the 3′ ORF for the RNA-dependent RNA polymerase (Pol or RdRp). Both ORFs are overlapped in 16–130 nt and the viral polymerase is expressed as a fusion CP-Pol protein (predicted size of 170 kDa) generated by a ribosomal-1 frameshifting [3–5]. Some strains of S. cerevisiae have an additional dsRNA molecule of 1.6–1.8 kbp called M-dsRNA, that encoded fort a killer toxin and self-immunity; this “satellite virus” depends on the viral proteins encoded by L-A (the “helper virus”) for its encapsidation and replication [6]. The ScV-L-A virions can encapsidated independently one L-A or two M-dsRNAs, and their protein composition is estimated in 1–2 CP-Pol and 120 CP proteins [7]. The CP protein participate in cap-snatching mechanism in which m(7)Gp from cap structure of cellular mRNAs is transferred to viral transcripts, required for its stability and efficient translation [8–10]. The existence of extrachromosomal dsRNA elements of variable length and cytoplasmic elongated icosahedral VLPs have been described in strains of the basidiomycetous yeast Xanthophyllomyces dendrorhous (formerly Phaffia rhodozyma) [11–13]. Recently, three putative totiviruses called XdV-L1A, XdV-L1B, and XdV-L2 were identified in the strain UCD 67–385 of X. dendrorhous, and also two dsRNA molecules (XdRm-S1 and XdRm-S2) that could be satellite viruses [14]. The XdV-L1A and XdV-L1B viruses are found in almost all dsRNA-harboring strains of X. dendrorhous, while the presence of L2- and minor size dsRNAs is quite variable among isolates from different regions of the earth [14, 15]. The presence of virus-like particles (VLPs) has been reported in some strains of X. dendrorhous and the encapsidation of dsRNAs into VLPs has been suggested according to results obtained from RNase protection assays [14]. In this work, we separated by sucrose gradient the VLPs of different strains of X. dendrorhous and characterized them in relation to its dsRNA and protein composition. Furthermore, the CPs were identified in VLPs of strain UCD 67-385 allowing confirming the predicted ORFs of the three totiviruses present in X. dendrorhous.
Materials and methods
Strains and culture conditions
Four strains of X. dendrorhous with different dsRNAs profiles were used: UCD 67-385 (L1-, L2-, S1- and S2-), UCD 67-202 (L1- and S1-), VKMY-2786 (L1-), and UCD 68-653C (L2-). The yeast were routinely grown at 22 °C in YM medium (0.3 % yeast extract, 0.3 % malt extract, 0.5 % peptone) supplemented with 2 % glucose. The cellular pellets used for dsRNA and VLPs extractions were obtained from late log-phase cultures by centrifugation at 7,000×g for 10 min.
Extraction and separation of VLPs
Samples of VLP were obtained from cultures of each X. dendrorhous strain as described previously [14]. Briefly, the yeast pellet (4–6 g) was washed and suspended in 20 ml of TBS buffer (10 mM Tris, 100 mM NaCl, 1 mM MgCl2, 0.1 mM EDTA, pH 7.4) supplemented with “CompleteTM Protease Inhibitor Cocktail Tablets” (Roche) according to the manufacturer’s instructions. Three grams of glass beads (0.5 mm in diameter) were added, and the samples were shaken for 30 s in a Mini beadbeater-16 homogenizer (Bio Spec, Bartlesville, USA), and subsequently cooled on ice for 1 min; the shaking and cooling steps were repeated four times. The supernatant was harvested after centrifugation at 12,000×g for 20 min and subjected to an additional centrifugation step at 120,000×g for 90 min (both centrifugations were performed at 4 °C), and the pellet, enriched of VLPs, was suspended in 1.5 ml of TBS buffer. For VLPs separation, 1 mL of each sample was loaded onto linear 10–40 % (w/v) sucrose gradients. After centrifugation at 120,000×g at 4 °C for 3 h, fractions of 500 μL were carefully collected from the tops of tubes. Aliquots of each fraction were used to the determinations of dsRNAs and protein content as described below.
RNase protection assay
250 µl of VLPs samples were digested with 50 µg/ml RNAse A for 30 min at 37 °C, before or after organic extraction with 1 volume of phenol (pH 4.0). Then the samples were extracted with one volume of chloroform:isoamylic alcohol (24:1) and the presence of dsRNA molecules was evaluated by agarose gel electrophoresis.
Isolation of total RNA
RNAs were isolated by organic extraction according to previously described method [16]. Briefly, 300 µl of each fraction were mixed with 1 volume acid phenol (pH 4.0):chloroform:isoamyl alcohol (25:24:1) and vortexed for 3 min. After centrifugation at 14,000×g, the aqueous phase was extracted with one volume of chloroform:isoamyl alcohol (24:1). Two volumes of isopropanol were added to the aqueous phase and incubated at −20 °C for 2 h. Samples were centrifuged at 14,000×g for 10 min and the pellet was suspended in 20 μL of nuclease-free water. Each sample was treated with DNase I and nuclease S1 (according to manufacturer’s instructions) and analyzed by agarose gel electrophoresis.
Protein analysis
For the determinations of protein content in each fraction of sucrose gradients, 30 μL was mixed with 10 μL of loading buffer (50 mM Tris–HCl pH 6.8, 2 % SDS, 10 % glycerol, 1 % 2-mercaptoethanol, 12.5 mM EDTA, 0.02 % bromophenol blue) and incubated at 100 °C for 5 min. Samples were separated using 10 % SDS-PAGE and stained with Coomassie blue G-250. For protein purification, the fractions that contained dsRNAs were pooled and mixed with 7 mL of TBS and centrifuged (120,000×g, 90 min, 4 °C). The pellet was suspended in 200 μL of TBS, then 30 μL of sample was electrophoresed on 10 % SDS-PAGE and stained with Coomassie brilliant blue G-250. Protein bands were excised and analyzed by MALDI-TOF MS at Alphalyse Inc. (USA) and at Cambridge Centre for Proteomics (University of Cambridge, UK). Peptide masses were identified by FindPept (http://web.expasy.org/findpept/) [17, 18] using theoretical peptides calculated from L1A (YP_007697650), L1B (AFH09413), and L2 (AFH09415) predicted CP sequences of X. dendrorhous viruses. The mass tolerance of sequence ions was set to ±0.5 Da and proteins were cleaved with porcine trypsin (C-term to K/R, before P). In silico analysis of X. dendrorhous CPs was done with MS-Digest, a part of Protein Prospector (http://prospector.ucsf.edu; enzymatic digestion:trypsin; 1 mass range 400–5,000 Da, and zero missed cleavages).
Secondary structure prediction
The secondary structure prediction was done with GeneSilico fold prediction metaserver [19], taking in account as final results the consensus of several prediction methods in each case: HMMSTR, SPARROW, *SPARROW, sspro4, sspal, cdm, jnet, sspred, sable, prof, porter, nnssp, netsurfp, ssp, pssfinder, soprano, spineX, spine, psipred, fdm, gor, proteus, psspred, raptorxss.
Results
Separation and analysis of VLPs
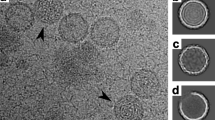
The samples of VLPs were obtained from cultures of X. dendrorhous strains that differ in their content of dsRNA molecules: UCD 67-385 (L1-dsRNA, L2-dsRNA, S1-dsRNA, and S2-dsRNA), VKM Y-2786 (L1-dsRNA), UCD 68-653C (L2-dsRNA), and UCD 67-202 (L1-dsRNA and S1-dsRNA). The integrity of VLPs in each sample was evaluated by RNAse protection assay as described in “Materials and methods” section, selecting for more analysis the samples in which the dsRNAs were resistant to digestion with RNase A (not shown). The VLPs were separated by buoyant density using ultracentrifugation in linear sucrose gradients, and the dsRNAs and proteins present in each fractions obtained were determined. The samples of dsRNAs were treated with Nuclease S1 and DNAse I before analysis in agarose gel electrophoresis. The results of analysis of VLPs from four strains of X. dendrorhous are shown in Fig. 1. In all samples, protein bands were observed in SDS-PAGE only in fractions positive for dsRNAs. In the analysis of VLPs from strains harboring only one type of dsRNA, a protein band of approx. 76 kDa was observed in fractions having L1-dsRNA (strain VKM Y-2786), while several protein bands were observed, being the largest one of approx. 37 kDa, in fractions having L2-dsRNA (strain UCD 68-653C). In the VLPs samples from the strain UCD 67-202, which has L1-dsRNA and S1-dsRNA, only a protein band of approx. 76 kDa was observed in all fractions having dsRNAs, independent of the dsRNAs content of the fraction. Fractions containing only L1-dsRNA were observed at higher densities, while both dsRNAs were observed in the other fractions. In the case of VLPs from the strain UCD 67-385, the fractions at higher densities have only L1-dsRNA, while the fractions at lower densities contain mainly the L2-dsRNA. The fours types of dsRNAs were observed in fractions at middle densities. A unique protein band of approx. 76 kDa was observed in fractions that have only L1-dsRNAs, and in the fractions that have L2-dsRNA an additional protein band of approx. 37 kDa was observed.
Separation and analysis of VLPs from different X. dendrorhous strains. VLPs were separated by ultracentrifugation in 10–40 % sucrose gradients. Fractions of 0.5 mL were collected from the top of the tubes and the dsRNA and protein content was determined. In each panel is indicated the X. dendrorhous strain, and the analysis of dsRNAs (agarose gel electrophoresis) and protein (PAGE-SDS) are shown at top and bottom, respectively. The fraction numbers are indicated at the top of each photo. Asterisk and double asterisk denoted the protein bands of 76 and 37 kDa, respectively. K 1 Kb DNA marker, L lambda HindIII DNA marker, MP protein marker
Identification and analysis of viral capsid proteins
The proteins observed in the analysis of VLPs samples probably are the CP proteins, because their estimated molecular weights match with those of putative CPs encoded by the totiviruses XdV-L1A (76.6 kDa), XdV-L1B (76.2 kDa), and XdV-L2 (38.8 kDa). To corroborate this, the proteins of VLPs samples from strain UCD 67-385 were purified and analyzed by MALDI-TOF–MS peptide mass fingerprinting (PMF). The peptide masses data obtained were analyzed and compared with the predicted aminoacid sequences of viral CP proteins. As shown in Fig. 2, several peptides matched with CP of XdV-L1A, XdV-L1B, and XdV-L2, with coverages of 26, 23, and 19 %, respectively. This result strongly suggests that the three viral CP proteins are expressed in the strain UCD 67-385 of X. dendrorhous, and that the predicted gag ORFs, and probably the pol ORFs, are correctly annotated. In the comparison of aminoacid sequences of CP proteins of several fungal totiviruses (Fig. 3) is observed that the residues described to be important for the cap-snatching mechanism of Sc-V-L-A and L-BC viruses [8, 10, 20] are conserved in CP proteins of XdV-L1A and XdV-L1B. However, the CP protein of XdV-L2 lacks four (Y150, D152, Y154, and Y615) of the eight residues important for cap-snatching, similar to the CP protein of Zygosaccharomyces bailii virus Z.
Comparison of theoretical and experimental peptides masses of capsid proteins from strain UCD 67-385. The protein bands were excised from SDS-PAGE and analyzed by MALDI-TOF MS. Peptide masses were identified by FindPept using theoretical peptides calculated from predicted capsid protein sequences of X. dendrorhous viruses. Matches between theoretical and experimental data are underlined
Comparison of primary structure of capsid proteins of different totiviruses. The residues important for cap-snatching mechanism are denoted by arrows. GenBank accession numbers: ScV-L-A (AAA50506), ScV-L-BC (NP_042580.1), XdV-L1A (YP_007697650), XdV-L2 (AFH09415), XdV-L1B (AFH09413), BRVF (YP_001497150), ZbV (NP_624324), SsVL (AGG68770), TaV1 (ADQ54105)
Discussion
Fractions enriched concomitantly in dsRNAs and proteins were obtained in sucrose gradient analysis of VLPs samples of the four X. dendrorhous strains studied, allowing to affirm that the dsRNAs present in each strain are encapsidated in particles. These virions are formed by a main protein of 76 kDa in the strains VKM Y-2786 and UCD 67-202, while in strain UCD 68-653C by a main protein of 37 kDa. In strain UCD 67-385, it is possible to affirm that virions at higher densities are formed by a 76 kDa protein, but in the fractions at minor densities the protein bands of two sizes were observed together two or four dsRNAs. With our results it is not possible to conclude if in these fractions correspond to virions formed by two proteins or to virions formed by different proteins but have similar buoyant densities. The smaller dsRNAs S1- and S2- were encapsidated in the different X. dendrorhous strains supporting the hypothesis that they are satellite elements. The satellite dsRNAs in the viral system of S. cerevisiae are encapsidated independently of helper virus, generating virions with one L-A molecule or two M molecules [5–7, 21–23]; in some case can encapsidated 1 to 8 X dsRNA, a molecule of 0.52 kb spontaneously arose in a non killer strain [24]. More analyses are necessary to evaluate if this exclusion mechanism of dsRNA encapsidation exists in totiviruses of X. dendrorhous, or if the fractions showing multiple dsRNAs in the sucrose gradients correspond to virions that encapsidated more than one type of dsRNAs. In relation to the protein content of virions, our results strongly suggest that the two protein bands observed in the electrophoretic analysis of VLPs from strain UCD 67-385 are the viral CPs. The upper band would correspond to two CPs encoded by XdV-L1A (76.6 kDa) and XdV-L1B (76.2 kDa), and the lower band to a CP encoded by XdV-L2 (38.8 kDa). In this way, the gag ORFs predicted in the totiviruses of X. dendrorhous are confirmed and consequently the pol ORFs, because both ORFs are overlapped. Only the CPs were detected in PAGE-SDS analysis of all VLPs samples indicative of a low proportion of viral polymerase in virions, similar to ScV-L-A virions [21]. This is in accordance with the synthesis of the viral polymerase as a fusion with CP by a -1 ribosomal frameshifting, event that occur at low frequency [3].
XdV-L2 can be considered a replicative intermediate of XdV-L1A, because it is identical in sequence but without 989 bp at the 5′- end. However, according to our results this molecule is expressed in X. dendrorhous and for that, taking in account the replicative model of totiviruses of S. cerevisiae [25, 26], a XdV-L2 (+) ssRNA must be recognized by CP-Pol, the virion completed by the polymerization of CP proteins, then Pol synthesizes the complimentary strand and a (+) ssRNA that is extruded from the particle, and translated by host machinery. This strongly suggests that XdV-L2 behaves as a totiviruses, but we cannot discard the possibility that it uses proteins encoded by XdV-L1A and/or XdV-L1B for its replication/expression in the strain UCD 67-385 of X. dendrorhous. By example, the Sc-L-A virus furnishes its mRNA with cap structure derived from cellular mRNAs, transferring m7Gp from the cap donor to the 5′ end of the viral transcript, mechanism known as cap-snatching [8]. The 5′ cap structure (m7GpppX-) is essential for stability and efficient translation of mRNAs in eukaryotic cells. In the case of capsids protein encoded by XdV-L1A and XdV-L1B, all residues important for the cap-snatching mechanism [27] are conserved allowing to assume that these totiviruses use the same mechanism. By contrast, the CP encoded by XdV-L2 lacks 334 residues from amino-terminal region compared to CP protein of XdV-L1A, including four important residues for cap-snatching such as the His-154 that is essential for decapping of host mRNA [8]. There is a sequence in XdV-L2 CP (8-TDMDAHD-14) that differs from consensus at position 10, an aspartic acid instead of tyrosine. This rise the possibility that XdV-L2 depend of the proteins encoded by the other X. dendrorhous totiviruses fir its efficient translation in the host. The complete sequencing of L2-dsRNA and CP characterization in the strain UCD 68-653C will help to elucidate this question, because this strain the L2-dsRNA is stable maintained and expressed alone.
Recently, it was described that the core region in the crystal structure of ScV-L-A CP composed by helix 3–5 (aa 85–139) and helix 12–13 (aa 345–383) correspond closely in size and location to helices 2–3 (aa 84–122) and H10–12 (aa 350–397) of Helminthosporium victoriae virus 190S (HvV190S) CP, both members of different genera totiviruses. This centrally located stretch of residues with predominantly helical content was observed too in predicted secondary structures of CPs of representative members of Victorivirus, Giardiavirus, Leishmaniavirus, and Trichomonasvirus genera, suggesting a conservation of this helical core in Totiviridae family [28]. We compared the predicted secondary structure of CP of several totiviruses (Fig. 4), observing a very close conservation of this helix core among Sc-V-L-A, ScV-L-BC, Tuber aestivum virus 1 (TaV1), Scheffersomyces segobiensis virus L (SsVL), Black raspberry virus F (BRVF), and XdV-L1A. Stretches of helices were observed too for the shorter CPs of XdV-L2 and Zygosaccharomyces bailii virus Z (ZbV). In the case of CP of XdV-L1B, the conserved stretch of three helices in the amino region, which correspond to helix 3–5 (aa 85–139) in Sc-V-L-A and to helix 2–4 (aa 87–139) in XdV-L1A, was not observed. It must be taken into account that the results shown in Fig. 4 are the consensus of eighteen predictor software, but six of them predicted helices of four to nine residues in length at position ranging from aa 105–112 of the CP of XdV-L1B, according to which this helix core would exist.
Secondary structure of capsid proteins of totiviruses. The structure of ScV-L-A CP identified in the crystal structure [29] was compared with the predicted CP structures of other eight totiviruses. The α-helix and β-strand are represented by cylinder and arrow, respectively. The helices forming part of the helical core in ScV-L-A CP are denoted (H). All predictions were done using several methods at GeneSilico fold prediction metaserver, and the consensus results are shown. The predicted stretches of helices that may form part of structural core are boxed. GenBank accession numbers as in Fig. 3
References
P. Mertens, Virus Res. 101, 3–13 (2004)
W. Magliani, S. Conti, M. Gerloni, D. Bertolotti, L. Polonelli, Clin. Microbiol. Rev. 10, 369–400 (1997)
J.D. Dinman, T. Icho, R.B. Wickner, Proc. Natl. Acad. Sci. USA. 88, 174–178 (1991)
T. Fujimura, J.C. Ribas, A.M. Makhov, R.B. Wickner, Nature 359, 746–749 (1992)
R.B. Wickner, C.C. Wang, J.L. Patterson, in Virus Taxonomy: classification and Nomenclature of Viruses: eighth Report of the International Committee on the Taxonomy of Viruses, ed. by C.M. Fauquet, M.A. Mayo, J. Maniloff, U. Desselberger, L.A. Ball (Elsevier Academic Press, San Diego, 2005), pp. 571–580
M.J. Schmitt, F. Breinig, Nat. Rev. Microbiol. 4, 212–221 (2006)
R. Esteban, R.B. Wickner, Mol. Cell. Biol. 6, 1552–1561 (1986)
T. Fujimura, R. Esteban, Proc. Natl. Acad. Sci. USA. 108, 17667–17671 (2011)
T. Fujimura, R. Esteban, J. Biol. Chem. 287, 12797–12804 (2012)
T. Fujimura, R. Esteban, J. Biol. Chem. 288, 23716–23724 (2013)
A. Castillo, V. Cifuentes, Curr. Genet. 26, 364–368 (1994)
I. Pfeiffer, J. Kucsera, J. Varga, A. Parducz, L. Ferenczy, Curr. Genet. 30, 294–297 (1996)
I. Pfeiffer, J. Litter, J. Kucsera, Acta Biol. Hung. 52, 299–306 (2001)
M. Baeza, N. Bravo, M. Sanhueza, O. Flores, P. Villarreal, V. Cifuentes, Virol J 9, 140 (2012)
M. Baeza, M. Sanhueza, O. Flores, V. Oviedo, D. Libkind, V. Cifuentes, Virol J 6, 160 (2009)
M. Baeza, M. Fernández-Lobato, V. Cifuentes, in Microbial Carotenoids from Fungi, ed. by J.L. Barredo (Humana Press, New York, 2012), pp. 195–205
A. Gattiker, W.V. Bienvenut, A. Bairoch, E. Gasteiger, Proteomics 2, 1435–1444 (2002)
E. Gasteiger, C. Hoogland, A. Gattiker, M.R. Wilkins, R.D. Appel, A. Bairoch, in The Proteomics Protocols Handbook, ed. by J.M. Walker (Humana Press, New York, 2005), pp. 571–607
M.A. Kurowski, Nucleic Acids Res. 31, 3305–3307 (2003)
D.J. Taylor, J. Bruenn, BMC Biol. 7, 88 (2009)
K.A. Bostian, J.A. Sturgeon, D.J. Tipper, J. Bacteriol. 143, 463–470 (1980)
D.J. Tipper, M.J. Schmitt, Mol. Microbiol. 5, 2331–2338 (1991)
D.J. Thiele, E.M. Hannig, M.J. Leibowitz, Mol. Cell. Biol. 4, 92–100 (1984)
R. Esteban, R.B. Wickner, J. Virol. 62, 1278–1285 (1988)
J.R. Caston, B.L. Trus, F.P. Booy, R.B. Wickner, J.S. Wall, A.C. Steven, J. Cell Biol. 138, 975–985 (1997)
R.B. Wickner, Microbiol. Rev. 60, 250–265 (1996)
D.J. Taylor, M.J. Ballinger, S.M. Bowman, J.A. Bruenn, PeerJ 1, e50 (2013)
S.E. Dunn, H. Li, G. Cardone, M.L. Nibert, S.A. Ghabrial, T.S. Baker, PLoS Pathog. 9, e1003225 (2013)
H. Naitow, J. Tang, M. Canady, R.B. Wickner, J.E. Johnson, Nat. Struct. Biol. 9, 725–728 (2002)
Acknowledgments
This work was supported by a Grant from Fondecyt (11060157), and a CONICYT Scholarships to Oriana Flores.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by Thomas Hohn.
Rights and permissions
About this article
Cite this article
Flores, O., Alcaíno, J., Fernandez-Lobato, M. et al. Characterization of virus-like particles and identification of capsid proteins in Xanthophyllomyces dendrorhous . Virus Genes 50, 253–259 (2015). https://doi.org/10.1007/s11262-015-1171-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-015-1171-3