Abstract
Purpose
Diabetic nephropathy (DN) is one of the most serious complications of diabetes that leads to decline of renal function. Although numerous studies have revealed that microRNAs (miRNAs) play essential roles in the progression of DN, whether miR-365 is involved remains elusive.
Methods
The successful construction of DN model was confirmed by ELSIA, hematoxylin–eosin (HE) and Masson staining assay. The expression of miR-365 was detected through RT-qPCR. The levels of BDNF, p-TrkB, α-smooth muscle actin (SMA), collagen IV (Col.IV), transforming growth factor-β1 (TGF-β1), tumor necrosis factor α (TNF-α), and interleukin-6 (IL-6) were evaluated by western blot, IF or ELISA assays. Luciferase reporter assay was used to detect the interaction between miR-365 and BDNF.
Results
The DN mice model was induced by streptozotocin (STZ). Then miR-365 expression was found to upregulate in tissues of DN rat. Furthermore, elevated expression of miR-365 was found in high glucose (HG)-treated HK-2 cells. Silencing of miR-365 suppressed the accumulation of ECM components and secretion of inflammatory cytokines in HK-2 cells. In addition, it was demonstrated that miR-365 could target BDNF. The protein levels of BDNF and p-TrkB were negatively regulated by miR-365 in HK-2 cells. Moreover, inhibition of miR-365 suppressed the levels of SMA, Col.IV, TGF-β1, TNF-α, and IL-6, indicating the renal fibrosis was inhibited by miR-365 knockdown.
Conclusion
MiR-365 could regulate BDNF-TrkB signal axis in STZ induced DN fibrosis and renal function. The results of the current study might provide a promising biomarker for the treatment of DN in the future.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Diabetes mellitus is a prevalent metabolic disorder leading to various chronic complications including peripheral neuropathy and retinopathy [1]. Diabetes can cause chronic renal dysfunction and diabetic nephropathy (DN) [2]. Nearly 30–40% of patients with diabetes will have DN [3]. Moreover, the incidence of DN is still on rise all over the world. It is the major reason for end-stage renal disease (ESRD) [4]. DN is characterized by glomerular basement membrane (GBM) thickening, extracellular matrix (ECM) accumulation, increasing mesangial matrix, and ultimately glomerulosclerosis and tubulointerstitial fibrosis [5]. DN can cause renal fibrosis and further impair the function of kidney [6]. Although modern therapies are widely used to control the glycemic and blood pressure in patients, progressive renal damage is still on-going [7]. Therefore, it is of great significance to identify novel biomarkers for improving the treatment of DN.
Abundant researches have shown that miRNAs contribute to the pathogenesis of various human deceases. For example, miR-135a-5p restrains the proliferation of head and neck squamous cell carcinoma cells via regulating HOXA10 [8]. MiR-27a and miR-142-5p aggravates the development of nonalcoholic fatty liver disease through modulation of Nrf2 signaling pathway [9]. MiR-454-3p is involved in biological behaviors of trophoblast cells in preeclampsia via targeting THBS2 [10]. MiR-330-5p regulates cell growth and death in acute myeloid leukemia [11]. More importantly, abundant evidences demonstrate that miRNAs play essential parts in regulating the progression of DN. For instance, miR-24-3p serves as a sponge of circRNA circ_0080425 to regulate the proliferation of mesangial cells and renal fibrosis in DN through targeting fibroblast growth factor 11 [12]. DN progression is affected by miR-218 through binding with IKK-β and regulating inflammation induced by NK-κB [13]. MiR-101a elevation inhibits renal fibrosis in DN though targeting KDM3A and suppressing the YAP/TGF-β/Smad signaling pathway [14]. MiR-675 is associated with the progression of DN by targeting EGR1 and depressing vitamin D receptor level in CIHP-1/HEK 293 cells [15]. MiR-365 is located on chromosome 16p13.12, a region that participates in various biological processes [16]. Moreover, miR-365 has been formerly recognized as one of the mechano-responsive miRNAs (mechanomiRs) potently stimulating inflammatory signals [17]. Nonetheless, the role of miR-365 in DN remains elusive.
In the present work, the functions as well as the underlying mechanism of miR-365 in DN were carefully investigated. The data of this work indicated that miR-365 regulated HFD/STZ induced DN fibrosis and renal function through targeting BDNF-TrkB signal axis. The results of the study might provide a new insight into exploring effective treatment method for DN.
Materials and methods
Cell culture, transfection, and DN cell model
Human kidney tubular epithelial (HK-2) cells were bought from the American Type Culture Collection (Manassas, VA, USA) and maintained in low-glucose DMEM supplemented with 10% FBS, 100 mg/ml streptomycin, and 100 IU/ml penicillin at 37 °C with 5% CO2. MiR-365 mimic, miR-365 inhibitor, sh-BDNF, mimic NC, inhibitor NC, and sh-NC were obtained from GenePharma (Shanghai, China). HK-2 cells were grown in six-well plates and the oligonucleotides above were introduced into HK-2 cells via Lipofectamine 2000 (Invitrogen). After transfection, HK-2 cells were incubated with HG (30 mM) or normal glucose (NG; 5 mM) for 48 h to induce DN cell model.
DN rat model
The 8-week-old male Sprague–Dawley rats (200–220 g) were employed for this study. Rats (n = 20) were intraperitoneal injected with STZ (55 mg/kg in 0.1 mol/L citrate buffer) to induce diabetes. For control group, rats were subjected to citrate buffer (n = 20). After 72 h, blood glucose levels > 11.1 mmol/L suggested the successfully building of DN models. After collecting blood and urine samples, the rats were sacrificed and tissue samples were harvested and stored at − 80 °C. All animal studies were performed under the approval of the Animal Research Committee of Haining People's Hospital.
Immunofluorescence (IF)
The rat renal tissues were formalin-fixed and paraffin-embedded. Then the slices were put into a container and incubated with EDTA (1 mM; pH 8.0) at 95 °C for 20 min for antigen retrieval. After blocking in bovine serum albumin (5%) for half an hour, the sections were incubated with primary antibodies against collagen IV (Col.IV) (ab235296, Abcam, Shanghai, China) at 4 °C overnight. After rinsing with PBS, the sections were incubated with secondary antibodies for 50 min in darkness. At last, the sections were counterstained with DAPI for 10 min in a dark room. Images were captured by confocal microscopy.
Quantitative real-time polymerase chain reaction (RT-qPCR)
TRIzol reagent (TaKaRa) was used to extract total RNAs from renal tissues or HK-2 cells. Reverse transcription of RNA samples (1 μg) was performed via TaqMan RT reagents (TaKaRa). Quantitative real-time PCR was executed by a SYBR Premix Ex Taq II (TaKaRa). Reaction conditions for PCR were: 95 °C for 1 min, 40 cycles of 95 °C for 15 s, 55 °C for 30 s, and 72 °C for 30 s. U6 or GAPDH was used as internal control for miR-365 and BDNF, respectively. Relative expression of miR-365 and BDNF was analyzed via the 2−ΔΔCt method. Each reaction was done in triplicate. The primers were displayed as below: miR-365, Forward: 5′-CGTAA TGCCC CTAAA AAT-3′, Reverse: 5′-GTGCA GGGTC CGAGGT-3′; BDNF, Forward: 5′‑CCAGG TGAGA AGAGT GATG-3′, Reverse: 5′‑AGTGA TGTCG TCGTC AGA‑3′; U6, Forward: 5′-GCTTC GGCAG CACAT ATACT AAAAT-3′, Reverse: 5′-CGCTT CACGA ATTTG CGTGT CAT-3′; GAPDH, Forward: 5′-TGCAC CACCA ACTGC TTAGC‑3′, Reverse: 5′-GGCAT GGACT GTGGT CATGAG-3′.
Enzyme-linked immunosorbent assay (ELISA)
The concentrations of inflammatory cytokines such as tumor necrosis factor α (TNF-α) and interleukin-6 (IL-6) were evaluated via the Mouse TNF-α ELISA Kit, Mouse IL-6 ELISA Kit, Human TNF-α ELISA Kit, and Human IL-6 ELISA kit (Beyotime, Shanghai, China). Briefly, supernatants were incubated with enzyme-specific substrates (100 μM) at 37 °C for 4 h. The absorbance at 450 nm was evaluated via a microplate reader (Corona Electric, Ibaraki, Japan). Serum creatinine (Cr) was detected based on the sarcosine oxidase method via Creatinine Assay kit (Jiancheng Bioengineering Institute, Nanjing, China). Blood urea nitrogen (BUN) was evaluated according to urease method by a Urea Assay Kit (Jiancheng Bioengineering Institute, Nanjing, China) in accordance with previous studies [18]. The urinary albumin was detected by ELISA kits (Cell Biolabs, INC., India).
Hematoxylin and eosin (HE) staining
Renal tissues were paraffin-embedded and cut into 5 mm thick slices. After dewaxing and dehydrating, the slices were stained with hematoxylin and eosin (HE) for 5 min and ammonia water for 30 s. Then the slices were rinsed by 70% and 90% alcohol and counterstained in eosin–phloxine solution for 2 min.
Masson staining
After dewaxing and rehydrating, the sections were stained with hematoxylin solution for 8 min. Next the sections were rinsed in running water for 8 min, and stained in ponceau acid fuchsin solution for 5 min. After 5 min’s differentiation in phosphomolybdic–phosphotungstic acid, the slices were transferred into aniline blue solution for 5 min. Thereafter, 0.2% acetic acid was used for the differentiation for 2 min. Sections were dehydrated and Nikon microscope (Japan) was employed to capture images.
Western blot analysis
RIPA Lysis Buffer (Beyotime Institute of Biotechnology, Shanghai, China) was employed for extracting proteins from renal tissues and HK-2 cells. A BCA assay (Dingguo, Beijing, China) was utilized for assessing the concentrations of proteins. Then the proteins were subjected to 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and electrophoretically transferred onto polyvinylidene difluoride membranes (PVDF; Millipore, MA, USA). Blocked the membranes with 5% nonfat milk in tris-buffered saline containing 0.1% Tween 20 (TBST) for 1 h at room temperature, then incubated with primary antibodies against α-SMA (ab5694, Abcam), Col. IV (ab86042, Abcam), TGF-β1 (ab215715, Abcam), BDNF (ab108319, Abcam), p-TrkB (ab229908) or GAPDH (ab8245, Abcam) at 4 °C for a night. Then, incubated with HRP-conjugated secondary antibody (ab6728, Abcam) for 2 h at room temperature. Subsequently, a chemiluminescence detection kit (Amersham Pharmacia Biotech, NJ, USA) was applied for detecting the protein signals.
Luciferase reporter assay
The 3′ UTR of wild-type (WT) and mutant (MUT) BDNF were cloned (BDNF-WT and BDNF-MUT) and inserted into the pmirGLO vector (Promega, Madison, USA). Then BDNF-WT or BDNF-MUT pmirGLO vectors were transfected with mimic NC or miR-365 mimic via Lipofectamine 2000, respectively. After 48 h, the luciferase activity was evaluated by a Dual-Luciferase Reporter System (Promega).
Immunohistochemistry (IHC)
Paraffin-embedded sections of renal tissues were subjected to IHC to evaluate the expression of Col. IV. Briefly, the paraffin-embedded sections were deparaffinized, rehydrated, and then incubated with a pH 6.0 citrate buffer solution for 15 min at 121 °C. After washing with PBS, the tissues were then incubated in a 3% hydrogen peroxide solution for 10 min at indoor temperature. After blocking with 1% BSA solution for 15 min, the tissues were stained with primary antibody of Col. IV (ab6586, Abcam) at 4 °C overnight. Then the sections were incubated with HRP-conjugated secondary antibody (ab6728, Abcam) at indoor temperature for 1 h. Next, the sections were stained with a DAB solution and counterstained with hematoxylin for 30 s. Furthermore, 0.1% sodium bicarbonate was used to blue the sections for 1 min. Finally, IHC images were captured by a microscope.
Statistical analysis
SPSS 24.0 (IBM, USA) was employed for statistical analysis in this study. Data were shown as mean ± standard deviation (SD). Differences between or among groups were analyzed by Student’s t test or one-way analysis of variance (ANOVA) followed by Tukey’s test. All experiments were performed with at least three separated experiments. P < 0.05 was considered to be statistically significant.
Results
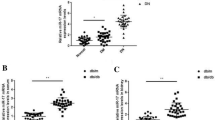
MiR-365 was highly expressed in the renal tissues of DN rats
To explore the mechanism of miR-365 in DN fibrosis and renal function, a DN rat model was established. The results from ELISA delineated that the level of blood glucose was obviously increased in DN model group compared with that in control group (Fig. 1a). Moreover, the levels of serum creatinine (Cr) and blood urea nitrogen (BUN) were evidently higher in model group than that in control group (Fig. 1b, c). Moreover, the elevated level of urinary albumin after 24 h was observed in model group in comparison with control group (Fig. 1d). Besides, the concentrations of TNF-α and IL-6 were evaluated in ELISA assay. The data delineated that TNF-α and IL-6 concentrations were markedly increased in model group (Fig. 1e, f). The results from HE staining vividly showed the pathological features of rat renal tissues, delineating that the renal tissues were more seriously damaged in model group compared with that in control group (Fig. 1g). In addition, the area of renal fibrosis was bigger in the model group than that in control group according to the data from Masson staining (Fig. 1h). These findings implied that the DN rat model was successfully built in this study. Furthermore, the expression of miR-365 was remarkably increased in the model group in comparison with the control group (Fig. 1i), suggesting miR-365 might exert important functions in the development of DN.
MiR-365 was highly expressed in the renal tissues of DN rats. a The blood glucose level was evaluated by ELISA in control and model groups. b–d The level of serum Cr BUN and urinary albumin in control and model groups was assessed via ELISA assay. e–f The levels of TNF-α and IL-6 were determined by ELISA assay in control and model groups. g The pathologic changes in control and model groups were observed by HE staining. h The renal fibrosis in control and model groups was viewed by Masson staining. i MiR-365 level was detected via RT-qPCR. *P < 0.05
Knockdown of miR-365 reduced the accumulation of ECM components and secretion of inflammatory cytokines in HG-induced HK-2 cells
Next, the role of miR-365 in high glucose-induced HK-2 cells was investigated. First, the miR-365 level was identified to elevate in HG-triggered HK-2 cells compared with NG-triggered HK-2 cells (Fig. 2a). Afterwards, miR-365 was knocked down in HG-induced HK-2 cells via transfection of miR-365 inhibitor (Fig. 2b). Then the protein levels of fibrosis related proteins (α-SMA, Col.IV, and TGF-β1) were detected by western blot analysis. The results revealed that the increased protein levels of α-SMA, Col.IV, and TGF-β1 in HG-induced HK-2 cells were suppressed by the inhibition of miR-365 (Fig. 2c). Similarly, IF staining displayed that the level of Col.IV was dramatically reduced by silencing of miR-365 in HG-induced HK-2 cells (Fig. 2d). Moreover, the concentrations of TNF-α and IL-6 were downregulated in the HG + miR-365 inhibitor group compared with that in HG + inhibitor NC group (Fig. 2f, g). Taken together, knockdown of miR-365 reduced the accumulation of ECM components and secretion of inflammatory cytokines in HG-induced HK-2 cells.
Knockdown of miR-365 reduced the accumulation of ECM components and secretion of inflammatory cytokines in HG-induced HK-2 cells. a The miR-365 level was measured by RT-qPCR in HG and NG induced HK-2 cells. b The expression of miR-365 in NG, HG + inhibitor NC, and HG + miR-365 inhibitor groups was shown by RT-qPCR. c The protein expression of α-SMA, Col.IV and TGF-β1 in NG, HG + inhibitor NC, and HG + miR-365 inhibitor groups was evaluated by western blot analysis. d The levels of Col.IV in NG, HG + inhibitor NC, and HG + miR-365 inhibitor groups were viewed by IF staining. e–f The concentrations of TNF-α and IL-6 in NG, HG + inhibitor NC, and HG + miR-365 inhibitor groups were assessed by ELISA. *P < 0.05 compared with NG group, #P < 0.05 compared with HG + inhibitor NC group
BDNF was downregulated by miR-365 in renal tissue of DN rat model and HG-induced HK-2 cells
Furthermore, the underlying mechanisms of miR-365 in DN were explored. Targetscan (http://www.targetscan.org/vert_72/) predicted that miR-365 had potential binding sites for BDNF (Fig. 3a). Dual-luciferase reporter assay uncovered that the increase of miR-365 could obviously lower the luciferase activity of BDNF-WT vector than that in the mimic NC group (Fig. 3c). Besides, the luciferase activity of the BDNF-MUT vector showed no significant changes in both miR-365 mimic and mimic NC groups (Fig. 3c). Additionally, the expression of BDNF was confirmed to be markedly downregulated in tissues of the model group and HG-induced HK-2 cells (Fig. 3c, d). More importantly, the expression of BDNF was reduced by miR-365 overexpression and elevated by miR-365 silencing (Fig. 3e). Besides, increase of miR-365 expression prominently repressed the protein expression of BDNF and p-TrkB while restraint of miR-365 expression strikingly elevated the protein expression of BDNF and p-TrkB (Fig. 3f). Altogether, BDNF was downregulated by miR-365 in renal tissue of DN rat model and HG-induced HK-2 cells.
BDNF was downregulated by miR-365 in renal tissue of DN rat model and HG-induced HK-2 cells. a Targetscan was used to predict the potential binding sites for miR-365 and BDNF. b Luciferase reporter assay was applied for detecting the binding ability of miR-365 and BDNF. *P < 0.05 compared with mimic NC group. c The protein expression of BDNF in control and model groups was detected by western blot analysis. *P < 0.05 compared with control group. d Western blot analysis was used to detect the protein expression of BDNF in HG and NG induced HK-2 cells. *P < 0.05 compared with NG group. e The miR-365 level was measured through RT-qPCR. *P < 0.05 compared with mimic NC group, #P < 0.05 compared with inhibitor NC group. f The protein levels of BDNF and p-TrkB were detected by western blot analysis. *P < 0.05 compared with mimic NC group, #P < 0.05 compared with inhibitor NC group
BDNF upregulation by miR-365 knockdown reduced the accumulation of ECM components and secretion of inflammatory cytokines in HG-induced HK-2 cells
Thereafter, the functions of miR-365 in HG-induced HK-2 cells were measured. The miR-365 level was decreased in the HG + miR-365 inhibitor + sh-NC and HG + miR-365 inhibitor + sh-BDNF groups compared with HG + inhibitor NC + sh-NC and HG + inhibitor NC + sh-BDNF groups (Fig. 4a). Moreover, the protein expression of BDNF and p-TrkB was decreased by downregulation of miR-365. The protein expression of BDNF and p-TrkB was increased by knockdown of miR-365, but these effects were reversed by silencing of BDNF (Fig. 4b). The protein levels of α-SMA, Col.IV, and TGF-β1 were decreased in HG + inhibitor NC + sh-BDNF group compared with HG + miR-365 inhibitor + sh-NC group, and increased in HG + miR-365 inhibitor + sh-NC group compared with HG + miR-365 inhibitor + sh-NC group, but these effects were counteracted by the suppression of BDNF (Fig. 4c). In addition, the level of Col.IV was reduced in HG + inhibitor NC + sh-BDNF group compared with HG + miR-365 inhibitor + sh-NC group. Besides, BDNF downregulation abolished the elevated level of Col.IV in HG + miR-365 inhibitor + sh-NC group (Fig. 4d). The data from ELISA unveiled that BDNF depletion resulted in increasing of TNF-α and IL-6 concentrations compared with HG + inhibitor NC + sh-NC, while miR-365 silencing increased the concentrations of TNF-α and IL-6, and these effects were abrogated by BDNF depression (Fig. 4e, f). To sum up, BDNF upregulation by miR-365 knockdown reduced the accumulation of ECM components and secretion of inflammatory cytokines in HG-induced HK-2 cells.
Silencing of miR-365 suppressed the accumulation of ECM components and secretion of inflammatory cytokines in HG-induced HK-2 cells by increasing BDNF expression. a The miR-365 level was evaluated by RT-qPCR. b Western blot analysis was applied to determine the protein levels of BDNF and p-TrkB. c The protein levels of α-SMA, Col.IV, and TGF-β1 werer assessed by western blot analysis. d The expression level of Col.IV was shown by IF staining. e, f The concentrations of TNF-α and IL-6 were determined through ELISA. *P < 0.05
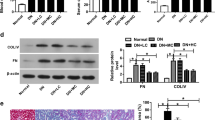
Downregulation of miR-365 promoted BDNF expression to inhibit fibrosis in DN rat model
Subsequently, we investigated whether miR-365 regulated DN fibrosis via regulating of BDNF. RT-qPCR data manifested that the miR-365 level was decreased in Model + miR-365 antagomir + sh-NC and Model + miR-365 antagomir + sh-BDNF groups (Fig. 5a). The protein expression of BDNF and p-TrkB was increased due to downregulation of miR-365 in renal tissues but BDNF silencing reversed these effects (Fig. 5b). What’s more, the Cr, BUN, and urinary albumin levels were reduced by the inhibition of miR-365 but these effects were abolished by BDNF silencing (Fig. 5c–e). Additionally, the decreased levels of TNF-α and IL-6 caused by miR-365 downregulation were abrogated by BDNF interference (Fig. 5f, g). HE staining exhibited that the damaged renal tissues were alleviated by miR-365 downregulation but counteracted by the suppression of BDNF (Fig. 5h). Besides, miR-365 downregulation decreased the area of renal fibrosis but this effect was offset by the downregulation of BDNF (Fig. 5i). IHC staining disclosed that the level of Col.IV was reduced as a result of miR-365 downregulation but BDNF knockdown reversed this effect (Fig. 5j). In conclusion, downregulation of miR-365 promoted BDNF expression to inhibit fibrosis in DN rat model.
Silencing of miR-365 inhibited renal fibrosis in a DN rat model by upregulating BDNF. a, b The expression of miR-365 was determined by RT-qPCR or western blot analysis. c–e The levels of serum Cr BUN and urinary albumin were tested by ELISA. f, g The concentrations of TNF-α and IL-6 was evaluated by ELISA. h HE staining was utilized to the damaged renal tissues. i The renal fibrosis was delineated by Masson staining. j The expression of Col.IV was evaluated by IHC. *P < 0.05 compared with Model + antagomir NC + sh-NC group, #P < 0.05 compared with Model + miR-365 antagomir + sh-NC group
Discussion
DN is a serious kidney disease caused by diabetes [19]. Human health is threated by DN and it has been considered as a vital public issue now. As an ultimate manifestation of DN, renal fibrosis is characterized by tubulointerstitial fibrosis and glomerulosclerosis as well as excessive accumulation of extracellular matrix component [20]. It is reported that renal fibrosis affects 10% people all over the world and among them 50% are patients over 70 years old [21]. According to a study from Zhang et al., the mesangial and fibroblast cell activation as well as monocyte, macrophage, and T cell mediated inflammation and apoptosis are associated with renal fibrosis [22]. Multiple studies confirm the relationship between renal fibrosis and DN. For instance, renal fibrosis is alleviated by wogonin in DN though suppressing NF-κB and TGF-β1-Smad3 signaling pathways [23]. Renal fibrosis is ameliorated by the upregulation of GPR120 in podocytes, thus influences the progression of DN [24]. In early DN, renal fibrosis is prevented by human umbilical cord-derived mesenchymal stem cells [25]. Therefore, renal fibrosis is an effective index to evaluate the serious of DN. Increasing number of patients were influenced by renal fibrosis now, which implies the need to identify novel biomarkers to improve the treatment method for DN.
Previously, increasing studies have unveiled that miR-365 is involved in a variety of diseases. For example, miR-365 promotes the progression of cerebral ischemia injury by downregulating of IGF1R [26]. MiR-365 suppresses proliferation and contributes to apoptosis of synoviocytes in rheumatoid arthritis mice model via inhibition of IGF1 expression through PI3K/AKT/mTOR pathway [27]. MiR-365 suppresses cadmium-mediated hepatoxicity by serving as a sponge of long noncoding RNA MT1DP and targeting Nrf2 [28]. Herein, to assess the role of miR-365 in DN, a DN rat model was established and miR-365 level was found to be upregulated in renal tissues of DN rat model. In addition, HK-2 cells were incubated in HG to build DN cell model. MiR-365 expression was also elevated in HG-induced HK-2 cells. Additionally, miR-365 silencing restrained the protein expression of renal fibrosis associated proteins (α-SMA, Col.IV, and TGF-β1) and inflammation related proteins (TNF-α and IL-6). In summary, the level of miR-365 was increased in renal tissues of DN mice mode and HG-induced HK-2 cells, and downregulation of miR-365 could suppress renal fibrosis and inflammation in DN, suggesting miR-365 suppression could restrain the progression of DN.
MiRNAs are widely recognized to bind with the 3′-untranslated region (3′-UTR) of target mRNA to regulate gene expression at post-transcription level [29]. There are growing numbers of studies showing that miRNAs are involved in the pathogenesis of diseases through targeting specific mRNAs including DN. For example, miR-34b-5p targets progranulin in an LPS-induced acute lung injury mouse model via regulating inflammation and apoptosis [30]. MiR-146b aggravates the progression of osteoarthritis through binding to alpha-2-macroglobulin [31]. MiR-34a-5p and miR-125b-5p inhibits Aβ-induced neurotoxicity in Alzheimer's disease by decreasing the expression of BACE1 [32]. MiR-320a triggers renal dysfunction in DN by targeting MafB [33]. In addition, miR-365 is also validated to regulate some diseases via targeting mRNAs. MiR-365 targets HDAC4 to suppress dexamethasone-mediated amelioration of osteogenesis MC3T3-E1 cells [34]. MiR-365 has been reported to regulate the progression of coronary atherosclerosis by targeting IL-6 [35]. Overexpression of miR-365 accelerates the growth of breast cancer cells by regulating ADAMTS-1 [36]. However, whether miR-365 could target such mRNA to modulate the progression of DN remains to be elucidated. In this work, miR-365 was identified to have potential binding sites for BDNF and this was confirmed by luciferase reporter assay. In a word, BDNF served as a target of miR-365 in DN.
BDNF is a member of neurotrophin family proteins [37]. Tyrosine kinase B (TrkB) is a high affinity receptor of BDNF [38]. BDNF has been reported to involve in the progression of depressive disorder and depression [39]. In addition, a 3-year follow up study shows that BDNF is associated with the development of autism spectrum disorder in children [40]. The levels of BDNF in plasma and serum are associated with neurocognition in patients with chronic schizophrenia patients [41]. Nevertheless, whether BDNF played a role in the progression of DN is still unknown. In the current work, BDNF was validated as a downstream target of miR-365. The expression of BDNF and p-TrkB were significantly downregulated in HG-induced HK-2 cells. In addition, the expression of BDNF and p-TrkB were negatively associated with miR-365 expression. More importantly, BDNF suppression abolished the decreased Cr and BUN levels triggered by miR-365 inhibition. Besides, downregulation of BDNF could reverse the downregulation of miR-365-mediated renal fibrosis and inflammation in HG-induced HK-2 cells and DN rat model. All in all, miR-365 regulated DN fibrosis and renal function via targeting BDNF-TrkB signal axis.
In this study, the mechanism of miR-365 in regulating BDNF-TrkB signal axis in STZ induced DN fibrosis and renal function was explored. It was the first evidence to uncover the mechanism of miR-365 and BDNF in regulating the development of DN. The results delineated that miR-365 aggravated renal fibrosis and inflammation of DN via regulating BDNF-TrkB signal axis. The findings of this study might offer a promising target for improving the treatment of DN.
Availability of data and material
All data generated or analyzed during this study are included in this published article, so the code availability is not applicable.
References
Fan Y, Li X, Xiao W, Fu J, Harris RC, Lindenmeyer M, Cohen CD, Guillot N, Baron MH, Wang N, Lee K, He JC, Schlondorff D, Chuang PY (2015) BAMBI elimination enhances alternative TGF-β signaling and glomerular dysfunction in diabetic mice. Diabetes 64:2220–2233
An L, Ji D, Hu W, Wang J, Jin X, Qu Y, Zhang N (2020) Interference of Hsa_circ_0003928 alleviates high glucose-induced cell apoptosis and inflammation in HK-2 cells via miR-151-3p/Anxa2. Diabetes Metab Syndr Obes 13:3157–3168
Helou N, Dwyer A, Shaha M, Zanchi A (2016) Multidisciplinary management of diabetic kidney disease: a systematic review and meta-analysis. JBI Database Syst Rev Implement Rep 14:169–207
Dai H, Liu Q, Liu B (2017) Research progress on mechanism of podocyte depletion in diabetic nephropathy. J Diabetes Res 2017:2615286
Loeffler I, Liebisch M, Wolf G (2012) Collagen VIII influences epithelial phenotypic changes in experimental diabetic nephropathy. Am J Physiol Ren Physiol 303:F733-745
Tang F, Hao Y, Zhang X, Qin J (2017) Effect of echinacoside on kidney fibrosis by inhibition of TGF-β1/Smads signaling pathway in the db/db mice model of diabetic nephropathy. Drug Des Dev Ther 11:2813–2826
Gross JL, de Azevedo MJ, Silveiro SP, Canani LH, Caramori ML, Zelmanovitz T (2005) Diabetic nephropathy: diagnosis, prevention, and treatment. Diabetes Care 28:164–176
Guo LM, Ding GF, Xu W, Ge H, Jiang Y, Chen XJ, Lu Y (2018) MiR-135a-5p represses proliferation of HNSCC by targeting HOXA10. Cancer Biol Ther 19:973–983
Hoch MJ, Bruno MT, Faustin A, Cruz N, Crandall L, Wisniewski T, Devinsky O, Shepherd TM (2019) 3T MRI whole-brain microscopy discrimination of subcortical anatomy, part 1: brain stem. Am J Neuroradiol 40:401–407
Ye Y, Li M, Chen L, Li S, Quan Z (2020) Circ-AK2 is associated with preeclampsia and regulates biological behaviors of trophoblast cells through miR-454-3p/THBS2. Placenta 103:156–163
Yi L, Zhou L, Luo J, Yang Q (2020) Circ-PTK2 promotes the proliferation and suppressed the apoptosis of acute myeloid leukemia cells through targeting miR-330-5p/FOXM1 axis. Blood Cells Mol Dis 86:102506
Liu H, Wang X, Wang ZY, Li L (2020) Circ_0080425 inhibits cell proliferation and fibrosis in diabetic nephropathy via sponging miR-24-3p and targeting fibroblast growth factor 11. J Cell Physiol 235:4520–4529
Li M, Guo Q, Cai H, Wang H, Ma Z, Zhang X (2020) miR-218 regulates diabetic nephropathy via targeting IKK-β and modulating NK-κB-mediated inflammation. J Cell Physiol 235:3362–3371
Ding H, Xu Y, Jiang N (2020) Upregulation of miR-101a suppresses chronic renal fibrosis by regulating KDM3A via blockade of the YAP-TGF-β-Smad signaling pathway. Mol Ther Nucleic Acids 19:1276–1289
Fan W, Peng Y, Liang Z, Yang Y, Zhang J (2019) A negative feedback loop of H19/miR-675/EGR1 is involved in diabetic nephropathy by downregulating the expression of the vitamin D receptor. J Cell Physiol 234:17505–17513
Zhu Y, Wen X, Zhao P (2018) MicroRNA-365 inhibits cell growth and promotes apoptosis in melanoma by targeting BCL2 and cyclin D1 (CCND1). Med Sci Monit 24:3679–3692
Yang X, Guan Y, Tian S, Wang Y, Sun K, Chen Q (2016) Mechanical and IL-1β responsive miR-365 contributes to osteoarthritis development by targeting histone deacetylase 4. Int J Mol Sci 17:436
Wang J, Zhou T, Liu J, Shangguan J, Liu X, Li Z, Zhou X, Ren Y, Wang C (2020) Application of (1)H-MRS in end-stage renal disease with depression. BMC Nephrol 21:225
Tsai YC, Kuo MC, Hung WW, Wu LY, Wu PH, Chang WA, Kuo PL, Hsu YL (2020) High glucose induces mesangial cell apoptosis through miR-15b-5p and promotes diabetic nephropathy by extracellular vesicle delivery. Mol Ther 28:963–974
Cho MH (2010) Renal fibrosis. Korean. J Pediatr 53:735–740
Humphreys BD (2018) Mechanisms of renal fibrosis. Annu Rev Physiol 80:309–326
Zhang M, Guo Y, Fu H, Hu S, Pan J, Wang Y, Cheng J, Song J, Yu Q, Zhang S, Xu JF, Pei G, Xiang X, Yang P, Wang CY (2015) Chop deficiency prevents UUO-induced renal fibrosis by attenuating fibrotic signals originated from Hmgb1/TLR4/NFκB/IL-1β signaling. Cell Death Dis 6:e1847
Zheng ZC, Zhu W, Lei L, Liu XQ, Wu YG (2020) Wogonin ameliorates renal inflammation and fibrosis by inhibiting NF-κB and TGF-β1/Smad3 signaling pathways in diabetic nephropathy. Drug Des Dev Ther 14:4135–4148
Wei TT, Yang LT, Guo F, Tao SB, Cheng L, Huang RS, Ma L, Fu P (2020) Activation of GPR120 in podocytes ameliorates kidney fibrosis and inflammation in diabetic nephropathy. Acta Pharmacol Sin 42:252–263
Xiang E, Han B, Zhang Q, Rao W, Wang Z, Chang C, Zhang Y, Tu C, Li C, Wu D (2020) Human umbilical cord-derived mesenchymal stem cells prevent the progression of early diabetic nephropathy through inhibiting inflammation and fibrosis. Stem Cell Res Ther 11:336
Chen Y, Liu L, Xing YY, Li Q, Zhao GH, Lu YY, Yang JJ (2020) Down-regulation of miR-365 suppresses cerebral ischemia injury by targeting IGF1R. J Biol Regul Homeost Agents 34:1857–1862
Wang X, Gong S, Pu D, Hu N, Wang Y, Fan P, Zhang J, Lu X (2020) Up-regulation of miR-365 promotes the apoptosis and restrains proliferation of synoviocytes through downregulation of IGF1 and the inactivation of the PI3K/AKT/mTOR pathway in mice with rheumatoid arthritis. Int Immunopharmacol 79:106067
Gao M, Li C, Xu M, Liu Y, Cong M, Liu S (2018) LncRNA MT1DP aggravates cadmium-induced oxidative stress by repressing the function of Nrf2 and is dependent on interaction with miR-365. Adv Sci (Weinh) 5:1800087
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Xie W, Lu Q, Wang K, Lu J, Gu X, Zhu D, Liu F, Guo Z (2018) miR-34b-5p inhibition attenuates lung inflammation and apoptosis in an LPS-induced acute lung injury mouse model by targeting progranulin. J Cell Physiol 233:6615–6631
Liu X, Liu L, Zhang H, Shao Y, Chen Z, Feng X, Fang H, Zhao C, Pan J, Zhang H, Zeng C, Cai D (2019) MiR-146b accelerates osteoarthritis progression by targeting alpha-2-macroglobulin. Aging (Albany NY) 11:6014–6028
Li P, Xu Y, Wang B, Huang J, Li Q (2020) miR-34a-5p and miR-125b-5p attenuate Aβ-induced neurotoxicity through targeting BACE1. J Neurol Sci 413:116793
He M, Wang J, Yin Z, Zhao Y, Hou H, Fan J, Li H, Wen Z, Tang J, Wang Y, Wang DW, Chen C (2019) MiR-320a induces diabetic nephropathy via inhibiting MafB. Aging (Albany NY) 11:3055–3079
Xu D, Gao Y, Hu N, Wu L, Chen Q (2017) miR-365 ameliorates dexamethasone-induced suppression of osteogenesis in MC3T3-E1 Cells by targeting HDAC4. Int J Mol Sci 18:977
Lin B, Feng DG, Wang F, Wang JX, Xu CG, Zhao H, Cheng ZY (2016) MiR-365 participates in coronary atherosclerosis through regulating IL-6. Eur Rev Med Pharmacol Sci 20:5186–5192
Li M, Liu L, Zang W, Wang Y, Du Y, Chen X, Li P, Li J, Zhao G (2015) miR-365 overexpression promotes cell proliferation and invasion by targeting ADAMTS-1 in breast cancer. Int J Oncol 47:296–302
Numakawa T, Suzuki S, Kumamaru E, Adachi N, Richards M, Kunugi H (2010) BDNF function and intracellular signaling in neurons. Histol Histopathol 25:237–258
Barbacid M (1994) The Trk family of neurotrophin receptors. J Neurobiol 25:1386–1403
Kallies G, Rapp MA, Fydrich T, Fehm L, Tschorn M, Terán C, Schwefel M, Pietrek A, Henze R, Hellweg R, Ströhle A, Heinzel S, Heissel A (2019) Serum brain-derived neurotrophic factor (BDNF) at rest and after acute aerobic exercise in major depressive disorder. Psychoneuroendocrinology 102:212–215
Francis K, Dougali A, Sideri K, Kroupis C, Vasdekis V, Dima K, Douzenis A (2018) Brain-derived neurotrophic factor (BDNF) in children with ASD and their parents: a 3-year follow-up. Acta Psychiatr Scand 137:433–441
Heitz U, Papmeyer M, Studerus E, Egloff L, Ittig S, Andreou C, Vogel T, Borgwardt S, Graf M, Eckert A, Riecher-Rössler A (2019) Plasma and serum brain-derived neurotrophic factor (BDNF) levels and their association with neurocognition in at-risk mental state, first episode psychosis and chronic schizophrenia patients. World J Biol Psychiatry 20:545–554
Funding
This research was supported by the Zhejiang Health Science and Technology Project (2021KY1130).
Author information
Authors and Affiliations
Contributions
PZ: methodology, software, validation, and data curation. XL: methodology, software, validation, and writing-original draft. YL: software, validation, and methodology. JZ: software, methodology, and writing—review and editing. YS: conceptualization, methodology, investigation, and supervision. JH: conceptualization, methodology, investigation, and supervision. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
There are no conflicts of interest concerning this article.
Ethics approval
The experiments involved in animals were performed according to the National Institutes of Health guide for the care and use of Laboratory animals.
Consent for publication
All authors agree to publish this article to this journal.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhao, P., Li, X., Li, Y. et al. Mechanism of miR-365 in regulating BDNF-TrkB signal axis of HFD/STZ induced diabetic nephropathy fibrosis and renal function. Int Urol Nephrol 53, 2177–2187 (2021). https://doi.org/10.1007/s11255-021-02853-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11255-021-02853-3