Abstract
Leptospirosis is a zoonosis of global distribution, caused by the infection of pathogenic Leptospira, a group of bacteria capable of infecting both domestic and wild animals. Mink (Neovison vison) in southern Chile is recognized as a wild and synanthropic rodent predator (among various other prey), and Leptospira infection in them can be acquired through contact with the pathogen in the environment or by eating infected prey. Thus, the aim of this study was to provide more specifics regarding the source of the infection for the American mink under the conditions of Southern Chile. Minks were captured in the Los Ríos region, southern Chile, in an area with well-developed dairy farming. Two areas were selected for mink trapping, one with a high degree of dairy farming and a second with a low degree of dairy farming. Within them, 16 study sites were visited, and 45 American mink were trapped and euthanized to obtain kidney tissue and blood serum samples for bacteria isolation and determination of antibodies titers, respectively. Molecular characterization of the isolated strains was performed. Three minks from sites of high-dairy farming industry and only one from sites with low-degree dairy farming were detected as infected through molecular confirmation. This study shows evidence that confirms previous findings made in southern Chile, regarding mink as host of Leptospira interrogans serovar Hardjo-prajitno associated to cattle-farming areas. However, typing information (Leptospira interrogans Copenhageni and Icterohaemorrhagiae) suggests that the consumption of rodents may also be a potential source of infection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Anthropogenic changes to the landscape increasingly threaten the survival of wild species, and recently, more records exist of the negative impacts of introduced alien species on endemic populations through the co-introduction of parasites or pathogens in a process termed “pathogen pollution” (Daszak et al. 2000; Cunningham et al. 2003; Medina-Vogel 2010). A good example of pathogen pollution in aquatic ecosystems is leptospirosis, one of the most important zoonotic bacterial diseases in animals and the most widely distributed in the world (Adler and de la Peña 2010). This infection is caused by spirochetes of the genus Leptospira and is characterized by fever, renal and hepatic insufficiency, lung diseases, and reproductive disorders (Guerra 2009; Adler and de la Peña 2010).
Water or moist soil contaminated with the urine of infected animals is known to be the main vehicle of inter- and intra-species transmission of pathogenic Leptospira (Faine 1994). The main transmission routes are through injured skin and via long periods of exposure to contaminated water or soil. The possibilities for transmission through direct contact within animal populations include sexual contact or artificial insemination (Faine 1994). However, predation of infected hosts (mostly rodents) could also be a transmission route (Reilly et al. 1970; Shophet and Marshall 1980; Cosson et al. 2014). Therefore, it has been suggested that the reduction of wildlife habitats favors spillback and spillover processes at the wildlife-livestock-human interface possibly leading to the emergence of leptospirosis in some geographic areas (Matthias et al. 2005; Matthias et al. 2008). Confirming the latter, Derne et al. (2011) observed that, in islands with higher biodiversity and lower human intervention, there were less cases of human leptospirosis. On the opposite, there are some geographic areas that promote the presence and the persistence of Leptospira in the ecosystem. Such is the case of the Tuscany area, as well as all of Central Italy, wherethe close contact between domestic and wild animals plus hunting activity and a wet environment makes pathogenic Leptospira spread favorable. An example of the latter is the situation of wild boar in the epidemiology of leptospirosis among wildlife in Central Italy (Cilia et al. 2020). Thus, the role of human intervention on ecosystems and its effect on the ecology of diseases is an ongoing discussion.
Considering the epidemiological aspects of this infection, wild animals with semi-aquatic habits would have a greater exposure to pathogenic Leptospira from a source that may be associated with domestic animals (Gaydos et al. 2007; Barros et al. 2014). Among others, the American mink (Neovison vison) meets this condition. American mink in Chile is considered an invasive species, and its current distribution is throughout a great part of the Patagonian territory with an apparent northern distribution now reaching the Malleco River approximately at 39° 60′ South parallel. Its introduction to Argentina and Chile occurred in the 1930s in Patagonia because of the liberation of individuals from the fur industry (Rozzi and Sherriffs 2003). The first infectious link between this invader and pathogenic Leptospira was reported in a study of European mink (Mustela lutreola) and other small carnivores in southwestern France (Moinet et al. 2010), where they found high seroprevalence and pathogen detection using molecular tools. In Chile, molecular detection of pathogenic Leptospira was also reported from renal tissue of captured American minks (Barros et al. 2014), suggesting two sources of infection: rodents, as they are a regular item of the mink diet in the region (Medina 1997; Medina-Vogel et al. 2013), and livestock production due to the proximity to cattle herds. American mink could then be an important link in the ecology of Leptospira, mainly due to its wide distribution, semi-aquatic life, and diet (Medina 1997; Millán et al. 2009; Moinet et al. 2010; Sepulveda et al. 2014). Considering that Leptospira infection rate of cattle in southern Chile has been reported as high, estimating up to 75% of dairy herds infected in the study region, and that it has clear negative effects on the cattle’s health (Salgado et al. 2014; Salgado et al. 2015), the present study aimed to provide more specifics regarding the source of the infection for the American mink under the conditions of Southern Chile.
Materials and methods
Study area
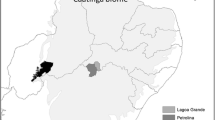
Between March and September 2017, a trapping campaign to catch American minks was performed in the Los Rios region, Chile, which extends approximately from the 39°17′ south up to the 40° 41′ south parallel, and the 73° 43′ west, up to the 71° 35’ West. Through geoprocessing (geospatial information system, GIS), the location of cattle herds registered on the Chilean agriculture and livestock service database was associated with the area from where each mink was captured. This methodology allowed for the classification of five trapping sites as areas with a high degree of bovine herd concentration (mainly represented by a high density of dairy cattle operations), while the other 14 sites were assessed as having a low degree of dairy farming (Fig. 1), characterized for subsistence-farming activities with less human intervention and significantly smaller numbers of domestic animals, in comparison with the first described hunting area.
GIS that represents the mink capture area in the Los Ríos District. Mink capture areas associated with high degree of bovine herd concentrations (white squares) and low degree bovine herds (white circles). Bovine herds are represented with white rhombus. Native forests, exotic forest exploitations, and mixed forests are represented by dark gray polygons for spatial representation and graphical interpretation to explain the concentration of the bovine herds within the central valley of the district
Trapping permits
The mink trapping was associated with a regional American Mink control initiative executed by the “Servicio Agricola y Ganadero” (Agriculture and Livestock Service) which is undertaking a regional effort to control this invasive species which is considered as “detrimental to agriculture and biodiversity,” therefore considered by Chilean law as a detrimental species that can be hunted on all the Chilean territory without quotas or hunting seasons (Chilean law N° 19.473, article 6 of the current regulation). The animals used on this research were culled under the regional mink control initiative, and the carcasses were delivered for research purposes; therefore, no special permits are required for research purposes according to the current regulation. Minks were trapped alive with modified Tomahawk traps with a single entrance.
Sampling
Each trapped animal was euthanized on site (to reduce stress) with an overdose of ketamine hydrochloride (100 mg per animal) via intra-cardiac injection following full sedation obtained by a previous intra-muscular injection of a combination of 100 mg of ketamine hydrochloride and 10 mg of xylazine hydrochloride (standard dose per individual). After this procedure, 5 ml of blood was obtained directly from the heart with a sterile blood collection tube. The trapped animals were then transported directly to the Instituto de Patología at the Universidad Austral de Chile (UACh) for necropsy, and during that procedure, the one kidney was extracted from each euthanized mink and deposited in a sterile 50-ml centrifuge tube. All samples were labeled with the animal’s number, date, and place of origin. Samples were kept at room temperature until they were transferred (less than 2 h) to the Infectious Diseases Lab, at the Instituto de Medicina Preventiva Veterinaria, UACh. In most cases, samples were immediately processed for the tissue culture protocol. A smaller number of samples (7) were refrigerated for a maximum of 2 days until processing. Blood samples were centrifuged upon arrival at the lab to extract the serum and then kept refrigerated until processed on the same day. Animals were handled strictly according to the recommendations in the university’s guide on the use of animals for research (https://www.uach.cl/organizacion/vicerrectoria-investigacion-desarrollo-y-creacion-artistica/utiles/subcomite-en-uso-de-animales-en-investigacion); however, it is important to note that the mink trapping was associated with a regional American Mink control initiative.
Diagnostic approach
Sera were analyzed using MAT (microscopic agglutination test) for the presence of antibodies anti-six Leptospira serovars according to a published protocol (Salgado et al. 2014). Kidney tissue was processed according to Faine et al. (1999). Briefly, 200 μl of the processed sample in four replicates was cultured in EMJH (Ellinghousen and McCullough and modified by Johnson and Harris) medium at 29 °C. The cultures were checked once a week for 13 weeks, and a positive result was reported with a typical Dinger ring growth. Thereafter, positive cultures were submitted for the DNA extraction-purification protocol using the High Pure PCR Template Preparation Kit (Roche), following the manufacturer instructions. DNA templates obtained from the above protocol were analyzed in a qPCR system (Roche LightCycler 2.0), using a TaqMan probe and targeting the LipL32gen (Stoddard et al. 2009), which is specific only for pathogenic Leptospira species. The amplification mixture for each sample: 0.7-μM primers, 0.15-μM probe, 10-μl Master Mix TaqMan Universal (Roche), and 5-μl DNA template, in a total volume of 20 μl. Samples were amplified with the following program: initial denaturation at 95 °C for 2 min, followed by 40 cycles of denaturation for 5 s at 95 °C and annealing/elongation for 30 s at 58 °C. The PCR protocol considered a negative and positive control to survey the proficiency of the reaction as well as negative and positive controls for the DNA extraction.
The “Multi Locus Sequence Typing” (MLST) method was used to molecularly characterize strains obtained from positive cultures. MLST was performed according to a protocol described by Thaipadungpanit et al. (2007), since it demonstrates the utility of this genotyping method for characterizing Leptospira isolates from a clinical perspective. Specifically, this protocol used the following housekeeping genes: glmU, pntA, sucA, tpiA, pfkB, mreA, and caiB. Briefly, PCR amplification was carried out using bacterial (Leptospira) DNA obtained from positive mink kidney cultures, where the initial denaturation step was 94 °C for 5 min, followed by 35 cycles of 94 °C for 30 s; annealing/elongation was at 50 °C for 60 s for all genes; extension was at 72 °C for 50 s; and then the final extension was at 72 °C for 7 min. The PCR products were purified using the Geneaid™ “Gene/DNA Genetic Extraction” Kit, and sequencing was done in both directions with primers initially used for PCR amplification. Sequencing was performed using the sequencer kit “BigDye Terminator v. 3.1 Cycle Sequencing Kit (ABI)”, and the automated DNA sequencer “ABI Prism 3130xl Genetic.” The sequences were analyzed using the Chrommas and BioEdit programs, and these were derived to the international database of free use (https://pubmlst.org/leptospira/) to obtain the allelic profile and assign the sequence type (ST).
Statistical analysis
Data was not normally distributed, so frequencies of positive culture results for each study area and sex were compared using Fisher exact test. The level of significance was alpha < 0.05.
Results
Twenty-five (55.6%) minks were trapped in low-density dairy farming and 20 (44.4%) in the area of high-density dairy farming. Twenty-nine (64.4%) were males, and 16 (35.5%) were females (Table 1).
None of the hunted animals showed serological positive results to any Leptospira spp. serovars. Four (8.8%) out 45 kidney samples clearly showed the characteristic Dinger ring indicative of Leptospira bacterial growth, and in all these, the presence of the LipL32 gene was confirmed by qPCR.
Although a higher proportion of animals with positive culture was observed in high density of dairy farming (3) than the area of low-density dairy farming (1), this difference was not statistically significant (P = 0.28) (Table 2). In the first area, the typing information showed that the specific species were Leptospira interrogans serovar Hardjo-prajitno and Leptospira interrogans Copenhageni and Icterohaemorrhagiae. In the area of low-density farming activity, just one positive culture sample showed Leptospira interrogans Copenhageni and Icterohaemorrhagiae. Similarly, although more females were positive (3) than males (1), this difference was not significant (P = 0.09).
The MLST analysis of the four isolates revealed that three of them presented the ST-17 while one isolate belonged to the ST-20.
Discussion
The present study provided new information regarding the role of American mink in the epidemiology of Leptospira infection as sentinels of semiaquatic wildlife in Patagonia.
Only one of the two kidneys of each animal was subjected to bacterial culture, since to the authors’ knowledge, there is no published evidence about increasing sensitivity of culture to growth pathogenic Leptospira after using both the kidneys.
The success of capture of male individuals was higher than females, probably since the former has a wider home environment, associated with more excursive behaviors, associated with territoriality and looking for mating, compared with females (Medina-Vogel et al. 2015). Although males move around more than females, the difference is not large enough to make any conclusion. Female home range is above the 60% of male home range. Furthermore, mink have intersexual territoriality in a relative linear habitat, so movements depend on neighbors and the season, as during reproduction, the territorial system breaks down. Also, the landscape characteristics are very important. So, we do not believe that we have data enough to make any conclusion about the difference between females and males.
The fact that none of the animals showed a serological response could be due to an early stage of the infectious process, especially in the four infected animals. According to the biology of this infection, the presence of antibodies cannot be detected before 15 to 20 days after the infection has occurred (Zuerner 2015). Another reason to explain the obtained results is the possibility that other serogroups that could be involved are missed in our MAT battery. The serologic (MAT) diagnostic battery used in this study consisted of 6 serogroups used routinely for different animal species (Salgado et al. 2014). It is important to highlight that there are more than 24 serogroups that cover more than 250 serovars. Finally, the lack of knowledge on the mink immune system response against this infection should also be considered. For example, a dilution of 1:100 in MAT may overestimate the immune response and makes it insufficient for its detection by this serologic tool.
The lower detection rate of pathogenic Leptospira, in relation to what was reported in the study by Barros et al. (2014), can be explained by the fact that they used direct PCR for the diagnostic pathogen detection, which implies a higher diagnostic sensitivity, mainly due to a detection limit and contamination problems for culture. However, direct PCR could not confirm the presence of live Leptospira in urine specimen. Therefore, the interpretation of this finding is not conclusive or confirmatory of active infection. On the contrary, in the present study, one can infer that in those 4 animals with positive culture results, the infection was active since the pathogen could be isolated, and as such, there was the probability of dissemination by urine due to its viability in renal tissue.
The highest proportion of individuals with positive results to the bacteriological culture is reported in the macro area of greater human intervention. In this area, a large bovine dairy mass is concentrated specifically in the areas of Puerto Trumao and Cocule in the region of Los Ríos (see Fig. 1). The discussion based on this finding should alert us, despite the result of the statistical analysis, to the fact that, in this area, the reported infection rate for the dairy cattle population is considered high (Salgado et al. 2014). In addition, the trend of a higher proportion of positive results in this area would suggest that cattle could act as a source of pathogenic Leptospira infection for the region’s mink population and for other semiaquatic mammals too, such as the endangered Southern river otter (Lontra provocax). Complementing these findings, the MLST technique has reported Leptospira interrogans serovar Hardjo-prajitno and Leptospira interrogans serovars Copenhageni and Icterohaemorrhagiae. The first serovar is directly associated with cattle, being responsible for severe clinical disease in this animal species (Ellis 1994). The abortion rate after Leptospira borgpetersenii Hardjobovis infection is 3 to 10%, whereas the rate increases up to 30% for Leptospira interrogans serovar Hardjo-type Hardjo-prajitno (Li Hardjo-prajitno) infection (Koizumi and Yasutomi 2012). In Chile, the virulence and severity of Leptospira interrogans serovar Hardjo-pratijno have been demonstrated in cattle from the Los Ríos region (Salgado et al. 2015). The results of the present study also allow us to consider that, although the rate of positive results was lower in the animals caught in lesser intervention area, the presence of the infection was also confirmed there.
Interestingly, three out of four isolates informed in this study were typed as Leptospira interrogans serovars Copenhageni and Icterohaemorrhagiae belonging to ST-17, which opens the interpretation about the potential source of infection, taking into account that strains with this genotype in the PubMLST database have been isolated from rats, canines, porcines, and humans in countries of South America, Europe, and Asia (last accessed September 23, 2020). The latter suggests that predation of infected hosts (mostly rodents) could also be a transmission route (Reilly et al. 1970; Shophet and Marshall 1980; Cosson et al. 2014). Medina (1997) helps to support this discussion since minks practice predation of both synanthropic and wild rodents, which are recognized as reservoirs of pathogenic Leptospira species. Also, recently, ST-17 was the predominant genotype in dogs in a study conducted in Italy (Bertasio et al. 2020), and it was confirmed as the major cause of canine leptospirosis. Interestingly, the same MLST genotype was also informed in the PubMLST database in rats, mice, cats, and some farm animals. Free-roaming dogs possibly play an important role in the dissemination of pathogenic Leptospira to sympatric wildlife.
The demographic explosion of mink causes them to be in direct or indirect contact with different domestic and wild animals, and human populations suggest that the transmission of the pathogen is possible. They, therefore, constitute a risk to public health affecting the biodiversity of the native wildlife, being a probable focus of dissemination of this and other pathogens present in domestic animals towards wildlife and vice versa.
As far as we are aware, the STs described here correspond to the first Chilean genotypes deposited to the Leptospira MLST database, and it is hoped that this information will be a contribution for the organisms involved in health and environmental protection, so that they can be considered in future control plans for this species, as a tool to mitigate the negative effects that this alien species exerts in the region and in the country.
References
Adler, B., de la Peña, A. 2010. Leptospira and leptospirosis, Vet Microbiol,140, 287-296
Barros, M., Sáenz, L., Lapierre, L., Nuñez, C., Medina-Vogel, G. 2014. High prevalence of pathogenic Leptospira in alien American mink (Neovison vison) in Patagonia, Rev. Chil. Hist. Nat, 87,19
Bertasio, C. , Boniotti, M.B., Lucchese, L., Ceglie, L., Bellinati, L., Mazzucato, M., Furlanello, T., D’Incau, M. Natale, Alda. 2020 Detection of New Leptospira Genotypes Infecting Symptomatic Dogs: Is a New Vaccine Formulation Needed? Pathogens, 9, 484.
Cilia, G., Bertelloni, F., Angelini, M., Cerri, D., Fratini, F. 2020. Leptospira Survey in Wild Boar (Sus scrofa) Hunted in Tuscany, Central Italy. Pathogens, 9, 377.
Cosson, J.F., Picardeau, M., Mielcarek, M., Tatard, C., Chaval, Y. 2014. Epidemiology of Leptospira Transmitted by Rodents in Southeast Asia, PLoS Negl Trop Dis, 8, e2902
Cunningham, A.A., Daszak, P., Rodriguez, J.P. 2003. Pathogen pollution: defining a parasitological threat to biodiversity conservation, J Parasitol, 89, S78-S83
Daszak P., Cunningham, A.A., Hyatt, A.D. 2000. Emerging infectious diseases of wildlife-threats to biodiversity and human health, Science, 287, 443-449
Derne, B.T., Fearnley, E.J., Lau, C.L., Paynter, S., Weinstein, P. 2011. Biodiversity and leptospirosis risk: A case of pathogen regulation? Med Hypothesis, 77, 339-344
Ellis, W. 1994. Leptospirosis as a cause of reproductive failure, Vet Clin North America Food Animal Pract, 10, 463-78
Faine, S. 1994. Leptospira and Leptospirosis, CRC Press Inc.: Boca Raton
Faine, S., Adler, B., Bolin, C., Perolat, P. 1999. Leptospira and leptospirosis, 2nd ed. Melbourne: MediSc
Gaydos, J.K., Conrad, P.A., Gilardi, K.V., Blundell, G.M., Ben-david, M. 2007. Does human proximity affect antibody prevalence in marine-foraging river otters (Lontra canadensis)? J Wildl Dis, 43, 116-123
Guerra, M. 2009. Leptospirosis, J Am Vet Med Assoc, 234, 472-478
Koizumi, N., Yasutomi, I. 2012. Prevalence of leptospirosis in farm animals, Jpn J Vet Res, 60, 55-58
Matthias, M.A., Díaz, M.M., Campos, K.J., Calderon, M., Willig, M.R., Pacheco, V., Gotuzzo, E., Gilman, R.H., Vinetz, J.M. 2005. Diversity of bat-associated Leptospira in the Peruvian Amazon inferred by bayesian phylogenetic analysis of 16S ribosomal DNA sequences, Am J Trop Med Hyg, 73, 964-974
Matthias, M.A., Ricaldi, J.N., Cespedes, M., Diaz, M.M., Galloway, R.L., Saito, M., Steigerwalt, A.G., Patra, K.P., Ore, C.V., Gotuzzo, E., Gilman, R.H., Levett, P.N., Vinetz, J.M. 2008. Human leptospirosis caused by a new, antigenically unique Leptospira associated with a Rattus species reservoir in the Peruvian Amazon, PLoS Negl Trop Dis, 2, e213
Medina, G. 1997. A comparison of the diet and distribution of southern river otter (Lutra provocax) and mink (Mustela vison) in Southern Chile, J. Zool, 242, 291-297
Medina-Vogel, G. 2010. Ecología de Enfermedades Infecciosas Emergentes y Conservación de Especies Silvestres, Ecology of Emerging infectious diseases and Wildlife Conservation, Arch Med Vet, 42, 11-24
Medina-Vogel, G., Barros, M., Organ, J., Bonesi, L. 2013. Evidence of competition between the Southern river otter and the alien invasive North American mink in marine habitats of southern Chile, J. Zool, 290, 27-34
Medina-Vogel, G., Barros, M., Monsalve, R.J., Pons, D. 2015. Assessment of the efficiency in trapping North American Mink (Neovison vison) for population control, Rev. Chil. Hist. Nat, 88, 1-12
Millán, J., Candela, M.G., López-Bao, J.V., Pereira, M., Jiménez, M.A., León-Vizcañino, L. 2009. Leptospirosis in wild and domestic carnivores in natural areas in Andalusia, Spain Vector-Borne Zoonot, 9, 549-554
Moinet, M., Fournier-Chambrillon, C., Fontaine, G.A., Aulagnier, S., Mesplède, A., Blanchard, B. 2010. Leptospirosis in free-ranging endangered European mink (Mustela lutreola) and other small carnivores (Mustelidae, Viverridae) from southwestern France, J Wildlife Dis, 46, 1141-1151
Reilly, J.R., Hanson, L.E., Ferns, D.H. 1970. Experimentally induced predator chain transmission of Leptospira grippotyphosa from rodents to wild marsupialia and carnivore, Am J Vet Res, 31, 1443-1448
Rozzi, R., Sherriffs, M. 2003. El visón (Mustela vison schreber, carnívora: Mustelidae), un nuevo mamífero exótico para la isla Navarino, An. Inst. Patago, 31, 97-104
Salgado, M., Otto, B., Sandoval, E., Reinhardt, G., Boqvist, S. 2014. A cross sectional observational study to estimate herd level risk factors for Leptospira spp. serovars in small holder dairy cattle farms in southern Chile, BMC Vet Res, 10, 126
Salgado, M., Otto, B., Moroni, M., Sandoval, E., Reinhardt, G., Boqvist, S. 2015. Leptospira interrogans serovar Hardjoprajitno isolated from a calf with clinical leptospirosis, BMC Vet Res, 11, 66
Sepulveda, M.A., Singer, R.S., Silva-Rodríguez, E.A., Eguren, A., Stowhas, P., Pelican, T. 2014. Invasive American Mink: Linking Pathogen Risk Between Domestic and Endangered Carnivores, EcoHealth, 11, 409-419
Shophet, R., Marshall, R.B. 1980. An experimental induced predator chain transmission of Leptospira ballum from mice to cats, Br Vet J, 136, 265-270
Stoddard, R.A., Gee, J.E., Wilkins, P.P., McCaustland, K., Hoffmaster, A.R. 2009. Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the LipL32 gene, Diagn Microbiol Infect Dis, 64, 247-255
Thaipadungpanit, J., Wuthiekanun, V., Chierakul, W., Smythe, L., Petkanchanapong, W., Limpaiboon, R., Apiwatanaporn, A., Slack, A., Suputtamongkol, Y., White, N., Feil, E., Day, N., Peacock, S. 2007. A dominant clone of Leptospira interrogans associated with an outbreak of human leptospirosis in Thailand, PLoS Negl Trop Dis, 1,56
Zuerner, R.L. 2015. Host response to leptospira infection, Curr Top Microbiol Immunol, 387, 223-50
Acknowledgments
This study was possible thanks to the regional government of Los Ríos and the “Programa De Control Comunitario Del Visón en la Región De Los Ríos” (community mink control program in the Los Ríos Region) IDI 30376022 and the Agriculture and Livestock service of the Los Ríos region. Special thanks to Carla Marchese and Diego Gallardo (veterinarians of the mink control initiative) for mink capture and sampling that made this study possible.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Statement of animal rights
The authors declare that the present study does not contain clinical studies or patient data. Animals were handled strictly according to the recommendations in the Universidad Austral de Chile’s guide on the use of animals for research (https://www.uach.cl/organizacion/vicerrectoria-investigacion-desarrollo-y-creacion-artistica/utiles/subcomite-en-uso-de-animales-en-investigacion). Since American Mink in Chile is considered an invasive species, therefore, it can be hunted on all the Chilean territory without quotas or hunting seasons (Chilean law No. 19.473, article 6 of the current regulation).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Alfaro, M.A.S., Raffo, E., Bustos, M.I. et al. New insights on the infection of pathogenic Leptospira species in American mink (Neovison vison) in southern Chile. Trop Anim Health Prod 53, 2 (2021). https://doi.org/10.1007/s11250-020-02469-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11250-020-02469-2