Abstract
The aim of the current study is to diagnose Brucella spp. infection using methods such as serology, bacterial isolation, and molecular analysis in buffaloes bred in Maranhão State. In order to do so, 390 samples of buffalo serum were subjected to serological tests, to Rose Bengal Plate Test (RBPT) and to 2-mercaptoethanol (2-ME) combined with slow agglutination test (SAT). Vaginal swabs were collected from seropositive animals and subjected to bacterial isolation and to generic PCR. According to the serological test, 16 animals had a positive reaction to the confirmatory test (2-ME/SAT). As for bacterial isolation, three samples resulted in the isolation of Brucella spp.-characteristic colonies, which were confirmed through PCR. These results confirmed Brucella spp. infection in the buffalo herd from Maranhão State.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bovine Brucellosis is a zoonosis distributed worldwide. It has great impact on developing countries due to losses in animal production (Santos et al. 2013) and to the effects of this infection on human health (Franco et al. 2007; Galińska and Zagórski 2013). The disease is caused by facultative intracellular gram-negative coccobacilli, which are bacteria belonging to genus Brucella. The infection caused by Brucella abortus has been reported in domestic buffaloes (Bubalus bubalis) in Brazil (Chaves et al. 2012; Silva et al. 2014; Sousa et al. 2015) and worldwide (Abubakar et al. 2010; Fosgate et al. 2011; Martínez et al. 2014).
Buffaloes show rusticity and great adaptability to different topographies, soils, and climatic factors. In addition, these animals produce meat and milk, which represents a good breeding alternative for the species, mainly in tropical countries. The population of buffaloes in South America is estimated at approximately 4 million animals; 3.5 million of them are bred in Brazil. The contact between buffaloes and cattle or other domestic and wild animals, as well as their access to different ecosystems, has exposed the species to different infectious diseases such as brucellosis (Paulin and Ferreira Neto 2008).
Brucella spp. enter the body of mammals through the mucous membranes of the digestive, genital, or nasal tracts; through the ocular conjunctiva; or through solutions of continuity of the skin. Their main gateway in bovines is the oropharyngeal mucosa. These bacteria are carried from the upper digestive tract to the lymph nodes, and they are mainly phagocytized by macrophages, wherein they may remain quiescent for months (Poester et al. 2013).
The disease may be diagnosed through direct and indirect methods. The direct methods comprise isolating and identifying the etiological agent in the material obtained from the suspect animal, such as tissues from aborted fetuses, placenta, vaginal exudates, and milk (Brasil 2004; Mol et al. 2012). Isolating the agent is the safest diagnostic method; however, it presents difficulties concerning sample collection and conservation, as well as those concerning the implementation procedures of the technique. On the other hand, the indirect or serological methods consist in detecting antibodies in serum, milk, and seminal plasma (Brasil 2004). According to Molnár et al. (2002), the serological methods are one of the main bases used to support brucellosis control programs. The correct diagnosis of brucellosis gives support and assurance to the implementation of eradication programs.
Thus, it is worth gathering data on brucellosis in buffaloes in order to use them as subsidy in the implementation of appropriate programs to control the disease in this species. Thus, given the lack of studies on brucellosis in buffalo species, the aim of the current study is to set the diagnosis of Brucella spp. infection through serological, microbiological, and molecular methods applied to buffaloes in Maranhão State, Brazil.
Materials and methods
Samples
The study was conducted in nine counties (Arari, Cajapió, Cajari, Matinha, Palmeirândia, São Bento, São João Batista, Viana e Vitória) located in the northwestern region of Maranhão State, Brazil. The study site was selected according to the highest concentration of buffalo herds in these counties (Santos et al. 2016).
Drawings were conducted in order to select the herds to be sampled, which should present prerequisites such as extensive farming system, number of female buffaloes equal to or higher than 20, and no history of previous vaccination against brucellosis. Thirteen herds were selected according to pre-established criteria. The Win Episcope 2.0 software (Blas et al. 2004) was used to estimate the number of blood samples collected in the herds, with recommended prevalence of at least 4% (Brasil 2006) and 95% probability of detecting at least one seropositive animal. Thus, 390 female buffaloes were sampled according to these criteria. In addition, vaginal swabs were collected to isolate Brucella spp. and to perform molecular analysis in the animals whose serological tests came out positive. The experimental procedures adopted in the current study were approved by the Ethics Committee on Animal Experimentation (CETEA-UEMA), under the Protocol No. 018/08.
Serological tests
The Rose Bengal Plate Test (RBPT) was performed as a screening test in order to detect anti-B. abortus antibodies. It was done using an antigen produced by the Technology Laboratory of Paraná State – (TECPAR - Laboratório de Tecnologia do Paraná). The RBPT-reacting samples were simultaneously subjected to 2-mercaptoethanol (2-ME) and to slow agglutination test (SAT) using the antigen produced by TECPAR, according to the following titrations: 1:25, 1:50, 1:100, and 1:200. The results were interpreted according to the Brazilian legislation in place (Brasil 2004). The results of the serological tests were delivered to the breeders, who were informed about the need and the importance of conducting microbiological and molecular diagnoses. Those who expressed the interest in participating in this stage of the study had the vaginal swab of their seropositive animals collected.
Bacterial isolation
Vaginal swab samples were used for bacterial isolation. All samples were handled in a level-3 biosafety laboratory. The collected material was resuspended in 1 mL sterile PBS and centrifuged at 12,000g for 5 min. The supernatant was discarded and the pellet was resuspended in 300 μL PBS 1X. Subsequently, 100 μL of the homogenate was placed on trypticase soy agar (TSA) plates and 100 μL of it was placed in trypticase soy broth (TSB) tubes added with selective supplement for Brucella (Himedia, Brasil) containing polymyxin B sulfate (2,500 IU), bacitracin (12,500 IU), nystatin (50,000 IU), cycloheximide (50 mg), nalidixic acid (2.5 mg), and vancomycin (10 mg). Then, the tubes and the plates were incubated in a kiln at 37 °C, under 5% CO2. The subcultures in the TSA plates were conducted in duplicate, using the broth in the 7th, 14th, and 21st post-incubation days. All plates were incubated for up to 21 days. They were monitored every 48 h in order to assess the possible presence of colony-forming units (CFU) with Brucella-compatible features.
The bacterial growth with Brucella-compatible features found in the TSB broth and on the TSA plates was inactivated in water bath at 85 °C for 30 min and subsequently subjected to molecular analysis.
DNA extraction
The guanidine isothiocyanate technique was used to extract DNA from the bacterial cultures, according to the procedures described by Pitcher et al. (1989). Then, DNA purity and concentration were checked though spectrophotometry (Sambrook and Russel 2001) and the DNA was stored at −20 °C, until the polymerase chain reaction (PCR) was carried out.
Polymerase chain reaction
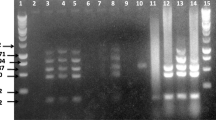
The current study used the B4 (5′-TGGCTCGGTTGCCAATATCAA-3′) and B5 (3′-CGCGCTTGCCTTTCAAGGTCTG-5′) primers, which amplify the bcsp 31 gene. Such a gene encodes a cell surface protein, which is found among the species from genus Brucella, according to Baily et al. (1992). The reaction was performed at the final volume of 25 μL containing 2 μL of extracted DNA (50 ng/μL); 0.25 μL of Taq DNA polymerase 5 U/μL (Phoneutria); 0.75 μL of MgCl2 (50 mM); 2.5 μL of each primer (10 μM); 2.5 μL of dNTP (2 mM); 2.5 μL of IB 10× Buffer [100 mM of Tris HCL; 15 mM of MgCl2; 500 mM of KCl] (Phoneutria); and 12 μL of H2O. The amplification was performed through Hot Start cycle at 94 °C for 3 min; denaturation at 94 °C for 30 s; annealing at 60 °C for 30 s; extension at 72 °C for 30 s; and final extension at 72 °C for 10 min. These primers promote the amplification of a product with 223 base pairs (bp), which may be analyzed through 1.5% agarose gel electrophoresis containing 5 μg/ml of ethidium bromide.
Results
According to the RBPT screening test, 21 (5.38%) out of the 390 analyzed serum samples were seropositive, and 16 (4.10%) of them were confirmed through 2-ME in combination with SAT (Table 1).
Seven vaginal swab samples from seropositive animals were subjected to bacterial isolation and three of them showed growth of round, convex, translucent, and whitish colonies characteristic of Brucella spp. (Alton et al. 1988). The PCR confirmed that these three isolates belonged to the genus Brucella. Alternatively, all samples showed bacterial growth in the TSB broth, and all vaginal swab samples tested positive for Brucella spp. (B4 / B5) when they were subjected to PCR (Fig. 1).
Table 2 summarizes the serological, microbiological, and molecular results found in the current study.
Discussion
This is the first report of Brucella spp. isolation in buffaloes bred in Maranhão State, Brazil. Buffaloes (B. bubalis) comprise an important domestic species in many tropical countries, and yet, the literature on brucellosis in this species remains very limited. According to the RBPT screening test conducted in the current study, 5.38% (21/390) of the animals were diagnosed as positive, and the diagnosis of 76.2% (16/21) of these seropositive animals was confirmed through 2 ME/SAT tests. Thus, the serological results showed five animals as false-positive through the 2-ME test, a fact that reinforced the importance of using both techniques to confirm the diagnosis of brucellosis. The false-positive reactions may due to the occurrence of non-specific antibodies found in infections caused by other bacteria, such as Yersinia enterocolitica O:9, Salmonella sp., Escherichia coli O:157, or Pseudomonas sp. (Nielsen et al. 2007; Bounaadja et al. 2009). In addition, they may result from the immunization with the B19 vaccine after the recommended age (Brasil 2004). However, all animals analyzed in the current study had not been vaccinated.
The seropositivity rate (4.10%) found in the current study was similar to the official data, which indicate between 4 and 5% prevalence of seropositive animals for brucellosis (Brasil 2006). The occurrence of brucellosis in buffaloes may due to the handling procedures implemented in the studied region. All the animals in the current study are extensively bred in flooded areas and have migratory and gregarious habits. They are transferred to other lands and put in contact with other animal groups, a fact that increases the possibility of spreading the disease, according to Wray (1975) and Marques and Cardoso (1997). These researchers stated that the use of large areas provides continuous access to different types of ecosystems and allows the animals to graze in ponds and weirs; thus, it increasingly exposes the species to certain microorganisms such as Brucella, since the bacterium survival rate in these environments is high.
Isolating the agent is the standard diagnostic method for brucellosis (Bricker 2002; Geresu and Kassa 2016). Thus, the bacteriological isolation of Brucella spp. was conducted in the current study in order to obtain the final and confirmatory diagnosis of seropositive animals. However, the isolation of Brucella spp. was just observed in three vaginal swab samples. In contrast, the PCR performed with the bacterial culture in broth resulted in amplified products in all samples (7/7), including those that showed no bacterial growth on TSA plates. Although the etiologic agent isolation is the gold standard technique used to diagnose brucellosis, the PCR is also useful to identify the presence of Brucella spp. infection without isolation; therefore, it may be used as alternative technique in such cases. In addition, PCR is able to detect the DNA of living and dead microorganisms, whereas the isolation technique just detects living Brucella (Hamdy and Amin 2002).
Likewise, Ali et al. (2014) were able to isolate Brucella spp. in four vaginal swabs collected from 35 female buffaloes with recent history of miscarriage. Samaha et al. (2008) isolated Brucella spp. in seven milk samples and in six tissue samples from 47 seropositive animals, according to the RBPT. Megid et al. (2005) isolated the biovar 1 of B. abortus in an aborted fetus in São Paulo State, Brazil. The authors highlighted the first biotype featuring in a B. abortus strain isolated from buffalo fetus in the country.
By comparing the PCR results and those of the culture isolation method used to detect Brucella in vaginal swab samples, the current study has found that the PCR technique was able to detect more positive samples than the bacterial isolation; 100% (7/7) and 42.8% (3/7), respectively. Similar results were found by Al-Azeem et al. (2012), who examined 18 milk samples using the isolation and PCR techniques and detected Brucella in 7 and 10 samples, respectively. On the other hand, Marianelli et al. (2008) used serological, bacteriological, and molecular methods and found that 37 out of the 53 seropositive milk samples (RBPT and Complement Fixation) were positive cultures. They also found that the PCR was able to detect the agent in just 25 of these assessed samples.
The results obtained in the current study were extremely important since they enabled the pioneering finding—through isolation and PCR techniques—of Brucella spp. infection in buffalo herds in Maranhão State. Such finding emphasizes the importance of properly implementing the National Program for the Control and Eradication of Animal Brucellosis and Tuberculosis (PNCEBT - Programa Nacional de Controle de Erradicação da Brucelose e Tuberculose Animal) in the state, since the herein investigated zoonosis is conveyed by food. Thus, the increased production of buffalo milk-derived products poses a risk to public health.
Finally, the herein exposed results confirmed Brucella spp. infection in the buffalo herd from Maranhão State. The isolation of Brucella spp. through vaginal swab, as well as the PCR technique, were used as complementary diagnosis alternatives that may support epidemiological studies about the distribution of the disease and/or its infection in buffalo herds.
References
Abubakar, M., JavedArshed, M., Hussain, M., Ehtisham-ul-Haq Ali, Q. 2010. Serological evidence of Brucella abortus prevalence in Punjab Province, Pakistan—a cross-sectional study. Transboundary and Emerging Diseases 57, 443–447.
Al-Azeem, M.W.A, Elmalt, L.M.; Abdein, A.E.D.Z.; Sayed, H.H. 2012. Molecular and serological studies on detection of Brucella species in cattle and buffaloes. Journal of Pharmaceutical and Biomedical Sciences 2(3), 16-24.
Ali, S., Ali, Q., Melzer, F., Khan, I., Akhter, S., Neubauer, H., Jamal, S.M. 2014. Isolation and identification of bovine Brucella isolates from Pakistan by biochemical tests and PCR. Tropical Animal Health and Production 46(1), 73-78.
Alton, G.G., Jones, L.M., Angus, R.D., Verger, J.M. (Eds.), 1988. Techniques for the Brucellosis Laboratory. Institut National de la Recherche´ Agronomique, INRA, Paris.
Baily, G.G., Krahn, J.B., Drasar, B.W., Stoker, N.G. 1992. Detection of Brucella melitensis and Brucella abortus by DNA amplification. Journal of Tropical Medicine and Hygiene 95(4), 271-275.
Blas, I.; Ortega, C.; Frankena, K.; Noordhuizen, J.; Thrusfield, M. WIN Episcope 2.0, EPIDECON, Borland® y Delphi®. http://www.clive.ed.ac.ed/winepiscope/. Accessed 15 jun 2004.
Bounaadja, L., Albert, D., Chénais, B., Hénault, S., Zygmunt, M.S., Poliak, S., Garin-Bastuji, B., 2009. Real-time PCR for identification of Brucella spp.: a comparative study of IS711, bcsp31 and per target genes. Veterinary Microbiology 137, 156–164.
Brasil, 2004. Empresa Brasileira de Pesquisa Agropecuária – EMBRAPA. Brucelose e Tuberculose bovina: epidemiologia, controle e diagnóstico. Embrapa informação tecnológica, Brasília, 94pp.
Brasil, 2006. Manual Técnico. MAPA/SDA/DAS, Ministério da Agricultura, Pecuária e Abastecimento. Programa Nacional de Controle e Erradicação da Brucelose e Tuberculose Animal (PNCEBT), Brasília. 184p.
Bricker, B.J., 2002. Diagnostic strategies used for the identification of Brucella. Veterinary Microbiology 90, 433–444.
Chaves, N.P., Bezerra, D.C., Santos, L.S., Sá, J.S., Santos, H.P., Pereira, H.M. 2012. Intercorrência entre leucose enzoótica e brucelose em búfalos (Bubalus bubalis) em sistema de produção extensivo. Pesquisa Veterinária Brasileira, 32(2), 131-134.
Fosgate, G.T., Diptee, M.D., Ramnanan, A., Adesiyun, A.A. 2011. Brucellosis in domestic water buffalo (Bubalus bubalis) of Trinidad and Tobago with comparative epidemiology to cattle. Tropical Animal Health and Production 43(8), 1479-1486.
Franco, M.P., Mulder, M., Gilman, R.H., Smits, H.L., 2007. Human brucellosis. The Lancet Infectious Diseases 7(12), 775-86.
Galińska, E.M., Zagórski, J., 2013. Brucellosis in humans—etiology, diagnostics, clinical forms. Annals of Agricultural and Environmental Medicine 20(2), 233-238.
Geresu, M.A., Kassa, G.M., 2016. A review on diagnostic methods of brucellosis. Journal of Veterinary Science & Technology, 7, 323.
Hamdy, M. E. R., Amin A. S., 2002. Detection of Brucella species in the milk of infected cattle, sheep, goats and camels by PCR. The Veterinary Journal 163, 299–305.
Marianelli, C., Martucciello, A., Tarantino, M., Vecchio, R., Iovane, G., Galiero, G., 2008. Evaluation of molecular methods for the detection of Brucella species in water buffalo milk. Journal of Dairy Science 9(110), 3779-86.
Marques, J.R.F.; Cardoso, L.S. 1997. A bubalinocultura no Brasil e no Mundo. In: SIMPÓSIO BRASILEIRO DE BUBALININOCULTURA, 1., 1997, Cruz das Almas. Anais. Cruz das Almas: Escola de Agronomia da Universidade Federal da Bahia, p.10-221.
Martínez, D., Thompson, C., Draghi, G., Canavesio, V., Jacobo, R., Zimmer, P., Elena, S., Nicola, A.M., de Echaide, S.T. 2014. Pheno- and genotyping of Brucella abortus biovar 5 isolated from a water buffalo (Bubalus bubalis) fetus: first case reported in the Americas. Veterinary Microbiology 173(1-2), 172-6.
Megid, J., Albert, D., Fagliari, J.J., Paes, A.C., Listoni, F.P., Pinto, M.R.A., Ribeiro M.G., Thiébaud M., Ueno T., Garin-Bastuji B., 2005. Isolation of Brucella abortus from cattle and buffalo in Brazil. Veterinary Record 156, 147-148.
Mol, J.P.S., França, S.A., Paixão, T.A., Santos, R.L., 2012. Laboratorial diagnosis of animal brucellosis. Revista Brasileira de Ciência Veterinária 19, 117-126.
Molnár, L., Molnár, E., Lima, E.S.C., Dias, H.L.T., 2002. Avaliação de seis testes sorológicos no diagnóstico da brucelose bubalina. Pesquisa Veterinária Brasileira 22(2), 41-44.
Nielsen, K., Smith, P., Yu, W.L., Halbert, G., 2007. Salmonella enterica serotype urbana interference with brucellosis serology. Journal of Immunoassay and Immunochemistry 28, 289–296.
Paulin, L. M. S., Ferreira Neto, J. S., 2008. Artigo de revisão – Brucelose em búfalos. Arquivo do Instituto Biológico 75(3), 389-401.
Pitcher, D.G., Saunders N.A., Owen, R.J., 1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Letters in Applied Microbiology 8, 151-156.
Poester, F.P., Samartino, L.E., Santos, R.L. 2013. Pathogenesis and pathobiology of brucellosis in livestock. Revue Scientifique et Technique - Office International des Épizooties 32, 105-115.
Samaha, H., Al-Rowaily, M., Khoudair, R.M., Ashour, H.H., 2008. Multicenter study of brucellosis in Egypt. Emerging Infectious Diseases 14(12), 1916-1918.
Sambrook, J., Russel, D.W., 2001. Molecular Cloning. A Laboratory Manual. 3rd ed. Cold Spring Harbor Laboratory Press, New York.
Santos, R.L., Martins, T.M., Borges, Á.M., Paixão, T.A., 2013. Economic losses due to bovine brucellosis in Brazil. Pesquisa Veterinária Brasileira 33, 759-764.
Santos, C.L.R., Santos Júnior, J.B., Cunha, M. C., Nunes, S.R.F., Bezerra, D.C., Torres Júnior, J.R.S., Chaves, N.P., 2016. Nível tecnológico e organizacional da cadeia produtiva da bubalinocultura de corte no estado do Maranhão. Arquivos do Instituto Biológico, 83 (e0022014), 1-8.
Silva, J.B., Rangel, C.P., da Fonseca, A.H., de Morais, E., Vinhote, W.M., da Silva Lima D.H., da Silva e Silva N., Barbosa J.D., 2014. Serological survey and risk factors for brucellosis in water buffaloes in the state of Pará, Brazil. Tropical Animal Health and Production 46(2), 385-389.
Sousa, M.G.S., Brito, M.F., Ubiali, D.G., Fonseca Jr, A.A., Silva, J.B., Reis, A.S., Oliveira, C.M.C., Barbosa, J.D. 2015. Detecção de Brucella abortus em linfonodos de búfalas (Bubalus bubalis) em diferentes fases da gestação. Pesquisa Veterinária Brasileira, 35(12), 951-955.
Wray, C., 1975. Survival and spread of pathogenic bacteria of veterinary importance within the environment. Veterinary Bulletin 45, 543-550.
Acknowledgements
This study was supported by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES - PROCAD-NF 680/2010), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), and Fundação de Amparo à Pesquisa e Desenvolvimento Científico do Maranhão (FAPEMA).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
dos Santos, L.S., Sá, J.C., dos Santos Ribeiro, D.L. et al. Detection of Brucella sp. infection through serological, microbiological, and molecular methods applied to buffaloes in Maranhão State, Brazil. Trop Anim Health Prod 49, 675–679 (2017). https://doi.org/10.1007/s11250-017-1238-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11250-017-1238-3