Abstract
Citrus is one of the major horticultural crops with high economic and nutraceutical value. Despite the fact that conventional research has developed numerous improved varieties, citriculture is still susceptible to various stresses and requires innovative solutions such as genetic engineering. Among all the currently available modern approaches, Agrobacterium-mediated transformation is the most efficient method for introducing desired traits in citrus. However, being a non-host for Agrobacterium, various citrus species, including Citrus aurantifolia and Citrus sinensis, are recalcitrant to this method. The available reports on Agrobacterium-mediated transformation of commercial citrus cultivars show very low transformation efficiency with poor recovery rates of whole transgenic plantlets. Here, we provide an efficient and reliable procedure of Agrobacterium-mediated transformation for both C. aurantifolia and C. sinensis. This protocol depends on providing callus-inducing treatment to explants before and during Agrobacterium co-cultivation, using optimum conditions for shoot regeneration and modifying in-vitro micrografting protocol to combat the loss of transgenic lines. As transgenic citrus shoots are difficult to root, we also developed the ideal conditions for their rooting. Using this protocol, the whole transgenic plantlets of C. aurantifolia and C. sinensis can be developed in about ~ 4 months, with transformation efficiency of 30% and 22% for the respective species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The world population is projected at 8 billion as of 2023 and is estimated to exceed 10 billion by the end of 2050 (Low et al. 2018). In between this increasing population, fruit production has consistently been inadequate to meet the required demand. Unbalanced consumption of fruits in the human diet leads to inadequate intake of micro- and macronutrients, which causes different forms of malnutrition (Mason-D'Croz et al. 2019). Citrus is one of the major fruit crops grown in more than 140 countries across the globe and has high nutritional value with an exotic taste. Despite its global availability, citrus is sensitive to various environmental stresses (Syvertsen and Garcia-Sanchez 2014). For the past many years, traditional breeding methods have been used to improve fruit yield and quality; however, in the case of citrus, it is an uphill task due to the long juvenile period, pollen/ovule sterility, apomixis, and absence of desired traits in related species. Through recent scientific developments in genetic engineering, adding desired traits from any source to target plants is now possible and thus, makes crop improvement easier in a limited period (De Clercq et al. 2002; Hensel et al. 2009; Febres et al. 2011).
The initial studies to genetically modify citrus were done by using protoplast as starting material. The first attempt was made by Kobayashi and Uchimiya, (1989), where protoplast-derived cultured cells showed the presence of foreign gene; however, Vardi et al. (1990) were the first to successfully develop transgenic citrus plants. Several methods, including Agrobacterium-mediated transformation, particle bombardment, electroporation, and RNA interference, were reported from different research labs within next 30 years (Sun et al. 2019; Conti et al. 2021). The genus citrus is not a natural host for Agrobacterium tumefaciens and is generally recalcitrant to Agrobacterium-mediated transformation (Poles et al. 2020; Conti et al. 2021). However, a widely used protocol, using this method, was developed by Orbović and Grosser, (2006) for stable genetic transformation of citrus cultivars. In the following years, other successful attempts of Agrobacterium-mediated transformation were also reported for different rootstock and scion cultivars (Dutt and Grosser 2009; Dutt et al. 2009). Further, Dutt et al. (2011) also reported substantial improvement in the transformation efficiency of Mexican lime by aiding in the callus development and shoot growth from infected epicotyl ends by an antioxidant, lipoic acid. Subsequently, the Agrobacterium-mediated transformation has become the most widely used method for generating transgenic citrus (Orbović and Grosser 2015; Sun et al. 2019). Among various explants, the Agrobacterium-mediated transformation of juvenile in-vitro epicotyl segments is considered more efficient and popular for the transformation of a wide range of citrus cultivars (Donmez et al. 2013; Orbović and Grosser 2015). Despite the refinement in transformation protocols, Citrus aurantifolia (Mexican lime) and Citrus sinensis (Sweet orange) remain recalcitrant and difficult to transform (Dutt and Grosser 2009; Dutt et al. 2011). To overcome this, we critically reviewed all the stages of Agrobacterium-mediated citrus transformation and introduced several needful and innovative improvements. For our study, we selected commercial cultivars, namely, ‘Pramalini’ (C. aurnatifolia) and ‘Mosambi’ (C. sinensis). We believe that the adoption of our protocol would increase the explant transformation and regeneration efficiency, reduce the loss of in-vitro micrografted shoots and enhance the rooting frequency in transgenic citrus shoots.
Results
Explant and Agrobacterium culture preparation
Appropriate epicotyl age is critical for highly efficient transformation. Epicotyls become suitable for transformation after 4–5 weeks of germination (Fig. 1a). Additionally, the type of cut at the epicotyl ends (oblique) is vital for transformation. Oblique cuts were made to expose the cambial ring (Fig. 1b).
Procedure for producing transgenic citrus by Agrobacterium-mediated transformation. a epicotyl preparation, b oblique cut on epicotyl end with exposed cambial ring, c development of the cambial callus after 10d on kanamycin selection, d initiation of shoot regeneration from cambial callus development after 25d of selection, e–f highly efficient regeneration of e C. aurantifolia and f C. sinensis on MRNB1 and ERB1 medium, respectively after 35d of selection, g–h screening of regenerated shoots; g no GUS activity was detected in non-transgenic shoot and h strong GUS activity was detected in transgenic shoot, i–k procedure of in-vitro micrografting; i elongated GUS positive shoot was cut to separate its apical portion and nodes, j a sharp V-shaped cut was made in scion and 0.5 cm longitudinal cut was made in rootstock and k scion and rootstock were fitted together to have cambial contact, l–m successful graft acceptance of l apical portion and m node, n–o hardening of successfully in-vitro micrografted plantlet derived from n apical portion and o node in greenhouse, p–q fully grown transgenic p C. aurantifolia and q C. sinensis in greenhouse, r–v rooting of transgenic shoots; r transgenic shoot cultured on rooting medium, successfully rooted plantlets of s C. aurantifolia and t C. sinensis, u positive GUS activity in rooted transgenic plantlet and v hardening of rooted transgenic plantlet in greenhouse condition
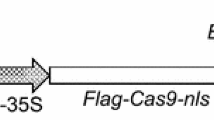
For the transformation, we used binary vector pCAMBIA2301, which contained gusA reporter gene with an intron in the coding region. The intron-GUS reporter system did not generate perceptible GUS activity in Agrobacterium cells but can be efficiently utilized by plant cells through intron splicing. The existence of the intron also increases the level of matured mRNA and provides obvious proof of a successful transformation (Ohta et al. 1990; Tanaka et al. 1990). This vector also contained nptII gene for plant selection and was introduced into Agrobacterium EHA105.
Epicotyl pre-treatment and co-cultivation
We assessed the transformation after infecting and co-cultivating the pre-treated epicotyls by GUS histochemical assay. We observed that the cambial region cells showed GUS activity and were successfully transformed (Fig. 2a). In this experiment, we stimulated the transformed cambial region to form a callus and observed strong GUS signals during different phases of cambial callus development and shoot regeneration. We successfully developed a cambial callus covering the entire cut end of epicotyl which regenerated to give transgenic shoots (Fig. 2b–d). A larger transformed area and a well-developed callus would regenerate to give a greater number of transformed shoots. Thus, this step increased our transformation efficiency.
Expression of gusA gene showing highly efficient Agrobacterium-mediated transformation. In citrus, cells located at the cambial region are competent for transformation. a Cambial region immediately after transformation. Arrow shows successfully transformed cambial region, development of a pronounced cambial callus after b 7d and c 14d on kanamycin selection. The cambial callus development is indicated by arrows and d the initiation of shoot regeneration after 18d of selection is indicated by the arrow. The process is represented by using C. aurantifolia epicotyls
Optimization of shoot regeneration and transformation efficiency
The proliferating callus was shifted to 16-h photoperiod for 7–10 days (d) and regeneration efficiency was calculated after 45d of epicotyl culture (Fig. 1c–f). For C. aurantifolia, we tested four media MRNB1, MRNB2, MRNB3 and MRNB4 (for composition, see materials and methods). We observed that the combination of ‘higher cytokinin with lower auxin levels’ gave the best regeneration, whereas the medium having only cytokinin gave poor results (Fig. 3a–d). We obtained the highest number of shoots per explant in the MRNB1 medium followed by the MRNB2 medium; however, MRNB4 gave the lowest number of regenerants (Table 1). In the case of C. sinensis, the combination of higher cytokinin along with lower auxin levels did not work, but the ‘lower cytokinin alone’ gave the maximum number of shoots per epicotyl (Fig. 4a–d). The four tested media were ERB1, ERB2, ERB3 and ERB4 (for composition, see materials and methods). ERB1 medium was superior to all the other screened media (Table 2). We also observed stages of somatic embryogenesis in the infected epicotyls after transformation, which could be the key pathway for explant regeneration (Fig. 5a–d) (Omar et al. 2016). The presence of malt extract in culture media is known to induce somatic embryogenesis in citrus (Pasquale et al. 1994).
Shoot regeneration on the explants of C. aurantifolia on different media. Regeneration frequency was observed after 45d of epicotyl culture on kanamycin selection. The antibiotic selection was maintained throughout the shoot regeneration process. Highly efficient regeneration on: a MRNB1, b MRNB2, c MRNB3 and d MRNB4 medium
Shoot regeneration on the explants of C. sinensis on different media. Regeneration frequency was observed after 45d of epicotyl culture on kanamycin selection. A strict antibiotic selection regime was maintained throughout the shoot regeneration process. Highly efficient regeneration on: a ERB1, b ERB2, c ERB3 and d ERB4 medium
Every regenerated shoot was separated from the mother explant. The longer (≥ 5–6 mm) and smaller shoots (< 5 mm) were processed as described in experimental procedures (see materials and methods-screening of transgenic shoots). Both longer and smaller shoots showed similar transformation efficiency. No GUS activity was observed in the non-transgenic shoot; however, a strong GUS activity was detected in the transgenic shoot (Fig. 1g, h). Expression of the gusA gene was used to calculate the transformation efficiency (%). Calculations were done using the formula: number of GUS positive shoots/total number of explants × 100. Agrobacterium-mediated transformation efficiency improves by increasing explant regeneration efficiency (Li et al. 2017). Similarly, we also obtained the highest transformation efficiency on MRNB1 and ERB1 media for C. aurantifolia and C. sinesis, respectively (Table 1 and 2). MRNB2 and ERB2, which were slightly different from MRNB1 and ERB1, respectively, also performed better with higher transformation efficiency than other media (MRNB3 and MRNB4; ERB3 and ERB4). Thus, based on our observations, we recommend using MRNB1 and ERB1 media for C. aurantifolia and C. sinensis, respectively for enhanced shoot regeneration and transformation efficiency.
In-vitro micrografting and rooting of transgenic shoots
To combat the loss in whole transgenic plant recovery, we did 2–3 separate in-vitro micrografting of single elongated transgenic shoot (see materials and methods–in-vitro micrografting and rooting of transgenic shoots). Utilizing our modified in-vitro micrografting procedure, we achieved a successful recovery of whole transgenic plants from ~ 92% of total GUS positive shoots. Additionally, we observed similar graft acceptance rates in shoot tips and nodes. A callus-like structure between scion and rootstock could be observed third day onwards in the in-vitro micrografting system. New leaves emerge from the scion after 10d of culture. We recommend changing the ML medium biweekly as it provides fresh nutrients to the micrograft system and facilitates graft union (Fig. 1i–m).
We observed 45% and 28% rooting in C. aurantifolia and C. sinensis, respectively (Fig. 1r–u). Rooting efficiency was calculated (%) as the total number of successfully rooted plantlets/total number of shoots cultured on rooting medium × 100.
Hardening of transgenic plantlets
We slowly acclimatized in-vitro developed plantlets by covering the pot with cling film using elastic bands around its rim to maintain high humidity. A few holes were made in the cling film to allow gas exchange. After 10–12d, we removed the elastic band and placed the cling film to cover the plantlet from the top but lifted it from one side. This was done to avoid sudden humidity changes for plantlets. After 2–3d, we completely removed the cling film. We observed almost 100% recovery rate of whole transgenic plantlets during greenhouse acclimatization (Fig. 1n–q, 1–v).
Confirmation of transgenic lines
To confirm transgenic lines, we isolated genomic DNA and total RNA from leaves of putative transgenic plantlets. We performed PCR from genomic DNA to check the presence of marker genes (Fig. 6a, b). For detecting the expression of nptII and gusA gene, we carried out semiquantitative RT-PCR from cDNA, prepared from total RNA taking cytochrome oxidase (cox) as the housekeeping gene (Fig. 7a–c). A brief description of the protocol is depicted below (Fig. 8).
PCR amplification of nptII and gusA gene from genomic DNA isolated from leaves of transgenic and wild-type (WT) citrus. Amplicons of 798 bps and 1029 bps of a nptII and b gusA genes, respectively, were obtained in transgenic lines. No amplification was observed in the WT line. M—1 kb DNA ladder (SD010; GeneDireX, Inc.), Lane 1–7—transgenic lines of C. aurantifolia, lane 8–14—transgenic lines of C. sinensis, lane 15—WT line, and lane 16—positive control. Binary vector pCAMBIA2301 was taken as the positive control; NTC no template control
Semiquantitative RT-PCR amplification from cDNA prepared by total RNA isolated from transgenic and WT citrus leaves. M—1 kb DNA ladder (SD010; GeneDireX, Inc.). We observed amplicons of 340 bps, 798 bps, and 1029 bps for a cox, b nptII, and c gusA genes, respectively, in transgenic lines. A 340 bps PCR product of the cox gene was obtained in WT line, but no amplicon of nptII and gusA gene was obtained. Lane 1–7—transgenic lines of C. aurantifolia, lane 8–15—transgenic lines of C. sinensis, lane 16—WT line and NTC no template control
Discussion
The efficiency of the Agrobacterium-mediated transformation in citrus is limited by various factors. In this report, we implemented several improvements in the protocol that contributed to our success in efficient development of transgenic citrus. Seed age and quality are some of the silent but significant aspects of the efficacy of this protocol. The overall health status of donor trees and fruits at the time of seed harvesting is also critical for the quality of raised explants and their regeneration (Yildiz 2012). We recommend the use of mature and freshly harvested seeds as we observed poor/delayed germination in older seeds, so longer storage must be avoided. We also recommend the use of 4–5 weeks older epicotyls for transformation experiments. Older epicotyls respond poorly to Agrobacterium-mediated transformation (Febres et al. 2011). Transformation efficiency of three-week-old epicotyl segments of ‘Washington navel’ orange was observed to be 87%, which decreases to 5 to 40% in 5–8 week-old explants (Bond and Roose 1998).
Many citrus cultivars have poor transformation and regeneration competency (Pena et al. 2004b; Poles et al. 2020). To develop an efficient transformation protocol, competency limitations must be addressed. In the case of citrus, the cells competent for transformation and regeneration are localized in the cambial region (Pena et al. 2004b; Guo et al. 2007). Additionally, shoots regenerated after prominent cambial callus development are more likely to be transgenic. So, the callus inducing treatment increases transformation frequency (Pena et al. 2004b). Consistent with this approach, we focused on providing treatments favouring the callus development during pre-culture and co-cultivation of epicotyls. We pre-cultured and co-cultivated epicotyl in the medium having the combination of auxin and cytokinin. Birch, (1997) has reported the importance of pre-culturing the explants in many plant species. The importance of pre-culturing in increasing the transformation efficiency has been reported by other authors as well (Villemont et al. 1997; Guo et al. 2007). During pre-culture, the cells start dividing and undergo DNA duplication and after adding Agrobacterium culture, the phytohormone-induced nuclei move to S-phase. In Petunia leaf mesophyll cells, the progression of the host cell into the S-phase is necessary for T-DNA transformation (Villemont et al. 1997). Similarly, the pre-culturing of citrus explants also showed enhanced transformation efficiency (Ainsley et al. 2001). Dutt and Grosser, (2009) showed increased transformation efficiency in citrus by pre-culturing epicotyls in hormone rich liquid medium for 3 h. In a study, a 6 h explant pre-culture increased the transformation efficiency of adult ‘Tarocco’ blood orange tissues (Peng et al. 2019). In many citrus cultivars including sour orange, sweet orange, lime, and Troyer citrange, the auxin supplemented co-cultivation medium increases the de-differentiation of cells and facilitates them to achieve a competent state for stable transformation (Ghorbel et al. 2000; Pena et al. 2004a; Rai 2006). Pena et al. (2004b) showed the advantage of BAP in co-cultivation medium in promoting multiple bud formation in Carrizo citrange. Together with auxin, cytokinin increases the competence of cells for transformation and contributes positively to callus development. (Sangwan et al. 1991; Hwang et al. 2017).
Gentle shaking was employed while infecting the epicotyl with Agrobacterium culture, to ensure proper exposure of every explant to the culture, thus increasing the transformation efficiency.
To establish an efficient transgenic development protocol, optimized shoot regeneration conditions are needed. Hence, we tested four different mediums for both, C. aurnatifolia and C. sinensis to get the best regeneration condition. For C. aurnatifolia, BAP along with NAA, gave the maximum number of shoots per explant. In many cultures, a high level of BAP and a low level of NAA have shown a positive impact on cell division and regeneration (Moreira-Dias et al. 2000; Pena et al. 2004b; Conti et al. 2021). For C. sinensis, BAP alone was sufficient to induce regeneration.
We screened longer shoots (≥ 5 mm) for GUS activity and elongated smaller regenerated shoots (< 5 mm) before GUS testing. The GUS positive shoots were further cultured on kanamycin free medium to minimize the possible negative effect of antibiotics on shoot elongation and recovery However, at this stage, we did not eliminate cefotaxime.
We standardized both methods of recovering whole transgenic plants in citrus, in-vitro micrografting and rooting. For in-vitro micrografting, we used three-week-old seedlings of C. jambhiri as rootstock. Older rootstock shows poor graft expectance. Generally, the success of shoot tip grafting in citrus was reported in between 30 and 50% (Chand et al. 2013). Every in-vitro micrografted shoot would not survive, leading to the loss of many transgenic shoots. To tackle this loss, we separately in-vitro micrografted the apical portion and nodes of the elongated transgenic shoot. Using our modified in-vitro grafting practice, almost every transgenic line survived, making this loss almost negligible. We successfully recovered whole transgenic plants from ~ 92% of the total GUS positive shoots. We also added a substantial amount of sucrose while culturing the scion-rootstock system. It has been reported that increasing sucrose concentration increases the number and size of new leaves and lateral roots (MacGregor et al. 2008). The use of liquid medium for culturing micrografts allowed easy culture handling and media replenishing, efficient nutrient absorption, and lesser oxidative stress on the explant (Shukla et al. 2020). For rooting, we performed three biweekly subcultures on the rooting medium.
Rapid water loss could lead to the death of transgenic plantlets while shifting to in vivo conditions. We maintained high humidity while shifting the in-vitro established plantlets to greenhouse conditions, facilitating their slow acclimatization. Confirmation of transgenic plantlets was done by PCR and semiquantitative RT-PCR of nptII and gusA reporter genes.
Various reports showing the transgene integration inability and recalcitrant behaviour of C. aurantifolia and C. sinensis by Agrobacterium-mediated transformation is available (Almeida et al. 2003; Dutt et al. 2009). Pena et al. (1997) obtained 14 GUS positive shoots from 304 transformed stem pieces of 6–12 month-old C. aurantifolia seedlings. Using the same source of explant, Domínguez et al. (2000) obtained 42 transgenic plants out of 1200 explants. Dutt et al. (2009) used etiolated epicotyl explants and reported C. aurantifolia as a recalcitrant species with maximum transformation efficiency of 8%. On the other hand, C. sinensis is reported to have cultivar specific transformation efficiency. For example, Dutt et al. (2009) showed the existence of differences in transformation efficiency between ‘Valencia’, which appeared recalcitrant, while ‘OLL8’ and ‘Hamlin’ were transformable with 20–25% efficiency. Later, Miyata et al. (2012) also reported that three cultivars ‘Hamlin’, ‘Valencia’, and ‘Pera’ had varied transformation efficiency that ranged from 1.54 to 6.08%. To our best knowledge, this is the report on optimizing in-vitro regeneration conditions and transformation of Cv. ‘Mosambi’, a commercial cultivar of sweet orange. By using this protocol, we found the transformation efficiency of C. aurantifolia Cv. ‘Pramalini’ and C. sinensis Cv. ‘Mosambi’ to be 30% and 22%, respectively. We found these citrus species compatible for transformation and in-vitro micrografting.
We minimized the loss of transgenic shoots/plantlets at different stages and developed several hundred transgenic plants by using this protocol. Our protocol is not only suitable for the development of disease/stress resistant citrus but also for functional genomics studies on citrus.
Materials and methods
Plant material
Mature and freshly harvested seeds of C. aurantifolia Cv. ‘Pramalini’ and C. sinensis Cv. ‘Mosambi’ were used in the study. Seeds were dipped in 70% v/v ethanol for one min before washing in sterile distilled water (SDW). After air-drying and removing the outer coat, the seeds were sterilized with 2% v/v sodium hypochlorite for 15 min. Disinfected seeds were rinsed three to four times in SDW and blotted dry on autoclaved filter paper. The sterilized seeds were cultured on SG medium [MS salts (Murashige and Skoog, (1962) supplemented with Gamborg B5 vitamin (Gamborg et al. 1968), 30 g l−1 sucrose and 3 g l−1 gelrite; pH 5.8] for germination and incubated for 3–4 weeks at 25 ± 2 °C in dark, followed by a 16-h photoperiod for another one week. The light green epicotyls with 12–15 cm height were ideal for transformation.
Epicotyl pre-culture
After removing the root, hypocotyl, cotyledon and apical hook from the etiolated seedlings, the light green epicotyls were obliquely cut into ~ 2 cm2 long segments. Epicotyls with the exposed cambial ring were pre-cultured into 60 ml of LM2NB medium (C. aurantifolia) [MS salts supplemented with Gamborg B5 vitamin, 50 g l−1 sucrose, 0.5 g l−1 malt extract, 1 mg l−1 2,4-d, 0.1 mg l−1 NAA and 2.5 mg l−1 BAP; pH 5.8]/LE2B medium (C. sinensis) [MS salts supplemented with EME vitamin (Grosser and Gmitter 1990), 50 g l−1 sucrose, 0.5 g l−1 malt extract, 1 mg l−1 2,4-d and 1 mg l−1 BAP; pH 5.8] for ~ 4 h with gentle shaking (100 r.p.m.) at RT.
Agrobacterium infection and co-cultivation of epicotyls
Agrobacterium strain EHA105 harboring binary vector pCAMBIA2301 was cultured on YEM agar [1 g l−1 yeast extract, 10 g l−1 of mannitol, 0.5 g l−1 K2HPo4, 0.2 g l−1 MgSo4, 0.1 g l−1 NaCl and 15 g l−1 agar] plates containing 50 mg l−1 kanamycin and 25 mg l−1 rifampicin for 2d at 28 °C in dark. On the day of transformation, one loop of Agrobacterium culture was inoculated into 60 ml of liquid YEM and cultured to OD600 of approximately 0.3 at 28 °C in dark with shaking at 200 r.p.m. At the time of inoculation, 100 µM of acetosyringone was also added along with appropriate antibiotics. Agrobacterium cells were harvested by centrifugation at 5000xg for 8 min at 25 °C and resuspended in MSA medium [MS salts supplemented with Gamborg B5 vitamin, 50 g l−1 sucrose and 100 µM acetosyringone; pH 5.8] to a final OD600 of 0.3 before use. Approximately 80–100 pre-cultured epicotyls were incubated with resuspended Agrobacterium culture for 15 min with gentle shaking at RT. Infected epicotyls were blotted dry on sterilized filter paper to remove the excess bacterial suspension. The dried epicotyls were co-cultivated on SMA2NB medium for 2d (C. aurantifolia) [MS salts supplemented with Gamborg B5 vitamin, 50 g l−1 sucrose, 0.5 g l−1 malt extract, 3 g l−1 gelrite, 1 mg l−1 2,4-d, 0.1 mg l−1 NAA, 2.5 mg l−1 BAP and 100 µM acetosyringone; pH 5.8]/SEA2B medium [MS salts supplemented with EME vitamin, 50 g l−1 sucrose, 0.5 g l−1 malt extract, 3 g l−1 gelrite, 1 mg l−1 2,4-d, 1 mg l−1 BAP and 100 µM acetosyringone; pH 5.8] for 3d (C. sinensis) at 25 ± 2 °C in dark with the oblique cut surface facing upward.
Determination of regeneration potential
The four different media used to evaluate the regeneration potential of co-cultivated epicotyls for C. aurantifolia were (1) MRNB1 medium—MS salts supplemented with 0.1 mg l−1 NAA and 2.5 mg l−1 BAP, (2) MRNB2 medium—MS salts supplemented with 0.5 mg l−1 NAA and 3 mg l−1 BAP, (3) MRNB3 medium—MS salts supplemented with 3 mg l−1 BAP, and (4) MRNB4 medium—MS salts supplemented with 1 mg l−1 BAP. Each medium was supplemented with Gamborg B5 vitamin, 50 g l−1 sucrose, 0.5 g l−1 malt extract, and 3 g l−1 gelrite; pH 5.8. The regeneration of C. sinensis epicotyls after Agrobacterium co-cultivation was evaluated on (1) ERB1 medium—MS salts supplemented with EME vitamin, 0.5 g l−1 malt extract and 1 mg l−1 BAP, (2) ERB2 medium—MT medium (Murashige and Tucker 1969) supplemented with 1 mg l−1 BAP, (3) ERB3 medium—MT medium supplemented with 2 mg l−1 BAP and (4) ERB4 medium—MT medium supplemented with 2 mg l−1 BAP and 0.2 mg l−1 IAA. Each medium was supplemented with 50 g l−1 sucrose and 3 g l−1 gelrite; pH 5.8. For the selection of transgenic shoots and control of Agrobacterium growth during regeneration, 100 mg l−1 kanamycin and 500 mg l−1 cefotaxime sodium salt, respectively were added to all the media. The infected epicotyls were cultured for 4 weeks at 25 ± 2 °C in dark for the callus development and shoot regeneration. After the emergence of shoots from proliferating cambial callus at the cut end, epicotyls were shifted to a 16-h photoperiod for 7–10d.
In-vitro germination of rootstock
After transferring the co-cultivated epicotyls to the regeneration medium, C. jambhiri (Rough lemon) seeds were in-vitro germinated (see materials and methods–plant material). Three-week-old C. jambhiri seedlings were used as rootstock for in-vitro micrografting of transgenic shoots.
Screening of transgenic shoots
A small Stem basal portion of the individually separated shoot (≥ 5–6 mm) was assayed histochemically for GUS activity (Jefferson et al. 1987; Moore et al. 1992). GUS positive shoots were further cultured for the recovery of whole transgenic plantlet and GUS negative shoots or escapes were discarded straight away. Smaller shoots (< 5 mm) were transferred to MEGK medium (C. aurantifolia) [MS salts and vitamin supplemented with 30 g l−1 sucrose, 3 g l−1 gelrite, 1 mg l−1 GA3 (Gibberellic acid), 100 mg l−1 kanamycin and 500 mg l−1 cefotaxime sodium salt; pH 5.8]/EEGK medium (C. sinensis) [MS salts supplemented with EME vitamin, 50 g l−1 sucrose, 0.5 g l−1 malt extract, 3 g l−1 gelrite, 1 mg l−1 GA3, 100 mg l−1 kanamycin and 500 mg l−1 cefotaxime sodium salt; pH 5.8] for 3–4d at 25 ± 2 °C with a 16-h photoperiod, for elongation before being tested for the GUS activity.
In-vitro micrografting and rooting of transgenic shoots
For in-vitro micrografting, GUS positive shoots were transferred for elongation, to MEG medium (MEGK without kanamycin; C. aurantifolia)/EEG medium (EEGK without kanamycin; C. sinensis) at 25 ± 2 °C with a 16-h photoperiod for 12–15d. A transgenic shoot with 2–3 well-developed nodes was placed on a sterilized 90 mm Petri dish, and a cut was made after the shoot tip and every node. Separated shoot tip and each node were treated as an individual scion and grafted individually on rootstocks. The cotyledon, apical hook, lateral secondary, and tertiary roots were removed from three-weeks-old rootstock seedlings (C. jambhiri). Also, the main taproot was shortened to a final length of 4–5 cm. A longitudinal slit of 0.5 cm was made in the decapitated rootstock. After the preparation of the rootstock, a sharp V-shaped cut was made in the scion, exposing its cambial ring. The scion was fitted in the slit of prepared rootstock and cultured in ML medium [MS salts supplemented with Gamborg B5 vitamin and 50 g l−1 sucrose; pH 5.8] for 3–4 weeks at 25 ± 2 °C with a 16-h photoperiod. The whole micrograft system was supported by M-shaped filter paper bridge having a hole at the center.
For rooting, the GUS positive shoots were transferred to MR medium (C. aurantifolia) [MS salts and vitamin supplemented with 30 g l−1 sucrose, 3 g l−1 gelrite and 500 mg l−1 cefotaxime sodium salt; pH 5.8]/EIR medium (C. sinensis) [MS salts supplemented with EME vitamin, 50 g l−1 sucrose, 0.5 g l−1 malt extract, 3 g l−1 gelrite, 1 mg l−1 IBA and 500 mg l−1 cefotaxime sodium salt; pH 5.8] for 3–4 weeks at 25 ± 2°c with a 16-h photoperiod. The transgenic shoots failed to develop roots even after three biweekly subcultures were in-vitro micrografted.
Hardening and analysis of transgenic plantlets
Successfully in-vitro micrografted or rooted plantlets were gently pulled out from the culture medium. Plantlets were transferred to 2″ pots filled with a 1:1 mixture of steam-sterilized garden soil and soilrite and acclimated under greenhouse conditions.
After 2 months of hardening, the genomic DNA and RNA were extracted from leaves of putative transgenic plantlets using DNeasy® plant mini kit (69104; Qiagen, Hilden, Germany) and RNeasy® plant mini kit (74904; Qiagen, Hilden, Germany) (follow manufacturer's protocol), respectively. cDNA was prepared using the iScript™ cDNA synthesis kit (1708841; Bio-Rad, Hercules, USA) according to the procedure recommended by the manufacturer. For the confirmation of transgenic plantlets, we detected the presence and expression of marker genes (nptII and gusA) by PCR and semiquantitative RT-PCR, respectively.
Statistical analysis
Statistical comparison among different regeneration media was compared by one-way ANOVA followed by Tukey’s HSD test (p < 0.05). Data analysis was done using Microsoft Office Excel 2019. Data represent means ± standard error (SE). Experiments were repeated 3 times.
Data availability
Data sharing is applicable to this article as datasets were generated or analysed during the current study.
References
Ainsley PJ, Collins GG, Sedgley M (2001) Factors affecting Agrobacterium mediated gene transfer and the selection of transgenic calli in paper shell almond (Prunus dulcis Mill). J Hort Sci Biotech 76:522–528. https://doi.org/10.1080/14620316.2001.11511403
Almeida WA, Mourão Filho FD, Mendes BM, Pavan A, Rodriguez AP (2003) Agrobacterium-mediated transformation of Citrus sinensis and Citrus limonia epicotyl segments. Scientia Agricola 60:23–29. https://doi.org/10.1590/S0103-90162003000100005
Birch RG (1997) Plant transformation: problems and strategies for practical application. Annu Rev Plant Biol 48(1):297–326. https://doi.org/10.1146/annurev.arplant.48.1.297
Bond JE, Roose ML (1998) Agrobacterium-mediated transformation of the commercially important citrus cultivar Washington navel orange. Plant Cell Rep 18:229–234. https://doi.org/10.1007/s002990050562
Chand L, Sharma S, Dalal R, Poonia AK (2013) In vitro shoot tip grafting in citrus species—a review. Agric Rev 34:279–287. https://doi.org/10.5958/j.0976-0741.34.4.013
Conti G, Xoconostle-Cázares B, Marcelino-Pérez G, Hopp HE, Reyes CA (2021) Citrus genetic transformation: an overview of the current strategies and insights on the new emerging technologies. Front Plant Sci. https://doi.org/10.3389/fpls.2021.768197
De Clercq J, Zambre M, Van Montagu M, Dillen W, Angenon G (2002) An optimized Agrobacterium-mediated transformation procedure for Phaseolus acutifolius A. Gray Plant Cell Rep 21:333–340. https://doi.org/10.1007/s00299-002-0518-0
Domínguez A, Guerri J, Cambra M, Navarro L, Moreno P, Pena L (2000) Efficient production of transgenic citrus plants expressing the coat protein gene of citrus tristeza virus. Plant Cell Rep Mar 19(4):427–33. https://doi.org/10.1007/s002990050751
Donmez D, Simsek O, Izgu T, Aka Kacar Y, Yalcin Mendi Y (2013) Genetic transformation in citrus. Sci World J. https://doi.org/10.1155/2013/491207
Dutt M, Grosser JW (2009) Evaluation of parameters affecting Agrobacterium-mediated transformation of citrus. Plant Cell Tissue Organ Culture 98:331–340. https://doi.org/10.1007/s11240-009-9567-1
Dutt M, Orbovic V, Grosser JW (2009) Cultivar dependent gene transfer into citrus using Agrobacterium. InProc Fla State Hortic Soc 122:85–89
Dutt M, Vasconcellos M, Grosser JW (2011) Effects of antioxidants on Agrobacterium-mediated transformation and accelerated production of transgenic plants of Mexican lime (Citrus aurantifolia Swingle). Plant Cell Tissue Organ Cult 107(1):79–89. https://doi.org/10.1007/s11240-011-9959-x
Febres V, Fisher L, Khalaf A, Moore GA (2011) Citrus transformation: challenges and prospects. In: Alvarez M (ed) Genetic transformation. IntechOpen, London, pp 101–122
Gamborg OL, Miller RA, Ojima K (1968) Nutrient requirements of suspension cultures of soyabean root cells Exp. Cell Res 50:151–158. https://doi.org/10.1016/0014-4827(68)90403-5
Ghorbel R, Domínguez A, Navarro L, Peña L (2000) High efficiency genetic transformation of sour orange (Citrus aurantium) and production of transgenic trees containing the coat protein gene of citrus tristeza virus. Tree Physiol 20(17):1183–1189. https://doi.org/10.1093/treephys/20.17.1183
Grosser JW, Gmitter FG Jr (1990) Protoplast fusion and citrus improvement. Plant Breed Rev 8:339–374. https://doi.org/10.1002/9781118061053.ch10
Guo W, Li D, Duan YX (2007) Citrus transgenics: current status and prospects. Transgenic Plant J 1(1):202–209
Hensel G, Kastner C, Oleszczuk S, Riechen J, Kumlehn J (2009) Agrobacterium-mediated gene transfer to cereal crop plants: current protocols for barley, wheat, triticale, and maize. Int J Plant Genomics. https://doi.org/10.1155/2009/835608
Hwang HH, Yu M, Lai EM (2017) Agrobacterium-mediated plant transformation: biology and applications. The Arabidopsis Book 15, U.S. National Library of Medicine
Jefferson AR (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5:387–405. https://doi.org/10.1007/BF02667740
Kobayashi S, Uchimiya H (1989) Expression and integration of a foreign gene in orange (Citrus sinensis Osb.) protoplasts by direct DNA transfer. Jpn J Genet 64:91–97. https://doi.org/10.1266/jjg.64.91
Li S, Cong Y, Liu Y, Wang T, Shuai Q, Chen N, Gai J, Li Y (2017) Optimization of Agrobacterium-mediated transformation in soybean. Front Plant Sci 8:246. https://doi.org/10.3389/fpls.2017.00246
Low LY, Yang SK, Kok DX, Ong-Abdullah J, Tan NP, Lai KS (2018) Transgenic plants: gene constructs, vector and transformation method. In: Celik O (ed) New visions in plant science, vol 19. IntechOpen, London, pp 41–61
MacGregor DR, Deak KI, Ingram PA, Malamy JE (2008) Root system architecture in Arabidopsis grown in culture is regulated by sucrose uptake in the aerial tissues. Plant Cell 20:2643–2660. https://doi.org/10.1105/tpc.107.055475
Mason-D’Croz D, Bogard JR, Sulser TB, Cenacchi N, Dunston S, Herrero M, Wiebe K (2019) Gaps between fruit and vegetable production, demand, and recommended consumption at global and national levels: an integrated modelling study. Lancet Planet Health 3:318–329. https://doi.org/10.1016/S2542-5196(19)30095-6
Miyata LY, Harakava R, Stipp LC, Mendes BM, Appezzato-da-Glória B, de Assis Alves Mourão Filho F (2012) GUS expression in sweet oranges (Citrus sinensis L. Osbeck) driven by three different phloem-specific promoters. Plant Cell Rep 31(11):2005–2013. https://doi.org/10.1007/s00299-012-1312-2
Moore GA, Jacono CC, Neidigh JL, Lawrence SD, Cline K (1992) Agrobacterium-mediated transformation of citrus stem segments and regeneration of transgenic plants. Plant Cell Rep 11:238–242. https://doi.org/10.1007/BF00235073
Moreira-Dias JM, Molina RV, Bordón Y, Guardiola JL, Garcia-Luis A (2000) Direct and indirect shoot organogenic pathways in epicotyl cuttings of Troyer citrange differ in hormone requirements and in their response to light. Ann Bot 85(1):103–110. https://doi.org/10.1006/anbo.2000.1001
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiol Plant 15:473–497. https://doi.org/10.1111/j.1399-3054.1962.tb08052.x
Murashige T, Tucker DPH (1969) Growth factor requirements of Citrus tissue culture. In: Chapman HD (ed) Proceedings of the1st international citrus symposium, vol 3. University of California Riverside, Riverside, pp 1155–116
Ohta S, Mita S, Hattori T, Nakamura K (1990) Construction and expression in tobacco of a β-glucuronidase (GUS) reporter gene containing an intron within the coding sequence. Plant Cell Physiol 31:805–813. https://doi.org/10.1093/oxfordjournals.pcp.a077982
Omar AA, Dutt M, Gmitter FG, Grosser JW (2016) Somatic embryogenesis: still a relevant technique in citrus improvement. In: In vitro embryogenesis in higher plants. Humana Press, New York, pp 289–327
Orbović V, Grosser JW (2006) Citrus: sweet orange (Citrus sinensis L. Osbeck ‘Valencia’) and Carrizo citrange [Citrus sinensis (L.) Osbeck × Poncirus trifoliata (L.) Raf.]. In: Wang K (ed) Agrobacterium protocols. Methods in molecular biology. Humana Press Inc., Totowa, pp 177–189
Orbović V, Grosser JW (2015) Citrus transformation using juvenile tissue explants. In: Wang K (ed) Agrobacterium protocols. Methods in molecular biology, 3rd ed. Humana Press Inc., USA, pp 245–257
Pasquale FD, Carimi F, Crescimanno FG (1994) Somatic embryogenesis from styles of different cultivars of Citrus limon (L.) Burm. Aust J Bot 42:587–594. https://doi.org/10.1071/BT9940587
Pena L, Cervera M, Juárez J, Navarro A, Pina JA, Navarro L (1997) Genetic transformation of lime (Citrus aurantifolia Swing.): factors affecting transformation and regeneration. Plant Cell Rep 16(11):731–7. https://doi.org/10.1007/s002990050311
Pena L, Cervera M, Fagoaga C, Perez R, Romero J, Juárez J, Pina JA, Navarro L (2004a) Agrobacterium-mediated transformation of citrus. Transgenic Crops of the World: Essential Protocols, pp 145–156
Pena L, Perez RM, Cervera M, Juarez JA, Navarro L (2004b) Early events in Agrobacterium-mediated genetic transformation of citrus explants. Ann Bot 94:67–74. https://doi.org/10.1093/aob/mch117
Peng A, Zou X, Xu L, He Y, Lei T, Yao L, Li Q, Chen S (2019) Improved protocol for the transformation of adult Citrus sinensis Osbeck ‘Tarocco’blood orange tissues. In Vitro Cell Dev Biol Plant 55:659–667. https://doi.org/10.1007/s11627-019-10011-9
Poles L, Licciardello C, Distefano G, Nicolosi E, Gentile A, La Malfa S (2020) Recent advances of in vitro culture for the application of new breeding techniques in citrus. Plants 9:938. https://doi.org/10.3390/plants9080938
Rai M (2006) Refinement of the Citrus tristeza virus resistance gene (Ctv) positional map in Poncirus trifoliata and generation of transgenic grapefruit (Citrus paradisi) plant lines with candidate resistance genes in this region. Plant Mol Biol 61:399–414. https://doi.org/10.1007/s11103-006-0018-7
Sangwan RS, Bourgeois Y, Sangwan-Norreel BS (1991) Genetic transformation of Arabidopsis thaliana zygotic embryos and identification of critical parameters influencing transformation efficiency. Mol Gen Genet 230:475–485. https://doi.org/10.1007/BF00280305
Shukla MR, Piunno K, Saxena PK, Jones AM (2020) Improved in vitro rooting in liquid culture using a two piece scaffold system. Eng Life Sci 20:126–132. https://doi.org/10.1002/elsc.201900133
Sun L, Ke F, Nie Z, Wang P, Xu J (2019) Citrus genetic engineering for disease resistance: past, present and future. Int J Mol Sci 20:5256. https://doi.org/10.3390/ijms20215256
Syvertsen JP, Garcia-Sanchez F (2014) Multiple abiotic stresses occurring with salinity stress in citrus. Environ Exp Bot 103:128–137. https://doi.org/10.1016/j.envexpbot.2013.09.015
Tanaka A, Mita S, Ohta S, Kyozuka J, Shimamoto K, Nakamura K (1990) Enhancement of foreign gene expression by a dicot intron in rice but not in tobacco is correlated with an increased level of mRNA and an efficient splicing of the intron. Nucleic Acids Res 18:6767–6770. https://doi.org/10.1093/nar/18.23.6767
Vardi A, Bleichman S, Aviv D (1990) Genetic transformation of Citrus protoplasts and regeneration of transgenic plants. Plant Sci 69:199–206. https://doi.org/10.1016/0168-9452(90)90118-8
Villemont E, Dubois F, Sangwan RS, Vasseur G, Bourgeois Y, Sangwan-Norreel BS (1997) Role of the host cell cycle in the Agrobacterium-mediated genetic transformation of Petunia: evidence of an S-phase control mechanism for T-DNA transfer. Planta 201:160–172. https://doi.org/10.1007/BF01007700
Yildiz M (2012) The prerequisite of the success in plant tissue culture: high frequency shoot regeneration. Recent advances in plant in vitro culture. In: Leva A, Rinaldi LMR (ed) Intech, recent advances in plant in vitro culture. IntechOpen, London
Acknowledgements
SS and ZT thanks MHRD for Junior and Senior research fellowships. This work was supported by a faculty initiation Grant (FIG-100677) by IITR to HC.
Author information
Authors and Affiliations
Contributions
SS and HC contributed to experimental design and conceived the study. SS and ZT performed the experiment. SS and HC wrote the manuscript. HC and AKS supervised the experiment and reviewed the manuscript. SK and DKS gave input during experiments. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have not disclosed any competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Singh, S., Tarannum, Z., Kokane, S. et al. Efficient transformation and regeneration of transgenic plants in commercial cultivars of Citrus aurantifolia and Citrus sinensis. Transgenic Res 32, 523–536 (2023). https://doi.org/10.1007/s11248-023-00367-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11248-023-00367-5