Abstract
AAL toxin, the major virulence factor of Alternaria alternata f. sp. lycopersici, is recognized to cause necrotic cell death in plants. Glutathione (GSH) is a noteworthy participant in plant defence. However, how GSH is involved in regulating the AAL treated cell death is yet to be explored. Here, Arabidopsis thaliana Col-0, and previously developed transgenic line AtECS1, were exogenously treated with AAL toxin and a proteomic profile (ProteomeXchange accession: PXD017124) was obtained by nano LC–MS/MS analysis. Few salicylic acid (SA) and ethylene (ET) responsive proteins, along with others were identified. Selected SA-responsive genes were noted to be up regulated in AAL treated AtECS1 compared to Col-0 by quantitative real time-PCR (qRT-PCR), beside the up regulation of ascorbate peroxidase 1 (APX1) and chaperone like heat shock protein (HSP), together with myrosinase. Interestingly, ET biosynthetic and signaling marker genes were down regulated in AAL treated AtECS1 compared to Col-0. Augmentation of SA content and proteins regulated by it, while, reduction of endogenous 1-aminocyclopropane-1-carboxylate (ACC) content and ET-related proteins was significant in AAL treated AtECS1 compared to Col-0. Collectively, these findings suggested that under necrotrophic attack as mimicked here by AAL treatment, GSH may be involved in resistance primarily by SA-mediated ET suppression in addition to various stress responsive molecules.
Key message
GSH mediated resistance to AAL toxin may be conferred in Arabidopsis by regulating SA and ET pathways along with other stress related molecules to reduce necrotic cell death.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
AAL toxin is a type of sphinganine analogue mycotoxins (SAMs) that inhibit eukaryotic sphinganine N-acyltransferase (acyl-CoA-dependent ceramide synthase), which is the key enzyme in the sphingolipid ceramide biosynthetic pathway. It is the major virulence effector molecule produced by toxigenic and necrotrophic Alternaria alternata f. sp. lycopersici. Among the five series (TA, TB, TC, TD, and TE) of AAL toxin, TA is the most common kind of AAL toxin (Xu and Du 2006). AAL-toxins affect organisms other than host plants and thus their toxicity is host selective but not host specific (Tsuge et al. 2013).
Glutathione (γ-glutamylcysteinylglycine), an abundant and ubiquitous low-molecular-weight tripeptide thiol, exists in two forms viz. oxidized, GSSG and reduced, GSH. It is found in millimolar concentration in all aerobic organisms and in almost all cell organelles. It has long been known that GSH played a significant role in plant defence (Dron et al. 1988; Wingate et al. 1988). The strategic position between oxidants viz. ROS and cellular reductants makes the GSH system perfectly configured for signaling functions, for cell cycle progression, and for regulating several epigenetic events (Foyer and Noctor 2005; Diaz Vivancos et al. 2010). Also, the role of GSH in the regulation of expression of several stress and defence genes, besides maintaining growth and development of plants is a well established fact (May et al. 1998; Ball et al. 2004). It is involved in conferring resistance against abiotic stresses like freezing, salinity stress, heavy metal tolerance, drought, detoxification of several xenobiotics, etc. (Ruiz and Blumwald 2002; Li et al. 2006; Cummins et al. 2011; Lata et al. 2011; Zagorchev et al. 2013; Gulyas et al. 2014; Sinha et al. 2015). In addition, several other evidences also reports the involvement of GSH in controlling biotic stresses induced by bacteria, fungi, and virus, by controlling oxido reduction of other thiols, by post translational modifications of protein or by inducing several plant defence genes through an intricate network of signaling molecules viz. SA, jasmonic acid (JA), ET, abscisic acid (ABA), reactive oxygen species (ROS), etc. (Parisy et al. 2007; Ghanta and Chattopadhyay 2011; Ghanta et al. 2011; Wang et al. 2011; Kuźniak et al. 2014; Cheng et al. 2015; Datta et al. 2015; Hernández et al. 2017).
Phytohormones like SA, JA, ET, ABA as well as ROS are known to play crucial roles as signaling molecules, in maintaining normal homeostasis in plants as well as participate in plant defence when threatened with a broad range of stresses. Crosstalk between these molecules has been known for decades to mitigate stress in plants (Chen et al. 1993; Thomma et al. 1998; Kunkel and Brooks 2002; Grant and Jones 2009). SA signaling is known to control the resistance to biotrophic infection, while ET and JA signaling is well-known to control necrotrophic infection in plants (Glazebrook 2005; Loake and Grant 2007). However, several studies are there where SA has been known to check the progression of necrotrophic infection in plants, as well (Murphy et al. 2000; Kouzai et al. 2018). Apart from the role of SA, ET, JA, etc. in defence response, they also play an important role in plant PCD (Van Breusegem and Dat 2006; Reinbothe et al. 2009; Wang et al. 2017). Several studies showed that JA and ET have important roles to play in AAL toxin induced PCD (Egusa et al. 2009; Zhang et al. 2016). Also, it has been found that in Arabidopsis LOH2 mutant, the ET pathway genes and the genes responsive to ROS were earliest to be up regulated on AAL treatment (Gechev et al. 2004). Another study showed that while, JA and ET conferred susceptibility of tomato plants to AAL, SA conferred resistance to A. alternata infection (Jia et al. 2013). Till now, though, evidences are there in understanding the A. alternata and AAL mediated necrotrophism and PCD through JA, ET, and SA pathway, but how GSH takes part in regulating AAL mediated necrosis and PCD has been unknown till date. To elucidate, we performed an initial proteomic study with wild type Col-0 and AtECS1, the transgenic A. thaliana line exhibiting enhanced GSH content (Datta et al. 2015), with or without AAL treatment. Selected SA and ET responsive identified protein species was further analyzed by qRT-PCR and western blot. Together, GSH seems to play an important role in conferring resistance to AAL toxin, possibly by interacting through SA and ET mediated pathways, besides influencing several other effector molecules related to stress resistance.
Materials and methods
Plant growth and AAL treatment
Wild type Arabidopsis thaliana plants of Columbia ecotype (Col-0; Nottingham Arabidopsis Stock Centre, N1093) and transgenic line viz. AtECS1, developed earlier (Datta et al. 2015), were grown in Murashige and Skoog (Murashige and Skoog 1962) medium and maintained in a growth chamber at 22 ± 1 °C under a 16 h light/8 h dark cycles as standardized before (Datta et al. 2013). Leaves of four weeks old plants were treated with 10 µM AAL toxin (Chemfaces, China) with needleless syringe as described previously (Willekens et al. 1997) and maintained for 4 days (96 h). Control plants were treated with water (mock treated). After four days, samples were harvested, for all subsequent studies.
Measurement of chlorophyll content
Chlorophyll content was measured following Lichtenthaler (1987). Briefly, leaves were crushed in liquid nitrogen and chlorophyll was extracted at 95% ethanol overnight and spectrophotometric absorption of the samples was noted.
Estimation of callose deposition
Estimation of callose deposition was done following Underwood et al. (2007). Briefly, leaves were cleared of pigment by vacuum-infiltration of alcoholic lactophenol and incubated at 65 °C for 30 min. The leaves were transferred to fresh alcoholic lactophenol solution and incubated at room temperature overnight. Cleared leaves were rinsed briefly in 50% ethanol, then in water, and stained with 0.01% aniline blue (Sigma, USA) in 150 mM K2HPO4 (pH 9.5). The fluorescence was observed with Olympus IX 81 microscope using DAPI filter.
DAB assay
Detection of hydrogen peroxide (H2O2) was performed by staining leaves with 3, 3′-diaminobenzidine (DAB) (Sigma, USA) following Thordal-Christensen et al. (1997) with a slight modification. Leaves were stained in DAB solution for about 8 h. Following incubation, the DAB solution was replaced by bleaching solution (ethanol:acetic acid:glycerol = 3:1:1) and boiled for 15 min. After replacing with fresh bleaching solution, leaves were allowed to stand for 30 min. Leaves were visualized for DAB staining.
Protein extraction and digestion
Total protein from mock treated and AAL treated samples were isolated using the phenol extraction method (Isaacson et al. 2006), with minor modification. The experiment was repeated thrice for all four samples. Briefly, about 500 mg of leaf tissues from each sample were grounded in liquid nitrogen and suspended in extraction buffer (700 mM sucrose, 500 mM Tris–HCl, pH 7.5, 50 mM EDTA, 100 mM KCl, 2% (w/v) β-mercaptoethanol, 1 mM phenyl methyl sulfonyl fluoride). After washing the pellet and drying, it was dissolved in 8 M urea in 10 mM Tris buffer. Protein concentration was determined using Bradford reagent (Bradford 1976) using BSA as standard. Approximately 100 µg of protein from each sample were digested with trypsin (Promega Trypsin Gold MS grade, USA) with protease to substrate ratio of 1:50 for 16 h at 37 °C. The digested proteins were lyophilized and finally dissolved at 0.1% formic acid in 50% acetonitrile. The samples were then desalted using Zip-Tip μ-C18 (Millipore, USA) and used for further analysis.
Protein identification using nano LC–MS/MS
Peptides were loaded onto EASY-nLC 1000 (Thermo Scientific™, USA) where initially peptides were separated in C18 trap column (C18 3 µm, 75 µm × 20 mm), an analytical column (C18 3 µm, 75 µm × 150 mm), using mobile phase A (0.1% v/v formic acid in water) and B (0.1% v/v formic acid in acetonitrile), using the following gradient: 5–35% B for 0–100 min, 35–50% B for 100–110 min, 50–95% B for 110–115 min, 95% B for 115–120 min, 95–5% B for 120–125 min and 5% B for 125–130 min with 300 nL/min flow rate. The MS/MS analysis was conducted using LTQ orbitrap XL (Thermo Fisher Scientific, USA). A full mass spectrometry (MS) scan (350–2000 m/z) was performed at the positive ion mode with a resolution of 50,000, with an AGC value of 1*e6, maximum IT of 100 ms and dynamic exclusion of 30 s. The raw LC–MS/MS files were searched by using Mascot 2.4 (Matrix Science https://www.matrixscience.com/help/apr2012.html) containing Proteome Discoverer1.4 (Thermo Fisher Scientific, USA), against the database of Arabidopsis thaliana available from Swissprot. Search parameters included carbamidomethylation of cysteine as a static modification, and oxidation of methionine as dynamic modification. The proteolytic enzyme was specified as trypsin, and maximum missed cleavage of 2 was allowed. Peptide mass tolerance was set at 10 ppm and fragment mass tolerance was set at 0.8 Da. An automatic decoy database search was performed as part of the search. False discovery rates (FDR) for peptide identification were < 1.0%. To improve the accuracy and sensitivity of peptide identification Mascot results were filtered through Mascot Percolator package. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (https://www.ebi.ac.uk/pride/archive/) via the PRIDE partner repository (Vizcaíno et al. 2013) with the accession: PXD017124. Among the identified proteins, those with at least 1 unique peptide and score 30 and above, with PSM values more than 1 were considered for further studies.
Functional categorization of identified proteins was performed using KEGG (Kyoto Encyclopedia of Genes and Genomes, https://www.genome.jp/kegg/) (Kanehisa and Goto 2000). PANTHER (https://pantherdb.org/) classification tool was used to group the identified proteins into biological process, molecular function and cellular components and also under several protein classes (Mi et al. 2019).
RNA isolation and PCR
Total RNA isolation and consecutive qRT-PCR from mock treated along with AAL treated samples were performed as standardized before (Datta and Chattopadhyay 2015; Datta et al. 2015). Total RNA was extracted using TRIzol method. cDNA was synthesized using the RevertAid H Minus First Strand cDNA Synthesis kit (Thermo Scientific, USA) using 1 μg of RNA from each sample and proceeded for qRT-PCR. The qRT-PCR analysis was performed using Light Cycler 96 System (Roche Applied Science, USA) with iTAQ ™ Universal SYBR® Green Supermix (BIO-RAD, USA) following manufacturer‘s instructions. PCR amplification was performed at 94 °C for 30 s, annealing for 15 s and 72 °C for 30 s, with a preincubation at 94 °C for 600 s. The primers used for qRT-PCR were listed in Supplemental Table S1. The constitutively expressed Elongation factor-1alpha (eEF-1 alpha) was used as the reference gene.
Western blot analysis
Western blot was done as standardized previously (Datta et al. 2015). Briefly, leaves were crushed in liquid nitrogen and proteins were extracted after homogenizing leaves in 50 mM potassium phosphate buffer, pH 7.8, containing 0.15% (v/v) Triton X-100. Protein samples were quantified by Bradford assay (Bradford 1976), resolved in 12% SDS PAGE gel and transferred onto polyvinylidene difluoride membrane (Millipore, USA) and blocked with 5% skimmed milk. The PR4 and PR5 protein bands were detected by using a rabbit polyclonal anti-PR4 (dilution 1:2000) and anti-PR5 antibody (dilution 1:10,000) as the primary antibody and an anti-rabbit IgG conjugated to horseradish peroxidase as the secondary antibody (Agrisera, Sweden). For detecting ACO1 proteins, goat polyclonal anti-ACO1 antibody (Santacruz Biotechnology, China; dilution 1:200) were used as the primary antibody and an anti-goat IgG conjugated to horseradish peroxidase as the secondary antibody. Tubulin (Agrisera; dilution 1:1000) was used as housekeeping control. SuperSignal West Pico chemiluminescent substrate (Pierce, USA) was used for visualizing immunoreactive proteins. The experiment was repeated thrice for all four samples.
HPLC analysis
Estimation of SA content
Extraction and quantification of SA was performed following Freeman et al. (2005). SA was quantified by HPLC (Waters, USA) with a fluorescence detector by using Symmetry C18 reverse-phase column (5 µm, 4.6 × 250 mm) at excitation and emission wavelength of 254 nm and 395 nm respectively. The elution condition was methanol gradient (solvent A, water and 1% formate; and solvent B, 100% methanol and 1% formate) of 10 to 40% B (10 min), 40 to 50% B (5 min), 50 to 100% B (2.5 min), 100 to 40% B (2.5 min), 40 to 10% B (1 min) and 10% B (1 min) with a flow-rate of 1 mL/min over 22 min.
Estimation of ACC content
Estimation of ACC was performed using o-phthaldialdehyde pre-column derivatization method as standardized before by Bushey et al. (1987) and Datta et al. (2015). The HPLC analysis was conducted using a 515 HPLC pump with a 2475 fluorescence detector as mentioned above; at a flow-rate of 0.6 mL/min. AccQ-Tag (3.9 × 9 × 150 mm) column with an excitation wavelength of 325 nm and an emission wavelength of 465 nm was used. The elution solvents were: solvent A, composed of 0.1 M sodium acetate at pH 5.8 and methanol as solvent B. Initially B was 10%. From 0 to 4 min, a linear gradient of 10–44% B was applied, from 4 to 10 min, a linear gradient of 44–50% B was applied, from 10 to 12 min a linear gradient of 50–80% B was applied, from 12 to 16 min, 80–100% B and from 16 to 20 min and finally B decreased to 0% linearly.
Statistical analysis
All experiments were repeated thrice. Mean and standard errors (SE) were calculated from three independent sets of biological replications as required. Statistical analysis was done using Student t test.
Results
Effect of AAL treatment on cell death, chlorophyll content and callose deposition in Col-0 and AtECS1 plants
As mentioned previously (Datta et al. 2015), Lycopersicon esculentum, γ-ECS gene cloned in pBI121 under CaMV35S promoter and introduced in Col-0 by Agrobacaterium tumefaciens mediated transformation using the floral dip method. Amongst various AtECS transgenic lines developed, AtECS1 line was selected for further analysis, since this line has the significant total GSH content viz. 2.24-fold higher GSH as compared to Col-0. Interestingly, no notable morphological differences between Col-0 and transgenic Arabidopsis was noted. It was also found that AtECS1 exhibited remarkable up regulation of ACS2, ACS6, and ACO1 at transcript as well as protein levels, while they were down regulated in the GSH-depleted mutant, phytoalexin deficient2-1 (pad2-1) (Datta et al. 2015).
Enhanced infectious lesions appeared in the wild type Col-0 compared to that of AtECS1, under AAL treated condition, in comparison to the mock treated plants (Fig. 1a–l). About 40% of the leaf area in Col-0 and 11% of leaf area in AtECS1 were found to be damaged as found 96 h post treatment of AAL (Supplemental Fig. S1). Thus, GSH diminished the appearance of necrotic lesion in A. thaliana under AAL treated condition.
Leaf chlorophyll content known to be one of the key indicators of photosynthetic capacity (Cannella et al. 2016), its impairment is also an indicator of stress in plants (Carter 1994; Datta and Chattopadhyay 2015). Measurement of chlorophyll revealed a drop in its content in Col-0 plants by about 44% after AAL treatment compared to AtECS1 where chlorophyll content dropped to 19%, after AAL toxin treatment (Supplemental Fig. S2). Total protein content was also found to be less in AAL treated Col-0 in comparison to the AtECS1 plants by about 29% (Supplemental Fig. S3).
Leaflets were stained with aniline blue to detect callose deposition, which appeared as bright spots on blue background. The callose deposits were much more in Col-0 than in AtECS1 after AAL treatment, which corroborated with the appearance of necrotic lesions (Fig. 2).
Study of in situ accumulation of H2O2 on AAL toxin treatment was done using DAB staining, where DAB is oxidized by H2O2 to produce a brown precipitate. More pronounced brown precipitate was noted in Col-0 plants as compared to the AtECS1, under AAL treated condition (Fig. 3).
Thus, GSH not only prevented loss of chlorophyll and total protein content, it also minimized the AAL induced cell death in plants as evidenced from decreased deposition of callose and H2O2 in AAL treated AtECS1 compared to the wild type Col-0.
Transcript analysis of selected stress marker genes
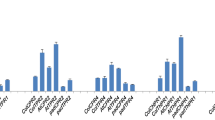
To study the response of plants to AAL toxin and to demonstrate the establishment of infection after AAL treatment, qRT-PCR was performed at five time points like 6 h, 24 h, 48 h, 72 h and 96 h, with selected transcripts viz. pathogenesis-related gene1 (PR1), pathogenesis-related gene2 (PR2) and 1-aminocyclopropane-1-carboxylate synthase 2 (ACS2), which are stress responsive (Gechev et al. 2004; Zhang et al. 2011). Up regulation of the expression of PR1 and PR2, in both AAL treated plants viz. Col-0 and AtECS1, indicated the establishment of the infection. In case of PR1 changes in expression between mock treated and AAL treated samples were first observed 48 h post infection, while in case of PR2 after 24 h the changes can be observed (Fig. 4a, b). Surprisingly, though the expression of ACS2 was up regulated in AAL treated Col-0 it was down regulated in AAL treated AtECS1 (Fig. 4c). The change in expression of ACS2 was found to be initiated 48 h post infection as well (Fig. 4c). PR1 and PR2 are established marker genes of SA-mediated defence pathway (Uknes et al. 1992; Mou et al. 2003) whereas ACS2 is the key enzyme of ET biosynthetic pathway (Yang and Hoffman 1984). Thus GSH might be playing a role in defence against AAL toxin by interacting with SA and ET pathways. Considering the time scale experiment, it can be noted that both at morphological as well as at molecular level, significant post-treatment changes were noted at 96 h. Hence, 96 h AAL post-treatment and mock treated was selected for further analysis.
Relative expression levels of three transcripts (a–c) at 6 h, 24 h, 48 h, 72 h and 96 h. Experiment was performed with both the mock and AAL treated Col-0 (Col C and Col T) and AtECS1 (At C and At T) to see establishment of infection. eEF-1 alpha was used as an internal control. Data are the mean ± SE for 3 individual experiments (n = 3) using 3 biological replicates independently. Stars indicate significant differences between samples at *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001 according to the Student t-test
Proteomic analysis of Col-0 and AtECS1 under mock treated and AAL treated conditions
Proteomics approach is often undertaken to obtain a mechanistic insight to the effect of several stresses on plants (Datta and Chattopadhyay 2015; Carella et al. 2016; Huang et al. 2016). To understand the effect of GSH on AAL toxin treatment and plant defence, gel-free proteomic study was performed with three biological replicates of mock treated and AAL treated samples of Col-0 and AtECS1, the transgenic line. A total of 263 proteins were identified in mock treated Col-0, 269 proteins in mock treated AtECS1, 251 proteins in AAL treated Col-0 and 298 in AAL treated AtECS1, having scores above 30 and with at least 1 unique peptide count. Among them proteins having PSM values more than 1 were represented in tables (Supplemental Table S2-S5).
Functional categorization of proteins
Using PANTHER, total proteins from all four samples were classified under: biological process, molecular function and cellular component. They were also assigned under several protein classes. Although no significant changes were observed in representative proteins in Col-0 mock treated and AAL treated samples, but proteins were to some extent over-represented in AAL treated AtECS1 samples, under all four categories viz. biological process, molecular function cellular component, and protein classes compared to other samples (Supplemental Fig. S4).
Functional categorization of the identified proteins in mock treated Col-0 showed that about 34% of the proteins were under stress and defence category, 37% of the proteins belonged to photosynthetic, respiratory, carbon and energy metabolism category and 29% of the remaining proteins belonged to other categories viz. protein and nucleotide metabolism, signaling pathways, nitrogen metabolism. In AAL treated Col-0 samples, about 35% proteins were under stress and defence category, 34% belonged to photosynthetic, respiratory; carbon and energy metabolism and 31% belonged to protein metabolism, developmental process, signaling pathways, nutrient metabolism. In mock treated AtECS1, about 32% were under stress and defence category, 33% under photosynthetic, respiratory, carbon and energy metabolism and 35% to other categories including nucleotide and protein metabolism, signaling pathways, amino acid metabolism. In AAL treated AtECS1 about 38% of proteins were under stress and defence category, 34% under photosynthetic, respiratory, carbon and energy metabolism and 28% under protein and nucleotide metabolism, signaling, lipid metabolism, amino acid metabolism category (Fig. 5 and Supplemental Tables S2–S5). Under stress and defence category, proteins like, glutathione-S-transferases, superoxide dismutases, catalases, peroxiredoxins, APX, HSPs, myrosinases, PR 2, 4, 5 were identified. Under photosynthetic, respiratory, carbon and energy metabolism category of proteins, ribulose bisphosphate carboxylase large and small chains, oxygen evolving enhancer proteins, RUBISCO activase, chlorophyll a/b binding proteins, glyceraldehyde-3-phosphatedehydrogenase, malate dehydrogenase, phosphoglycerate kinase, sedoheptulose 1,7 bisphosphatase, ATP synthase subunit alpha, beta, etc. were identified. Several ribosomal proteins, ribosome recycling factors involved in translation were identified. Also, proteins involved in lipid metabolism viz. non-specific lipid transfer proteins, signaling pathway proteins viz. calmodulins, cytoskeletal proteins like actin were identified. Proteins related to cell death process viz. several proteins belonging to HSP 70 family, cysteine proteinase, cytochrome c were also among the identified proteins. Among these identified proteins several stress responsive proteins viz. PR2, 5, carbonic anhydrase (CA), glutathione S-transferase 8, glutamine synthetase, peptidyl-prolyl cis–trans isomerase, cytochrome b6-f complex iron-sulfur subunit, were known to be effected by SA as well (Uknes et al. 1992; Cao et al. 1994; Slaymaker et al. 2002; Blanco et al. 2009; Zhang et al. 2016), while PR4 was also identified which was known to be ET responsive (Potter et al. 1993). A few from the above mentioned proteins were chosen for further studies to establish the role of GSH in mitigating AAL mediated stress in plants.
Expression analysis of selected stress related genes identified in proteomics
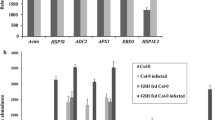
The expression analysis of some ET and SA responsive genes viz. PR4 (Uni Prot ID-P43082; Gene ID: 819632), PR5 (Uni Prot ID- P28493; Gene ID: 843842), CA1 (Uni Prot ID- P27140; Gene ID: 821134), was performed by qRT-PCR, which were identified in our proteomics data. The expression analysis of ET biosynthetic gene ACC oxidase (ACO1) (Yang and Hoffman 1984) was also done to see if ET biosynthesis can get influenced due to the toxin treatment. The expressions of the above mentioned genes were up regulated in AAL treated AtECS1 compared to AAL treated Col-0, except for PR4 and ACO1, which showed lower expression in AtECS1 compared to Col-0 after treatment with AAL. The expression of SA responsive PR1, were also noted as it was a strong marker of SA signaling, which got up regulated in AtECS1 compared to the Col-0, under AAL treated condition. Thus, there is an indication of the involvement of SA and ET in defence against AAL toxin. Higher expressions of APX1 (Uni Prot ID-Q05431; Gene ID: 837304), myrosinase (TGG1) (Uni Prot ID- P37702; Gene ID: 832669) and HSP70-1 (Uni Prot ID- P22953; Gene ID: 831020) were noted as well, in AtECS1 compared to Col-0 under AAL treated condition (Fig. 6).
Relative expression levels of transcripts. Experiment was performed in mock and AAL treated Col-0 (Col C and Col T) and AtECS1 (At C and At T). eEF-1 alpha was used as an internal control. Data are the mean ± SE for 3 individual experiments (n = 3) using 3 biological replicates independently. Stars indicate significant differences between samples at *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001 according to the Student t-test
Western blot analysis of selected stress responsive protein species identified in proteomics
To check if there were any changes in the expression, even at protein level, in addition to transcripts, western blot analysis was performed, where differential accumulation of PR5, PR4 and ACO1 were found in Col-0 and AtECS1. PR5 was up accumulated in AAL treated AtECS1 compared to AAL treated Col-0, while PR4 and ACO1, were down accumulated in AAL treated AtECS1 compared to AAL treated Col-0, though the levels of all proteins were higher in mock treated AtECS1 compared to Col-0 (Fig. 7).
Estimation of SA and ACC content in Col-0 and AtECS1 plants after AAL toxin treatment
To investigate if there was any direct effect of AAL toxin on the metabolite levels of SA and ACC, under differential GSH content, their estimation was performed by HPLC. It was noted that under AAL treated condition, the SA content was about 4 times higher in AtECS1 (Supplemental Fig. S5), compared to Col-0. However, ACC content was enhanced in Col-0 compared to AtECS1 under AAL treated condition, which corroborated with our previous transcript data and western blot analysis (Supplemental Fig. S6).
Discussion
GSH ameliorated AAL induced necrosis and cell death in plants
Enhanced GSH in AtECS1 caused decreased infectious lesion formation and its spreading, compared to Col-0 under AAL treated condition, as visualized in our morphological data. Since necrotrophic fungus kills the host tissues and feeds on them, it is advantageous for the host plants if there is a drop in infectious lesion formation and necrosis. Besides, presence of GSH also resulted in lower chlorophyll loss in leaves, of AAL treated AtECS1. Hence GSH improved the photosynthetic capacity of the plants even under stressed condition by maintaining the chlorophyll content, since photosynthetic rate and chlorophyll content are directly correlated (Fleischer 1935). This might help the plants in production of more carbon sources, which in turn helps in retaining the required biomass of plants, as well as provide the essential energy requirement for downstream defence reactions, since induction of host defence response is cost intensive (Swarbrick et al. 2006).
More pronounced degradation of protein was found in Col-0 compared to AtECS1 under AAL treated condition. Previous study reported that protein degradation is accelerated under carbon starvation to provide an alternative source of carbon for respiration (Araújo et al. 2011). Protein degradation may also increase under several pathogen induced stresses (Beers et al. 2000) and there was evidence as well for disruption of protein biosynthetic machinery under A. alternata infection (Sinha and Chattopadhyay 2011). In AtECS1 higher amount of chlorophyll, which indicated higher photosynthetic rate, could have compensated for the carbon starvation, and hence lowering protein degradation for respiration, under AAL treated condition. Moreover, GSH can help in lowering degradation of protein or protect them from getting damaged by activating other intrinsic defence mechanism (Park and Seo 2015; Kumar and Chattopadhyay 2018), under AAL treated condition.
Both callose deposition and the outburst of H2O2, which measures the extent of cell death, showed higher accumulation in Col-0 than in AtECS1 under AAL treated condition. H2O2 has been known as a signaling molecule which can help with stress acclimation, when present in low concentration, but also known to trigger cell death when present in excess amount (Gechev et al. 2002; Dat et al. 2003; Apel and Hirt 2004). Its overproduction can be a strategy to kill the host tissue in the initial phase of infection, in case of necrotrophic fungus (Tiedemann 1997). Previously it was known that A. alternata and AAL toxin as well, caused an oxidative outburst and concomitant deposition of callose in several plants including Arabidopsis (Gechev et al. 2004; Zhang et al. 2011; Chung 2012). GSH has been widely known for its antioxidative effect and as a scavenger of ROS molecules (Noctor and Foyer 1998). AtECS1 thus have gained an advantage over Col-0 in suppressing ROS induced cell death, triggered by AAL toxin.
Insight into GSH-mediated molecular changes to alleviate AAL induced stress in plants
The role of GSH in conferring stress tolerance in plants has been known. While external feeding of GSH or overexpressing lines having higher GSH content were always found to be more resistant to several stresses compared to wild type plants (Ghanta et al. 2011; Datta et al. 2015; Sinha et al. 2015), GSH depleted plants display susceptibilities to range of stresses (Dubreuil-Maurizi et al. 2011; Sobrino-Plata et al. 2014; Datta and Chattopadhyay 2015; Kumar et al. 2015). The resistance or susceptibilities of plants are direct manifestations of molecular changes happening inside the plants in response to stress. Proteomics studies revealed that while plants displaying higher GSH contents manifested enhanced expressions of stress and defence related proteins (Ghanta et al. 2011; Datta et al. 2015), a range of defence related proteins were down regulated in GSH depleted mutant pad2.1 (Datta and Chattopadhyay 2015; Kumar et al. 2015). Investigations on GSH depleted mutant viz. pad2.1 has been reported previously either in absence (Datta et al. 2015) or presence of both biotic/abiotic stresses (Datta and Chattopadhyay 2015; Kumar et al. 2015) and identified several down regulated defence related proteins. Considering this, presently we performed proteomic profiling of the transgenic line AtECS1 exhibiting enhanced GSH content to understand the mechanistic interaction of GSH with other molecules in mitigating fungal toxin induced stress in plants.
To investigate the role of GSH in plant defence against AAL toxin the initial proteomic analysis that was performed identified proteins under several categories. Among them, few proteins were found which were reported to be related to cell death in plants. PCD is controlled sequential events that lead to cell death (Lockshin and Zakeri 2004). It is crucial for multicellular development as well as in defence response by restricting the pathogens (Lam 2004). Fungal AAL toxin is known as one of the potent inducers of plant cell death (Wang et al. 1996). In the present proteomic study cysteine proteinase RD21A has been identified. It has been known to be involved in cell death in the transmitting tract and septum epidermis during flower development (Boex-Fontvieille et al. 2015). The protein has been known to function as a PCD promoting protease during infection by the necrotrophic fungal pathogen Botrytis cinerea and may participate in cell death in stressed or injured cells (Hayashi et al. 2001; Lampl et al. 2013). The protein was also found to be up regulated in tomato treated with AAL toxin (Zhang et al. 2016). Cytochrome C-1 has also been identified in our proteomics data. The release of cytochrome c from mitochondria is known to be an early event related to plant PCD (Balk et al. 1999; Balk and Leaver 2001). It has been reported to interact with a range of targets on PCD (Martínez-Fábregas et al. 2013). Along with the above mentioned proteins several members of HSP family proteins like, HSP70-1, HSP70-3, HSP70-2, HSP70-11 has also been identified in our proteomics data. While study reported that HSP can contribute to PCD during re-modelling of lace plant leaves (Rowarth et al. 2019) another report emphasized the role of HSP in inducing apoptosis in response to necrotrophic pathogen (Byth-Illing and Bornman 2014). Another study with tobacco protoplasts reported that HSP70 might play a role in suppression of apoptosis under increased temperature, during pathogen infection (Cronjé et al. 2004).
A set of proteins were SA and ET responsive. Previous comparative proteomic study in our lab between wild type and transgenic mint over expressing γ-ECS, having enhanced GSH content under A. alternata infection, identified differentially expressed proteins (Sinha et al. 2013). Corroborating with this and other evidences (Gechev et al. 2004; Wittstock et al. 2016) a few proteins from our LC–MS/MS proteome study were chosen for further downstream studies, to establish the relation of GSH with other molecules, under AAL treated condition:
GSH acts through SA and ET pathway to diminish the effect of AAL toxin in plants
Up regulation of PR4, the marker of ET signaling not only occurs under ET treatment but even under AAL toxin treatment as well (Gechev et al. 2004). In our proteomics data, hevein-like protein has been identified which corresponds to PR4 gene (Gene ID: 819632) was up regulated in AAL treated Col-0, which demonstrated that ET played an important role in AAL triggered response in plants. But, in case of AAL treated AtECS1 the expression of PR4 was lower than AAL treated Col-0, although the basal level of PR4 was higher in AtECS1 compared to Col-0, in mock treated plants. To study further the interaction of GSH and ET, in presence of AAL toxin, ACO1 gene expression was checked. Though basal level of ACO1 was greater in AtECS1 compared to Col-0, which led to the increased ET content in AtECS1, corroborating with the previous study (Datta et al. 2015), yet the expression profile of ACO1 was similar to PR4 under AAL treated condition i.e. higher in Col-0 than AtECS1. Also, our qRT-PCR data of ACS2 showed the same expression changes like PR4 and ACO1. The western blot analysis of both PR4 and ACO1 supported the transcript data. HPLC analysis of the ACC content showed that although basal level of ACC was higher in AtECS1 compared to Col-0, yet after AAL treatment the ACC content was much higher in Col-0 compared to AtECS1 corroborating with the transcript and protein levels of PR4 and ACO1. There are several reports that showed ET enhances cell death and necrosis in tomato, tobacco and pears under A. alternata infection or AAL treatment (Moore et al. 1999; Mase et al. 2012; Wang et al. 2017). Later in Arabidopsis it was also reported that application of ET inhibitor AVG reduced cell death by about 50% under AAL treated condition (Gechev et al. 2004). Thus, ET potentiates the establishment of the necrotic cell death in plants, under AAL treated condition.
Again, pathogenesis-related 5 protein or PR5 has been identified here which corresponds to PR5 gene (Gene ID: 843842), which is SA responsive gene, was found to be up regulated in AAL treated AtECS1 compared to Col-0. The expression of SA responsive PR1 gene also showed similar expression pattern as PR5. The western blot analysis of PR5 supported the transcript data as well. Since PR5 and PR1 both are SA responsive, their elevated expressions under enhanced GSH condition in AtECS1 indicated that GSH might had led to higher SA content. HPLC analysis showed that under AAL treated condition there was change in SA content. The basal level of SA was higher in AtECS1 as compared to Col-0. In our previous study, we also demonstrated that enhanced GSH level caused up regulation of SA content in plants under control condition (Ghanta et al. 2011). However, there was about fourfold more SA content in AtECS1 than in Col-0, under AAL treated condition. Previous reports in tomato showed that exogenous application of SA caused to acquire enhanced resistance against A. alternata (Esmailzadeh et al. 2008). In Arabidopsis also, microarray study after AAL treatment resulted in up regulation of SA pathway genes, PR5 and PR1 (Gechev et al. 2004), which implied involvement of SA in regulating AAL-induced stress in Arabidopsis.
ET was known to induce oxidative stress and ROS also can induce ET production through modulating expressions of the two ET biosynthetic pathway enzymes, ACC synthase and ACC oxidase (de Jong et al. 2002; Wang et al. 2002). Previous study showed that in absence of AAL, exogenous addition of ACC did increase the level of ET, but didn’t cause necrosis to appear (Moussatos et al. 1994). Thus, increase in ET is not the cause of necrosis and cell death, rather the presence of AAL caused a trigger of complex signaling that increased the biosynthesis and evolution of ET as well as ROS at toxic levels in the plants to cause the necrotic symptoms and cell death. So suppression of their level can be advantageous to plants, to arrest the effect of AAL toxin. Earlier study showed that exogenous application of SA caused suppression of ET pathway and hence decreased AAL mediated cell death (Jia et al. 2013). In present study, though SA level was found to be high after AAL treatment in Col-0, yet ET level was higher there as well and hence necrotic cell death was more there when compared to AtECS1 under AAL treated condition. Therefore, a threshold level of endogenous SA is required to be crossed, to transcend the toxic effect of ET presumably by suppressing its synthesis as well as its signaling, under AAL treated condition, as noted in AtECS1 with higher content of GSH. Thus, enhanced GSH caused SA mediated resistance by suppressing ET, when there is successful AAL mediated stress induction in plants.
GSH acts through various stress-responsive molecules to mitigate AAL induced stress
The role of CA in C3 plants had been known for decades in the conversion of HCO3− to CO2 to ensure maximum rates of fixation by Rubisco (Everson 1970; Poincelot 1972; Werdan and Heldt 1972). Besides this the role of CA as a salicylate binding antioxidant molecule has already been established (Slaymaker et al. 2002). In our present study we find that the beta carbonic anhydrase 1 protein was identified, which corresponds to CA1 (Gene ID: 821134) and the later showed higher expression in AAL treated AtECS1 compared to AAL treated Col-0. Thus in AtECS1, GSH can play a role in conferring resistance against AAL mediated stress, through up regulating CA, which not only helped in enhanced carbon fixation, but also plays a protective role against oxidative stress induced damage caused by the toxin.
Myrosinases (β‐thioglucoside glucohydrolase, TGG) are the group of enzymes that cleave glucosinolates, which are amino acid‐derived secondary metabolites (Barth and Jander 2006). In cruciferous plants like Arabidopsis, myrosinase-glucosinolate system forms a preformed chemical defence system against several insect, pathogens and herbivores (Bones and Rossiter 1996; Raybould and Moyes 2001). It was also reported that TGG-dependent hydrolysis of glucosinolate plays an important role in the response of Arabidopsis to Fumonisin B (FB1), a sphinganine analogue mycotoxin like AAL toxin (Abbas et al. 1994), since lack of TGG rendered plants more sensitive to FB1 (Zhao et al. 2015). Our proteomics data identified the protein myrosinase 1 which corresponds to TGG1 (Gene ID: 832669). Here, basal level expression of TGG1, was found to be higher in AtECS1 than Col-0, and it was highly induced in AtECS1 compared to Col-0 after AAL treatment. Thus, enhanced GSH level is capable to reduce AAL toxin mediated stress in AtECS1 by increasing myrosinase amount and hence catalyzing glucosinolate breakdown to pathotoxic products making the plants tolerant to the toxin.
One of the causal reasons of AAL toxin and A. alternata mediated cell death is ROS accumulation (Gechev et al. 2004; Prasad and Upadhyay 2010). The cytosolic L-ascorbate peroxidase 1 which has been identified in our proteomics data corresponds to APX1 (Gene ID: 837304), the later being found to give increased expression in AAL treated AtECS1 compared to Col-0. Increasing the expression of APX, a ROS scavenging antioxidant enzyme, may be a possible way by which GSH facilitated the reduction of toxic effect of ROS and hence necrotic cell death, under AAL mediated stress. This was in agreement with previous study with A. alternata infected plants (Sinha et al. 2013).
HSPs are a subset of proteins, acting as the molecular chaperones and known for their rapid induction under large numbers of stresses (Wang et al. 2004; Scarpeci et al. 2008). Lower expression of HSPs can cause increased susceptibility of plants to A. alternata induced stress (Zhu et al. 2017). Earlier it has been reported that GSH induced the expression of several HSPs (Kumar and Chattopadhyay 2018). In present proteomic study HSP70-1 protein was identified which corresponds to HSP70-1 gene (Gene ID: 831020). HSP70-1 gene was found to be up accumulated in AtECS1 compared to Col-0 under AAL treated condition, which corroborated with previous study where HSP was up regulated upon A. alternata infection (Sinha et al. 2013). Thus, enhanced GSH makes the plants more resistant, through the enhanced expression of HSPs.
Conclusions
In this study, 10 µM of AAL toxin was applied to the leaves of Arabidopsis, to establish the stress successfully. ET and ROS are the two key mediators in A. alternata as well as AAL induced cell death. GSH promoted mitigation of AAL induced stress via SA mediated suppression of ET. Moreover, being an antioxidant molecule itself, GSH was also found to act along with CA and APX as well to minimize the effect of toxic ROS that is a causal agent of necrotic cell death in AAL treated plants. GSH also can act through myrosinases in addition to chaperones like HSPs, to subside the effect of the toxin in plants as well. Thus our study with the model plant Arabidopsis suggests that GSH can act as a potential candidate in alleviating AAL induced distress in plants by acting through several stress and defence related molecules. This might help for future research on GSH mediated mitigation of environmental stress in economically important plants.
Abbreviations
- DAPI:
-
4′, 6-Diamidino-2-phenylindole
- PCD:
-
Programmed cell death
- PCR:
-
Polymerase chain reaction
- HPLC:
-
High performance liquid chromatography
- LOH2:
-
LAG one homologue 2
References
Abbas HK, Tanaka T, Duke SO, Porter JK, Wray EM, Hodges L, Sessions AE, Wang E, Merrill AH Jr, Riley RT (1994) Fumonisin- and AAL-toxin-induced disruption of sphingolipid metabolism with accumulation of free sphingoid bases. Plant Physiol 106:1085–1093. https://doi.org/10.1104/pp.106.3.1085
Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55:373–399. https://doi.org/10.1146/annurev.arplant.55.031903.141701
Araújo WL, Tohge T, Ishizaki K, Leaver CJ, Fernie AR (2011) Protein degradation: an alternative respiratory substrate for stressed plants. Trends Plant Sci 16:489–498. https://doi.org/10.1016/j.tplants.2011.05.008
Balk J, Leaver CJ (2001) The PET1-CMS mitochondrial mutation in sunflower is associated with premature programmed cell death and cytochrome c release. Plant Cell 13:1803–1818. https://doi.org/10.2307/3871320
Balk J, Leaver CJ, McCabe PF (1999) Translocation of cytochrome c from the mitochondria to the cytosol occurs during heat-induced programmed cell death in cucumber plants. FEBS Lett 463:151–154. https://doi.org/10.1016/s0014-5793(99)01611-7
Ball L, Accotto GP, Bechtold U, Creissen G, Funck D, Jimenez A, Kular B, Leyland N, Mejia-Carranza J, Reynolds H, Karpins S, Mullineaux PM (2004) Evidence for a direct link between glutathione biosynthesis and stress defense gene expression in Arabidopsis. Plant Cell 16:2448–2462. https://doi.org/10.1105/tpc.104.022608
Barth C, Jander G (2006) Arabidopsis myrosinases TGG1 and TGG2 have redundant function in glucosinolate breakdown and insect defense. Plant J 46:549–562. https://doi.org/10.1111/j.1365-313x.2006.02716.x
Beers EP, Woffenden BJ, Zhao C (2000) Plant proteolytic enzymes: possible roles during programmed cell death. Plant Mol Biol 44:399–415. https://doi.org/10.1007/978-94-010-0934-8_12
Blanco F, Salinas P, Cecchini NM, Jordana X, Van Hummelen P, Alvarez ME, Holuigue L (2009) Early genomic responses to salicylic acid in Arabidopsis. Plant Mol Biol 70:79–102. https://doi.org/10.1007/s11103-009-9458-1
Boex-Fontvieille E, Rustgi S, Reinbothe S, Reinbothe C (2015) A Kunitz-type protease inhibitor regulates programmed cell death during flower development in Arabidopsis thaliana. J Exp Bot 66(20):6119–6135. https://doi.org/10.1093/jxb/erv327
Bones AM, Rossiter JT (1996) The myrosinase–glucosinolate system, its organisation and biochemistry. Physiol Plant 97:194–208. https://doi.org/10.1034/j.1399-3054.1996.970128.x
Bradford MM (1976) A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein–dye binding. Anal Biochem 72:248–254. https://doi.org/10.1006/abio.1976.9999
Bushey DF, Law DM, Davies PJ (1987) High-performance liquid chromatography analysis of 1-aminocyclopropane-1-carboxylic acid using o-phthaldialdehyde precolumn derivatization. Anal Biochem 167:31–36. https://doi.org/10.1016/0003-2697(87)90130-8
Byth-Illing HA, Bornman L (2014) Heat shock, with recovery, promotes protection of Nicotiana tabacum during subsequent exposure to Ralstonia solanacearum. Cell Stress Chaperones 19:193–203. https://doi.org/10.1007/s12192-013-0445-8
Cannella D, Möllers KB, Frigaard NU, Jensen PE, Bjerrum MJ, Johansen KS, Felby C (2016) Light driven oxidation of polysaccharides by photosynthetic pigments and a metalloenzyme. Nat Commun 7:11134. https://doi.org/10.1038/ncomms11134
Cao H, Bowling SA, Gordon AS, Dong X (1994) Characterization of an Arabidopsis mutant that is non responsive to inducers of systemic acquired resistance. Plant Cell 6:1583–1592. https://doi.org/10.1105/tpc.6.11.1583
Carella P, Merl-Pham J, Wilson DC, Dey S, Hauck SM, Vlot AC, Cameron RK (2016) Comparative proteomics analysis of phloem exudates collected during the induction of systemic acquired resistance. Plant Physiol 171:1495–1510. https://doi.org/10.1104/pp.16.00269
Carter GA (1994) Ratios of leaf reflectances in narrow wavebands as indicators of plant stress. Int J Remote Sens 15:697–703. https://doi.org/10.1080/01431169408954109
Chen Z, Silva H, Klessig DF (1993) Active oxygen species in the induction of plant systemic acquired resistance by salicylic acid. Science 262:1883–1886. https://doi.org/10.1126/science.8266079
Cheng MC, Ko K, Chang WL, Kuo WC, Chen GH, Lin TP (2015) Increased glutathione contributes to stress tolerance and global translational changes in Arabidopsis. Plant J 83:926–939. https://doi.org/10.1111/tpj.12940
Chung KR (2012) Stress response and pathogenicity of the necrotrophic fungal pathogen Alternaria alternata. Scientifica 2012:1–17. https://doi.org/10.6064/2012/635431
Cronjé MJ, Weir IE, Bornman L (2004) Salicylic acid-mediated potentiation of Hsp70 induction correlates with reduced apoptosis in tobacco protoplasts. Cytometry A 61:76–87. https://doi.org/10.1002/cyto.a.20036
Cummins I, Dixon DP, Freitag-Pohl S, Skipsey M, Edwards R (2011) Multiple roles for plant glutathione transferases in xenobiotic detoxification. Drug Metab Rev 43:266–280. https://doi.org/10.3109/03602532.2011.552910
Dat JF, Pellinen R, Beeckman T, Van De Cotte B, Langebartels C, Kangasjärvi J, Inzé D, Van Breusegem F (2003) Changes in hydrogen peroxide homeostasis trigger an active cell death process in tobacco. Plant J 33:621–632. https://doi.org/10.1046/j.1365-313x.2003.01655.x
Datta R, Chattopadhyay S (2015) Changes in the proteome of pad2-1, a glutathione depleted Arabidopsis mutant, during Pseudomonas syringae infection. J Proteomics 126:82–93. https://doi.org/10.1016/j.jprot.2015.04.036
Datta R, Sinha R, Chattopadhyay S (2013) Changes in leaf proteome profile of Arabidopsis thaliana in response to salicylic acid. J Biosci 38:317–328. https://doi.org/10.1007/s12038-013-9308-9
Datta R, Kumar D, Sultana A, Hazra S, Bhattacharyya D, Chattopadhyay S (2015) Glutathione regulates 1-aminocyclopropane-1-carboxylate synthase transcription via WRKY33 and 1-aminocyclopropane-1-carboxylate oxidase by modulating messenger RNA stability to induce ethylene synthesis during stress. Plant Physiol 169:2963–2981. https://doi.org/10.1104/pp.15.01543
de Jong AJ, Yakimova ET, Kapchina VM, Woltering EJ (2002) A critical role for ethylene in hydrogen peroxide release during programmed cell death in tomato suspension cells. Planta 214:537–545. https://doi.org/10.1007/s004250100654
Diaz Vivancos P, Wolff T, Markovic J, Pallardo FV, Foyer CH (2010) A nuclear glutathione cycle within the cell cycle. Biochem J 431:169–178. https://doi.org/10.1042/bj20100409
Dron M, Clouse SD, Dixon RA, Lawton MA, Lamb CJ (1988) Glutathione and fungal elicitor regulation of a plant defense gene promoter in electroporated protoplasts. Proc Natl Acad Sci USA 85:6738–6742. https://doi.org/10.1073/pnas.85.18.6738
Dubreuil-Maurizi C, Vitecek J, Marty L, Branciard L, Frettinger P, Wendehenne D, Meyer AJ, Mauch F, Poinssot B (2011) Glutathione deficiency of the Arabidopsis mutantpad2-1 affects oxidative stress-related events, defense gene expression, and the hypersensitive response. Plant Physiol 157:2000–2012. https://doi.org/10.1104/pp.111.182667
Egusa M, Ozawa R, Takabayashi J, Otani H, Kodama M (2009) The jasmonate signaling pathway in tomato regulates susceptibility to a toxin-dependent necrotrophic pathogen. Planta 229:965–976. https://doi.org/10.1007/s00425-009-0890-x
Esmailzadeh M, Soleimani MJ, Rouhani H (2008) Exogenous applications of salicylic acid for inducing systemic acquired resistance against tomato stem canker disease. J Biol Sci 8:1039–1044. https://doi.org/10.3923/jbs.2008.1039.1044
Everson RG (1970) Carbonic anhydrase and CO2 fixation in isolated chloroplasts. Phytochemistry 9:25–32. https://doi.org/10.1016/s0031-9422(00)86610-8
Fleischer WE (1935) The relation between chlorophyll content and rate of photosynthesis. J Gen Physiol 18:573–597. https://doi.org/10.1085/jgp.18.4.573
Foyer CH, Noctor G (2005) Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell 17:1866–1875. https://doi.org/10.1105/tpc.105.033589
Freeman JL, Garcia D, Kim D, Hopf A, Salt DE (2005) Constitutively elevated salicylic acid signals glutathione-mediated nickel tolerance in Thlaspi nickel hyperaccumulators. Plant Physiol 137:1082–1091. https://doi.org/10.1104/pp.104.055293
Gechev T, Gadjev I, Van Breusegem F, Inzé D, Dukiandjiev S, Toneva V, Minkov I (2002) Hydrogen peroxide protects tobacco from oxidative stress by inducing a set of antioxidant enzymes. Cell Mol Life Sci 59:708–714. https://doi.org/10.1007/s00018-002-8459-x
Gechev TS, Gadjev IZ, Hille J (2004) An extensive microarray analysis of AAL-toxin-induced cell death in Arabidopsis thaliana brings new insights into the complexity of programmed cell death in plants. Cell Mol Life Sci 61:1185–1197. https://doi.org/10.1007/s00018-004-4067-2
Ghanta S, Chattopadhyay S (2011) Glutathione as a signaling molecule: another challenge to pathogens. Plant Signal Behav 6:783–788. https://doi.org/10.4161/psb.6.6.15147
Ghanta S, Bhattacharyya D, Sinha R, Banerjee A, Chattopadhyay S (2011) Nicotiana tabacum overexpressing γ-ECS exhibits biotic stress tolerance likely through NPR1-dependent salicylic acid-mediated pathway. Planta 233:895–910. https://doi.org/10.1007/s00425-011-1349-4
Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43:205–227. https://doi.org/10.1146/annurev.phyto.43.040204.135923
Grant M, Jones J (2009) Hormone (dis) harmony moulds plant health and disease. Science 324:750–752. https://doi.org/10.1126/science.1173771
Gulyás Z, Boldizsár A, Novák A, Szalai G, Pál M, Galiba G, Kocsy G (2014) Central role of the flowering repressor ZCCT2 in the redox control of freezing tolerance and the initial development of flower primordia in wheat. BMC Plant Biol 14:91. https://doi.org/10.1186/1471-2229-14-91
Hayashi Y, Yamada K, Shimada T, Matsushima R, Nishizawa NK, Nishimura M, Hara-Nishimura I (2001) A proteinase-storing body that prepares for cell death or stresses in the epidermal cells of Arabidopsis. Plant Cell Physiol 42:894–899. https://doi.org/10.1093/pcp/pce144
Hernández JA, Barba-Espín G, Diaz Vivancos P (2017) Glutathione-mediated biotic stress tolerance in plants. In: Hossain MA, Mostofa MG, Diaz-Vivancos P, Burritt DJ (eds) Glutathione in plant growth, development, and stress tolerance. Springer, Cham, pp 309–329
Huang Y, Jin D, Lu C, Lan X, Qiao P, Li H, Chen Y (2016) Proteomic responses associated with freezing tolerance in the callus of the Tibetan alpine plant Saussurea laniceps during cold acclimation. Plant Cell Tiss Organ Cult 124:81–95. https://doi.org/10.1007/s11240-015-0876-2
Isaacson T, Damasceno CMB, Saravanan RS, He Y, Catala C, Saladie M, Rose JK (2006) Sample extraction techniques for enhanced proteomic analysis of plant tissues. Nat Protoc 1:769–774. https://doi.org/10.1038/nprot.2006.102
Jia C, Zhang L, Liu L, Wang J, Li C, Wang Q (2013) Multiple phytohormone signalling pathways modulate susceptibility of tomato plants to Alternaria alternata f. sp. lycopersici. J Exp Bot 64:637–650. https://doi.org/10.1093/jxb/ers360
Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30. https://doi.org/10.1093/nar/28.1.27
Kouzai Y, Kimura M, Watanabe M, Kusunoki K, Osaka D, Suzuki T, Matsui H, Yamamoto M, Ichinose Y, Toyoda K, Matsuura T, Mori IC, Hirayama T, Minami E, Nishizawa Y, Inoue K, Onda Y, Mochida K, Noutoshi Y (2018) Salicylic acid-dependent immunity contributes to resistance against Rhizoctonia solani, a necrotrophic fungal agent of sheath blight, in rice and Brachypodium distachyon. New Phytol 217:771–783. https://doi.org/10.1111/nph.14849
Kumar D, Chattopadhyay S (2018) Glutathione modulates the expression of heat shock proteins via the transcription factors BZIP10 and MYB21 in Arabidopsis. J Exp Bot 69:3729–3743. https://doi.org/10.1093/jxb/ery166
Kumar D, Datta R, Hazra S, Sultana A, Mukhopadhyay R, Chattopadhyay S (2015) Transcriptomic profiling of Arabidopsis thaliana mutant pad2.1 in response to combined cold and osmotic stress. PLoS ONE 10:e0122690. https://doi.org/10.1371/journal.pone.0122690
Kunkel BN, Brooks DM (2002) Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol 5:325–331. https://doi.org/10.1016/s1369-5266(02)00275-3
Kuźniak E, Głowacki R, Chwatko G, Kopczewski T, Wielanek M, Gajewska E, Skłodowska M (2014) Involvement of ascorbate, glutathione, protein S-thiolation and salicylic acid in benzothiadiazole-inducible defence response of cucumber against Pseudomonas syringae pv lachrymans. Physiol Mol Plant Pathol 86:89–97. https://doi.org/10.1016/j.pmpp.2014.04.004
Lam E (2004) Controlled cell death, plant survival and development. Nat Rev Mol Cell Biol 5:305–315. https://doi.org/10.1038/nrm1358
Lampl N, Alkan N, Davydov O, Fluhr R (2013) Set-point control of RD21 protease activity by AtSerpin1 controls cell death in Arabidopsis. Plant J 74:498–510. https://doi.org/10.1111/tpj.12141
Lata C, Jha S, Dixit V, Sreenivasulu N, Prasad M (2011) Differential antioxidative responses to dehydration-induced oxidative stress in core set of foxtail millet cultivars [Setaria italica (L.)]. Protoplasma 248:817–828. https://doi.org/10.1007/s00709-010-0257-y
Li Y, Dankher OP, Carreira L, Smith AP, Meagher RB (2006) The shoot-specific expression of gamma-glutamylcysteine synthetase directs the long-distance transport of thiol-peptides to roots conferring tolerance to mercury and arsenic. Plant Physiol 141:288–298. https://doi.org/10.1104/pp.105.074815
Lichtenthaler HK (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. In: Packer L, Douce R (eds) Methods in enzymology, vol 148. Academic Press, Cambridge, pp 350–382
Loake G, Grant M (2007) Salicylic acid in plant defence—the players and protagonists. Curr Opin Plant Biol 10:466–472. https://doi.org/10.1016/j.pbi.2007.08.008
Lockshin RA, Zakeri Z (2004) Apoptosis, autophagy, and more. Int J Biochem Cell Biol 36:2405–2419. https://doi.org/10.1016/j.biocel.2004.04.011
Martínez-Fábregas J, Díaz-Moreno I, González-Arzola K, Janocha S, Navarro JA, Hervás M, Bernhardt R, Díaz-Quintana A, De la Rosa MÁ (2013) New Arabidopsis thaliana cytochrome c partners: a look into the elusive role of cytochrome c in programmed cell death in plants. Mol Cell Proteomics 12:3666–3676. https://doi.org/10.1074/mcp.M113.030692
Mase K, Mizuno T, Ishihama N, Fujii T, Mori H, Kodama M, Yoshioka H (2012) Ethylene signaling pathway and MAPK cascades are required for AAL toxin-induced programmed cell death. Mol Plant Microbe Interact 25:1015–1025. https://doi.org/10.1094/mpmi-02-12-0036-r
May MJ, Vernoux T, Leaver CJ, Van Montagu M, Inzé D (1998) Glutathione homeostasis in plants: implications for environmental sensing and plant development. J Exp Bot 49:649–667. https://doi.org/10.1093/jexbot/49.321.649
Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD (2019) PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res 47:419–426. https://doi.org/10.1093/nar/gky1038
Moore T, Martineau B, Bostock RM, Lincoln JE, Gilchrist DG (1999) Molecular and genetic characterization of ethylene involvement in mycotoxin-induced plant cell death. Physiol Mol Plant Path 54:73–85. https://doi.org/10.1006/pmpp.1998.0190
Mou Z, Fan W, Dong X (2003) Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113:935–944. https://doi.org/10.1016/s0092-8674(03)00429-x
Moussatos VV, Yang SF, Ward B, Gilchrist DG (1994) AAL-toxin induced physiological changes in Lycopersicon esculentum Mill: roles for ethylene and pyrimidine intermediates in necrosis. Physiol Mol Plant Path 44:455–468. https://doi.org/10.1016/s0885-5765(05)80101-8
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15:473–497. https://doi.org/10.1111/j.1399-3054.1962.tb08052.x
Murphy AM, Holcombe LJ, Carr JP (2000) Characteristics of salicylic acid-induced delay in disease caused by a necrotrophic fungal pathogen in tobacco. Physiol Mol Plant Pathol 57:47–54. https://doi.org/10.1006/pmpp.2000.0280
Noctor G, Foyer CH (1998) Ascorbate and Glutathione: keeping active oxygen under control. Annu Rev Plant Physiol Plant Mol Biol 49:249–279. https://doi.org/10.1146/annurev.arplant.49.1.249
Parisy V, Poinssot B, Owsianowski L, Buchala A, Glazebrook J, Mauch F (2007) Identification of PAD2 as a γ-glutamylcysteine synthetase highlights the importance of glutathione in disease resistance of Arabidopsis. Plant J 49:159–172. https://doi.org/10.1111/j.1365-313x.2006.02938.x
Park CJ, Seo YS (2015) Heat shock proteins: a review of the molecular chaperones for plant immunity. Plant Pathol J 31:323–333. https://doi.org/10.5423/ppj.rw.08.2015.0150
Poincelot RP (1972) Intracellular distribution of carbonic anhydrase in spinach leaves. Biochim Biophys Acta 258:637–642. https://doi.org/10.1016/0005-2744(72)90255-0
Potter S, Uknes S, Lawton K, Winter AM, Chandler D, DiMaio J, Novitzky R, Ward E, Ryals J (1993) Regulation of a hevein-like gene in Arabidopsis. Mol Plant Microbe Interact 6:680–705. https://doi.org/10.1094/mpmi-6-680
Prasad V, Upadhyay RS (2010) Alternaria alternata f.sp. lycopersici and its toxin trigger production of H2O2 and ethylene in tomato. J Plant Pathol 92:103–108
Raybould AF, Moyes CL (2001) The ecological genetics of aliphatic glucosinolates. Heredity 87:383–391. https://doi.org/10.1046/j.1365-2540.2001.00954.x
Reinbothe C, Springer A, Samol I, Reinbothe S (2009) Plant oxylipins: role of jasmonic acid during programmed cell death, defence and leaf senescence. FEBS J 276:4666–4681. https://doi.org/10.1111/j.1742-4658.2009.07193.x
Rowarth NM, Dauphinee AN, Denbigh GL, Gunawardena AH (2019) Hsp70 plays a role in programmed cell death during the remodelling of leaves of the lace plant (Aponogeton madagascariensis). J Exp Bot 71:907–918. https://doi.org/10.1093/jxb/erz447
Ruiz JM, Blumwald E (2002) Salinity-induced glutathione synthesis in Brassica napus. Planta 214:965–969. https://doi.org/10.1007/s00425-002-0748-y
Scarpeci TE, Zanor MI, Valle EM (2008) Investigating the role of plant heat shock proteins during oxidative stress. Plant Signal Behav 3:856–857. https://doi.org/10.4161/psb.3.10.6021
Sinha R, Chattopadhyay S (2011) Changes in the leaf proteome profile of Mentha arvensis in response to Alternaria alternata infection. J Proteomics 74:327–336. https://doi.org/10.1016/j.jprot.2010.11.009
Sinha R, Bhattacharyya D, Majumdar AB, Datta R, Hazra S, Chattopadhyay S (2013) Leaf proteome profiling of transgenic mint infected with Alternaria alternata. J Proteomics 93:117–132. https://doi.org/10.1016/j.jprot.2013.01.020
Sinha R, Kumar D, Datta R, Hazra S, Bhattacharyya D, Mazumdar AB, Mukhopadhyay R, Sultana A, Chattopadhyay S (2015) Integrated transcriptomic and proteomic analysis of Arabidopsis thaliana exposed to glutathione unravels its role in plant defense. Plant Cell Tissue Organ Cult 120:975–988. https://doi.org/10.1007/s11240-014-0651-9
Slaymaker DH, Navarre DA, Clark D, del Pozo O, Martin GB, Klessig DF (2002) The tobacco salicylic acid-binding protein 3 (SABP3) is the chloroplast carbonic anhydrase, which exhibits antioxidant activity and plays a role in the hypersensitive defense response. Proc Natl Acad Sci USA 99:11640–11645. https://doi.org/10.1073/pnas.182427699
Sobrino-Plata J, Meyssen D, Cuypers A, Escobar C, Hernández LE (2014) Glutathione is a key antioxidant metabolite to cope with mercury and cadmium stress. Plant Soil 377:369–381. https://doi.org/10.1007/s11104-013-2006-4
Swarbrick PJ, Schulze-Lefert P, Scholes JD (2006) Metabolic consequences of susceptibility and resistance (race-specific and broad-spectrum) in barley leaves challenged with powdery mildew. Plant Cell Environ 29:1061–1076. https://doi.org/10.1111/j.1365-3040.2005.01472.x
Thomma BP, Eggermont K, Penninckx IA, Mauch-Mani B, Vogelsang R, Cammue BP, Broekaert WF (1998) Separate jasmonate dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci USA 95:15107–15111. https://doi.org/10.1073/pnas.95.25.15107
Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB (1997) Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley—powdery mildew interaction. Plant J 11:1187–1194. https://doi.org/10.1046/j.1365-313x.1997.11061187.x
Tiedemann AV (1997) Evidence for a primary role of active oxygen species in induction of host cell death during infection of bean leaves with Botrytis cinerea. Physiol Mol Plant Pathol 50:151–166. https://doi.org/10.1006/pmpp.1996.0076
Tsuge T, Harimoto Y, Akimitsu K, Ohtani K, Kodama M, Akagi Y, Egusa M, Yamamoto M, Otani H (2013) Host-selective toxins produced by the plant pathogenic fungus Alternaria alternata. FEMS Microbiol Rev 37:44–66. https://doi.org/10.1111/j.1574-6976.2012.00350.x
Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J (1992) Acquired resistance in Arabidopsis. Plant Cell 4:645–656. https://doi.org/10.1105/tpc.4.6.645
Underwood W, Zhang S, He SY (2007) The Pseudomonas syringae type III effector tyrosine phosphatase HopAO1 suppresses innate immunity in Arabidopsis thaliana. Plant J 52:658–672. https://doi.org/10.1111/j.1365-313x.2007.03262.x
Van Breusegem F, Dat JF (2006) Reactive oxygen species in plant cell death. Plant Physiol 141:384–390. https://doi.org/10.1104/pp.106.078295
Vizcaíno JA, Côté RG, Csordas A, Dianes JA, Fabregat A, Foster JM, Griss J, Alpi E, Birim M, Contell J, O'Kelly G, Schoenegger A, Ovelleiro D, Pérez-Riverol Y, Reisinger F, Ríos D, Wang R, Hermjakob H (2013) The proteomics identifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res 41(D1):D1063–D1069. https://doi.org/10.1093/nar/gks1262
Wang H, Li J, Bostock RM, Gilchrist DG (1996) Apoptosis: a functional paradigm for programmed plant cell deathinduced by a host-selective phytotoxin and invoked during development. Plant Cell 8:375–391. https://doi.org/10.1105/tpc.8.3.375
Wang KL-C, Li H, Ecker JR (2002) Ethylene biosynthesis and signaling networks. Plant Cell 14:131–151. https://doi.org/10.1105/tpc.001768
Wang W, Vinocur B, Shoseyov O, Altman A (2004) Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci 9:244–252. https://doi.org/10.1016/j.tplants.2004.03.006
Wang SD, Zhu F, Yuan S, Yang H, Xu F, Shang J, Xu MY, Jia SD, Zhang ZW, Wang J, Xi DH, Lin HH (2011) The roles of ascorbic acid and glutathione in symptom alleviation to SA-deficient plants infected with RNA viruses. Planta 234:171–181. https://doi.org/10.1007/s00425-011-1391-2
Wang H, Lin J, Chang Y, Jiang CZ (2017) Comparative transcriptomic analysis reveals that ethylene/H2O2-mediated hypersensitive response and programmed cell death determine the compatible interaction of sand pear and Alternaria alternata. Front Plant Sci 8:195. https://doi.org/10.3389/fpls.2017.00195
Werdan K, Heldt HW (1972) Accumulation of bicarbonate in intact chloroplasts following a pH gradient. Biochim Biophys Acta 283:430–441. https://doi.org/10.1016/0005-2728(72)90260-5
Willekens H, Chamnongpol S, Davey M, Schraudner M, Langebartels C, Van Montagu M, Inzé D, Van Camp W (1997) Catalase is a sink for H2O2 and is indispensable for stress defence in C3 plants. EMBO J 16:4806–4816. https://doi.org/10.1093/emboj/16.16.4806
Wingate VMP, Lawton MA, Lamb CJ (1988) Glutathione causes a massive and selective induction of plant defense genes. Plant Physiol 87:206–210. https://doi.org/10.1104/pp.87.1.206
Wittstock U, Kurzbach E, Herfurth A-M, Stauber EJ (2016) Chapter six—glucosinolate breakdown. In: Kopriva S (ed) Advances in botanical research. Academic Press, Cambridge, pp 125–169
Xu L, Du L (2006) Direct detection and quantification of Alternaria alternata lycopersici toxins using high-performance liquid chromatography-evaporative light-scattering detection. J Microbiol Methods 64:398–405. https://doi.org/10.1016/j.mimet.2005.06.004
Yang SF, Hoffman NE (1984) Ethylene biosynthesis and its regulation in higher plants. Ann Rev Plant Physiol 35:155–189. https://doi.org/10.1146/annurev.arplant.35.1.155
Zagorchev L, Seal CE, Kranner I, Odjakova M (2013) A central role for thiols in plant tolerance to abiotic stress. Int J Mol Sci 14:7405–7432. https://doi.org/10.3390/ijms14047405
Zhang L, Jia C, Liu L, Zhang Z, Li C, Wang Q (2011) The involvement of jasmonates and ethylene in Alternaria alternata f. sp. lycopersici toxin-induced tomato cell death. J Exp Bot 62:5405–5418. https://doi.org/10.1093/jxb/err217
Zhang M, Koh J, Liu L, Shao Z, Liu H, Hu S, Zhu N, Dufresne CP, Chen S, Wang Q (2016) Critical role of COI1-dependent jasmonate pathway in AAL toxin induced PCD in tomato revealed by comparative proteomics. Sci Rep 6:28451. https://doi.org/10.1038/srep28451
Zhao Y, Wang J, Liu Y, Miao H, Cai C, Shao Z, Guo R, Sun B, Jia C, Zhang L, Gigolashvili T, Wang Q (2015) Classic myrosinase-dependent degradation of indole glucosinolate attenuates fumonisin B1-induced programmed cell death in Arabidopsis. Plant J 81:920–933. https://doi.org/10.1111/tpj.12778
Zhu L, Ni W, Liu S, Cai B, Xing H, Wang S (2017) Transcriptomics analysis of apple leaves in response to Alternaria alternata apple pathotype infection. Front Plant Sci 8:22. https://doi.org/10.3389/fpls.2017.00022
Acknowledgements
The authors are grateful to the Director, Council of Scientific & Industrial Research (CSIR)-Indian Institute of Chemical Biology, Kolkata for providing the necessary facilities. The authors are thankful to Dr. Arun Bandopadhyay, CSIR-IICB, Kolkata for his help and support to use nano LC–MS/MS facility. The authors would like to acknowledge Science Engineering Research Board (Grant No. EMR/2016/001121) and CSIR (Grant No. 31/02(1098)/2018-EMR-I), New Delhi for financial support.
Author information
Authors and Affiliations
Contributions
AS and SC designed the experiments. AS carried out the experimental and analytical works and drafted the manuscript. PB and KM helped in drafting the manuscripts and also in data analysis. SC supervised the analysis and prepared the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Manoj Prasad.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sultana, A., Boro, P., Mandal, K. et al. AAL-toxin induced stress in Arabidopsis thaliana is alleviated through GSH-mediated salicylic acid and ethylene pathways. Plant Cell Tiss Organ Cult 141, 299–314 (2020). https://doi.org/10.1007/s11240-020-01787-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-020-01787-5