Abstract
In vitro mitotic polyploidization using anti-microtubule agents has been commonly used for polyploid production. The present study was an attempt to develop an in vitro oryzalin chromosome doubling and stable tetraploid regenerating protocol for Dendrobium officinale. In this study, we successfully induced tetraploids by treating protocorms developed from the seeds of multiple genotypes. The highest frequency of polyploidy was 37.40%, achieved with 14.4 µM oryzalin treatment for 24 h. Among the obtained polyploid seedlings, 72 solid tetraploid, 33 mixoploid and 3 octoploid genotypes were identified via screening using flow cytometry (FCM). Three peaks were observed in the histograms of the diploid, tetraploid, and octoploid leaves, but four peaks were observed only in mixoploid leaves with FCM, indicating the existence of endopolyploid cells and the occurrence of conventional endoreduplication in the leaves of all ploidy levels. Recurrent ploidy identification of various tetraploid genotype regenerated plantlets obtained via protocorm-like bodies (PLBs) derived from axenic stem-node segments maintained stable tetraploid levels. Comparisons of phenotypic characteristics revealed that relative to the diploid plantlets, the tetraploid plantlets exhibited increased stem diameter, root diameter, labellum width and gynostemium width. Furthermore, the tetraploid plantlets showed lower plant height, leaf length and root length than the diploid plantlets. This efficient polyploid induction and ploidy-stable regeneration protocol can be used for the mass production of tetraploid D. officinale. The tetraploid genotypes regenerated in this study might be useful for nobile Dendrobium breeding in the future.
Key message
In vitro induction of various tetraploid genotypes in D. officinale and subsequent rapid micropropagation through induction of PLBs from axenic nodal segments were performed to obtain ploidy-stable regenerated plantlets
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Dendrobium orchids are one of the most popular potted plants worldwide due to their unique flower shape, color and size, floriferousness, relatively short production cycle, lengthy postharvest life, and year-round availability (Anderson 2006; Vendrame et al. 2008). The basic chromosome number of most Dendrobium orchids is 2n = 2x = 38, a few are 2n = 2x = 40 and a small number are 2n = 2x = 36 (Kamemoto et al. 1999). Because potted Dendrobium orchids in trade are mostly hybrids, wide crosses are routinely used to create new genetic variability at the diploid species level. It is well known that transferring desirable horticultural traits from wild species to commercial varieties has been shown to be an effective breeding means for rapidly developing new cultivars; however, the most popular commercial Dendrobium orchid varieties/hybrids are usually tetraploids, while most wild species are diploid (Kamemoto et al. 1999; Anderson 2006). Thus, there are some barriers to hybridization between these two groups. Therefore, converting diploid wild species into tetraploids to widen the source of germplasm for the development of new commercial orchid varieties for the market has become increasingly popular (Chen and Tang 2018).
Dendrobium officinale Kinura et Migo (2n = 2x = 38) is a highly evolved and specialized endangered species. It is mainly used in traditional medicine and as an ornamental plant in China (Yan et al. 2015). According to the official statistics from the website of the international orchid register, approximately 20 registered hybrids have been developed from intra- and intersectional hybridization of D. officinale, but there is no corresponding report on tetraploid germplasm for cross-breeding programs. Whether for boosting secondary metabolite production for traditional Chinese medicine applications or for improving ornamental traits, in vitro polyploid induction of D. officinale has received great attention by Chinese researchers (Jiang et al. 2014; Li and Xiang 2017; Pham et al. 2019). In previous studies, polyploidy induction in D. officinale has been achieved by in vitro treatments with colchicine in a mixed protocorm-like body (PLB) clone from multiplication subculture of protocorms with various genotypes, and flow cytometry (FCM) or chromosome counts have been mainly used in combination with morphological or anatomical parameters to determine polyploidy. However, there are few ways to concretely determine chromosome numbers and a unique endoreduplication phenomenon in many species of orchids was not observed during ploidy identification by FCM. In addition, obtaining putative tetraploids from a clone of the same genotype or from different genotypes has received little attention. Chen and Tang (2018) reported that a group of protocorms represents a range of genotypes of a population; however, PLBs from the same source are a clone that represents only a single genotype, and both their genetic constitutions are different; thus, the choice of protocorms or PLBs as initial materials for polyploid induction depends on the objectives of the experiment. Grosso et al. (2018) also found that genotype-specific metabolite fluctuations exist in various tetraploid genotype Dendrobium hybrids. For these reasons, an effective protocol was developed for rapid in vitro induction and concrete identification of various solid tetraploid genotypes by treating multigenotype protocorms.
Mitotic chromosome doubling of plant tissue in vitro is the pre-eminent method for inducing polyploidy. There is a general agreement that the artificial in vitro chromosome doubling protocol includes two steps: an induction phase and a confirmation phase. Because there is highly genotype-dependent efficiency among different plant species or within species and because of the complexity of the polyploidization protocol as reviewed by Dhooghe et al. (2011). Currently, no optimal broad-spectrum protocol is available; different explants require different methodologies, and several tests are necessary to determine the most suitable methods. However, there are some ways to optimize the induction protocol. The effects of oryzalin on chromosome doubling have been reported in Lilium (Van Tuyl et al. 1992), Anemone sylvestris (Zahumenická et al. 2018), Impatiens walleriana (Ghanbari et al. 2019), and Populus (Zeng et al. 2019). Due to the high affinity of oryzalin for microtubules of plant cells, this compound can be used at low concentrations, and it can yield higher conversion rates of regenerated polyploid plants than other compounds (Morejohn et al. 1987; Van Tuyl et al. 1992). However, literature regarding the use of oryzalin to induce plolyploids in Orchidaceae is still rare (Miguel and Leonhardt 2011; Chung et al. 2014; Hwang et al. 2015), and polyploid induction in D. officinale via the use of oryzalin has not been reported until now. In all previously reported oryzalin-induced protocols, the most efficient concentration for inducing tetaploid ranges from 8.7 to 57.7 µM and the optimal duration of treatment is 1–6 days depending on the plant/explant. The oryzalin that is used in most published reports is filter-sterilized prior to treatment (Miguel and Leonhardt 2011; Zahumenická et al. 2018; Zeng et al. 2019). Zhang et al. (2007) reported that autoclaving colchicine did not reduce its polyploidization capacity but did alleviate the toxicity to meristematic tissue in studies on in vitro chromosome doubling of Citrus sinensis. Colchicine can be sterilized by autoclaving, and the benefits of sterilization were demonstrated by Zhang et al. (2017). In our study, we attempt to autoclave oryzalin as an alternative to the sterilization method. In several species of Populus, in vitro preculture of original explants has been performed to identity suitable explant types before antimitotic agent application to produce polyploid plants (Cai and Kang 2011; Xu et al. 2016; Liu et al. 2018) because the preincubation period can synchronize the mitotic divisions and thus ameliorate the effect of the antimitotic agent (Chauvin et al. 2005). Moreover, the application method of oryzalin is crucial to increasing the polyploid induction rate. To our knowledge, aqueous oryzalin is generally added into a basal liquid or solid medium to induce explant polyploidization (Miguel and Leonhardt 2011; Zahumenická et al. 2018; Zeng et al. 2019), and few studies have reported direct immersion of explants into aqueous oryzalin solution.
Plant tissue culture provides a rapid and reliable system for the production of a large number of plantlets. Sometimes somaclonal variation may take place in the tissue culture process and is reported as a negative side effect in clonal propagation phases (Bairu et al. 2011). Therefore, finding a suitable explant type to obtain generically uniform plantlets in micropropagation is indispensable, and several explant types and developmental pathways have been tested in Dendrobium (Teixeira da Silva et al. 2015). Among these in vitro methods, induction of PLBs and complete plantlet regeneration through manipulation of axenic stem-node segments was shown to be efficient. Moreover, due to the characteristics of the short seedling cycle, stable genetic traits, and the superior traits of the female parent, stem-nodal sections were the most popular explants for in vitro polyploid induction (Dhooghe et al. 2011; Huy et al. 2019; Zeng et al. 2019). Thus, a similar micropropagation protocol for the induction of PLBs from axenic stem-node segments could be applied to our work for various tetraploid genotype regenerations.
The objective of this work was to develop an efficient protocol for the induction of various tetraploid genotypes from multigenotype protocorms of D. officinale using autoclaved oryzalin and its rapid propagation through the induction of PLBs from axenic stem-node segments. The phenotypic changes of the tetraploids relative to their diploid counterparts are also described.
Materials and methods
Plant material and in vitro protocorm establishment
As initial plant material, mature capsules of D. officinale were obtained from Langshan National Geological Park, Hunan. The seeds were stored in a desiccator containing silica gel for 4 days and kept in a refrigerator at − 20 °C.
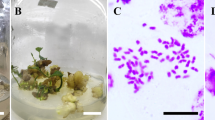
To produce in vitro protocorms, the seeds were processed using sterile filter paper packets and then were surface disinfected in 2% (v/v) Clorox solution with a drop of Tween-20 for 12 min. After rinsing three times with sterile distilled water, the seeds were spread onto the surface of Hyponex germination medium supplemented with Hyponex No.1 3 g l− 1, peptone 2 g l− 1, sucrose 20 g l− 1, potato homogenate 40 g l− 1 and agar 6.5 g l− 1 at pH 5.8. The cultures were incubated at 25 ± 2 °C under a 16/8 h light/dark photoperiod at a light intensity of 40 µmol m− 2s− 1 provided by white florescent tubes. Protocorms developed 6 weeks after sowing and were cultured for 3 weeks; subsequently, protocorms with shoot apical meristem were used as explants for subsequent polyploid induction (Fig. 1).
Polyploid induction
Stock solutions of oryzalin (1 mg of oryzalin in 1 ml of NaOH (1 M)) were prepared and diluted with sterilized distilled water to obtain working solutions at 8.7, 14.4 and 28.9 µM (w/v) and were autoclaved. Then, the glass jars with oryzalin solution were wrapped in tinfoil for protection against light and stored in a tissue culture room until used. For each treatment, 30 protocorms with shoot apical meristem were directly immersed in three different concentrations of oryzalin solution for 18, 24 and 30 h in the dark. Sterilized distilled water was used as one set of controls (0 µM oryzalin). A completely randomized 4 × 3 factorial treatment design was established with 30 protocorms per treatment, and each treatment was repeated two times. After exposure to oryzalin or sterilized water, the protocorms were rinsed three times using sterile distilled water and transferred onto fresh antimitotic-free seedling formation medium (growth conditions as mentioned above). Survival rates of protocorms were recorded at 7 weeks after oryzalin treatment. The surviving protocorms were transferred to a similar medium for further ploidy level analysis.
Flow cytometric analysis
Fresh leaves of 7-month-old seedlings were harvested after the oryzalin treatments for ploidy level identification with a CyFlow ploidy analyzer (Sysmex-Partec). The sample preparation was performed according to the CyStain UV Precise p kit manual with a minor modification as follows: samples were chopped in 400 µl nuclear extraction buffer for 30 s with a razor blade. Nuclei were mixed with 1600 µl of DAPI staining buffer and filtered through a 30 µm CellTrics filter into a sample tube. For each experimental set-up, samples of young leaves from the in vitro seedlings of untreated (control) diploid D. officinale were selected as the calibration standard to adjust the gain of the flow cytometer such that the first G0/G1 peaks were set at channel 50 on a 1000 channel scale. Histograms were generated after analyzing ≥ 5000 nuclei using the De Novo FCS Express 6 software. The number of polyploid plants was recorded based on FCM analysis.
Chromosome counting
The precise chromosome number of putative tetraploid plantlets was determined by conventional chromosome counting. Root tips approximately 2 cm in length were collected and pretreated with 200 mg l− 1 cycloheximide solution for 4 h at 20 °C. After fixation with Carnoy's solution (ethanol:glacial acetic acid = 3:1) overnight, the samples were washed twice with distilled water and then incubated in an enzyme mixture consisting of 1% cellulose (R-10, Yakult, Japan) and 1% pectolyase (Y-23, Yakult, Japan) in distilled water at 37 °C for 32–35 min. The root tips were rinsed twice with distilled water and soaked in distilled water for at least 10 min at room temperature. Then, the samples were squashed in modified phenol fuchsin solution and observed under a Leica DM3000 microscope (Germany).
Tetraploid regeneration and ploidy identification
In vitro plantlet regeneration was carried out from axenic stem-node segments of 30 elite tetraploid genotype seedlings, which exhibited normal growth and well-formed leaves and roots. Nodal segments (0.5–1.0 cm in length) excised from 7-month-old seedlings (with 1–2 internodes) were cultured on solid half-strength MS medium (Murashige and Skoog 1962) containing 2 mg l− 1 6-benzyladenine (6-BA) in combination with 0.5 mg l− 1 1-naphthaleneacetic acid (NAA) and 30 g l− 1 sugar and were tested for in vitro axillary bud induction. The axillary buds were removed from the stem-node segment, and the original stem-node segments were subcultivated in the same media to induce PLBs, followed by culture of many regenerated plantlets on H16 media (1 g l− 1 Hyponex No. 1 + 1 g l− 1 Hyponex No. 2 + 2 g l− 1 peptone + 2 g l− 1 activated charcoal + 1 mg l− 1 B5 + 0.5 mg l− 1 NAA + 15 g l− 1 sucrose + 50 g l− 1 banana homogenate + 10 g l− 1 apple homogenate + 6.5 g l− 1 agar). The cultures were incubated for 8 months under the conditions described previously. Plantlets were tested for tetraploid stability via FCM and chromosome counts. The ploidy identification process was the same as that described above.
Evaluation of phenotypic characteristics
Different tetraploid genotype plantlets were transplanted into plastic pots (9 cm wide × 7 cm high), containing tree bark as substrate and hardened in a greenhouse, and some diploid genotype regenerated plants subjected to the same growth conditions were selected as controls for comparison. In vitro acclimatization and ex vitro hardening of plantlets was conducted following the procedure reported by Hajong et al. (2013). Three months after culture, several phenotypic characteristics were evaluated, including plantlet height, stem diameter, leaf number, leaf length, leaf width, internode number, internode length, root number, root length, and root diameter.
D. officinale requires long period of growth before flowering. To induce in vitro flowering of plantlets, diploid and tetraploid plantlets of 2.5–4 cm height and with 3–4 leaves were cultured in solid Hyponex No. 1 medium containing 2 mg l− 1 6-BA, 0.1 mg l− 1 NAA, 2 g l− 1 peptone, 30 g l− 1 sucrose and 100 g l− 1 potato homogenate at pH 5.8. The culture conditions were same as those used for seed germination. Representative plantlets with normal flowers were chosen to compare flower size, labellum length, labellum width, gynostemium length, and gynostemium width between diploid and tetraploid plantlets. All the parameters were measured using a ruler or vernier caliper. Each experiment was performed twice with 30 greenhouse-grown plantlets per treatment or 20 well developed in vitro flowering plantlets per treatment.
Statistical analysis
The experimental data were statistically analyzed by two-way ANOVA with IBM SPSS 20.0 software (IBM Inc., New York, USA) followed by the LSD pairwise comparison test (p ≤ 0.05). Percentages were subjected to arcsine transformation before statistical analysis.
Results
Polyploid induction by oryzalin treatment
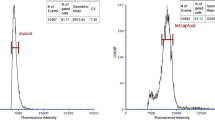
We detected 657 genotypes among the surviving seedlings from the oryzalin-treated protocorms by FCM at 7 months after induction. In total, 108 polyploids were obtained, including 72 tetraploids, 33 mixoploids and 3 octoploids. Since the octoploids showed stunted growth and were difficult to regenerate, they were excluded from data collection. The representative histogram presented in Fig. 2 shows that there were three peaks in the three ploidy levels, i.e., diploid, tetraploid and octoploid (Fig. 2a–c), whereas four peaks were observed in the mixoploid seedlings (Fig. 2d, e). This observation indicates the existence of endopolyploid cells in the leaves and whole-genome endoreduplication, in which the ratios between the positions of two neighboring DNA peaks equaled two.
Representative flow cytometry histogram after various genotype protocorm polyploidization treatments. a G0/G1 peak of the diploid standard (channel 50). b G0/G1 peak of the tetraploid plantlets (channel 100). c G0/G1 peak of the octoploid plantlets (channel 200). d, e Mixoploid plantlets (2−4x) and (4−8x)
FCM determination of the chromosome numbers of root tip cells of tetraploid (Fig. 3b) and control (Fig. 3a) seedlings confirmed that diploids had 38 chromosomes (Fig. 3c) and that the tetraploids had 76 chromosomes (Fig. 3d). The chromosomal counts were significantly correlated with the FCM analysis.
Diploids (top panels) and tetraploids (bottom panels). a, b In vitro culture of 7-month-old seedlings after oryzalin treatment (1000× magnification, Bars = 10 µm). c Chromosomes of root tip cells from a diploid 2n = 2x = 38. d Chromosomes of root tip cells from a tetraploid 2n = 4x = 76. Bars = 2 cm
Using the data from 12 treatments, the effects of oryzalin concentration and exposure time on survival rate and the frequency of polyploidy induction were investigated (Table 1). GLM-multivariate analysis indicated that the survival and tetraploid induction rates were significantly influenced by oryzalin concentration (F = 36.971, p = 0.000; F = 204.719, p = 0.000) and exposure time (F = 15.556, p = 0.001; F = 16.531, p = 0.000). Moreover, the frequency of polyploid induction was significantly affected by the interaction between oryzalin concentration and exposure time (F = 4.636, p = 0.012), whereas there was no significance interaction effect of oryzalin concentration and exposure time on survival rate. The optimum conditions for tetraploid induction was treatment with 14.4 µM oryzalin for 24 h, and the highest induction frequency of tetraploids was 37.40 ± 1.42.
Ploidy stability of regenerated tetraploid plantlets
The stem-node segments from 7-month-old seedlings (Fig. 3a, b) were cultured in induction medium for 14 days, and axillary buds were generated from stem nodes. Then, the axillary buds were excised, and the original stem-node segments were transferred to the same medium for two months, PLBs were generated from sections of axillary bud and differentiated into shoot tips. The induced PLBs were transferred to H16 media and then differentiated into cluster buds (Fig. 4a). A larger number of well-rooted seedlings were obtained after subculture (Fig. 4b). FCM analysis indicated that the first peak of the tetraploid histograms remained at the channel 100 position, and the tetraploid plants still had 76 chromosomes. Thus, the ploidy levels of regenerated plantlets from various genotypes were found to be stable.
Evaluation of phenotypic characters
The phenotypic characteristics of greenhouse-grown (90 days) and in vitro flowering (6 weeks) diploid and tetraploid plantlets were evaluated. Tetraploid exhibited marked differences from diploids in several phenotypic traits (Table 2; Figs. 5, 6). Plantlet heights, leaf length and root length were significantly lower in the tetraploid plantlets than in the diploid plantlets. Furthermore, stem diameter, root diameter, labellum width, and gynostemium width were much greater in tetraploids than in diploids. However, leaf number, leaf width, internode number, internode length, root number, flower size, labellum length, and gynostemium length were similar between the two ploidy levels.
Discussion
An efficient in vitro mitotic chromosome doubling protocol depends on many factors: species, explant type, antimitotic agents, exposure concentration and time, application method and confirmation technique (Dhooghe et al. 2011). Because the most popular commercial Dendrobium orchid varieties/hybrids are usually tetraploids (Anderson 2006), several diploid Dendrobium species, hybrids and cultivars have been converted into tetraploids though colchicine treatment, including D. scabrilingue (Sarathum et al. 2010), D. chrysotoxum (Atichart 2013), D. nobile (Vichiato et al. 2014), D. phalaenopsis × D. loddigesii hybrids (Grosso et al. 2018), Dendrobium santana × D. friedericksianum (Choopeng et al. 2019) and D.officinale (Jiang et al. 2014; Li and Xiang 2017) and cultivars (Pham et al. 2019). To date, the only tetraploid Dendrobium orchids to be produced using oryzalin are tetraploids of the Dendrobium cultivar ‘Gatton Sun Ray’ (Miguel and Leonhardt 2011). In the study by Miguel and Leonhardt (2011), PLBs were transferred onto solid modified VW media. In total, 9 tetraploids were screened by measuring stomatal guard cell lengths, achieved with 8.7, 14.4 and 57.7 µM oryzalin treatment for 6 days. The optimal treatment was identified as treatment with 14.4 µM for 6 days. In our study, protocorms with shoot apical meristem were directly soaked in one of three autoclaved oryzalin concentrations (8.7, 14.4 or 28.9 µM) for 18, 24 or 30 h. In total, 72 solid tetraploid genotypes were detected by FCM coupled with chromosome counting. The optimum treatment was treatment with 14.4 µM oryzalin for 24 h, and at the same oryzalin concentration, the most tetraploids were obtained at the lowest durations.
In orchids, numerous seeds develop within a single capsule. Orchid seeds germinate and form small, spherical, tuber-like bodies called protocorms. Protocorm development is sometimes considered a continuation of zygotic embryogenesis (Jones and Tisserat 1990; Ishii et al 1998). Miguel and Leonhardt (2011) reported the successful production of uniform polyploid plantlets from protocorms of Odontioda and Phalainopsis treated with oryzalin. Mixoploids are associated with tetraploid induction via in vitro chromosome doubling of multicellular organs (Cai and Kang 2011). In this context, in addition to obtaining a number of pure tetraploid plantlets, different types of mixoploids, such as diploid-tetraploid plantlets and tetraploid-octoploid plantlets, were also produced. The results indicated that there are various patterns of origin for protocorms, including single cell and multicellular origins.
Endopolyploidy is the existence of different ploidy levels among cells of an organism mainly due to endoreduplication (Leitch and Dodsworth 2017). This phenomenon is commonly observed in orchids, as cells in various somatic tissues of orchids undergo extensive endoreduplication, such as conventional whole-genome endoreduplication (with the ratio between two neighboring DNA content peaks equaling two) or progressive partial endoreduplication (with the ratio between two neighboring DNA content peaks being lower than two) (Trávníček et al. 2015). The true biological significance of this phenomenon is not yet fully understood (Larkins et al. 2001; Kron 2015). As detected by FCM, patterns of endopolyploidy depend on diverse ploidy levels, developmental stages and culture conditions (Chen et al. 2011). Our data showed that endopolyploidy occurred in the leaves, and three peaks were found in the leaves in both diploids and tetraploids. These results indicate that diploids and tetraploids have the same endopolyploidy pattern. Conversely, there were four peaks in the histograms of leaves from the diploid Phalaenopsis aphrodite subsp. formosana, whereas there were only three peaks for the tetraploids, and diploids displayed a higher tendency for endopolyploidy than did tetraploids (Chen et al. 2011). The endoreduplication pattern is mostly stable within a genus, and conventional whole-genome endoreduplication has been observed in somatic tissues of Dendrobium (Trávníček et al. 2015). Our histograms showed a similar pattern, but in the Dendrobium hybrids both conventional and partial endopolyploidy were encountered (Grosso et al. 2018).
PLBs structurally resemble protocorms and are induced from explants and/or calluses (Jones and Tisserat 1990; Chugh et al. 2009). Fang et al. (2016) found that protocorms and PLBs share similar transcriptomic signatures that differ extensively from those of zygotic embryos, PLB regeneration does not follow the embryogenesis program. PLBs can be successfully induced from various explants, including protocorms, shoot tips, axillary buds, leaf sections and nodal segments, as well as pseudobulb segments, as reviewed by Teixeira da Silva et al. (2015). The direct regeneration of PLBs from axenic stem-node segments from pregerminated seedlings was considered an efficient protocol for in vitro propagation of D. officinale (Wei et al. 2012; Shiau et al. 2005) and other Dendrobium orchids (Luo et al. 2008). In this study, recurrent ploidy level screening and chromosome count of various genotype regenerated plantlets derived from PLBs from axenic stem-node segments showed stable tetraploid levels. A similar micropropagation protocol for the induction of PLBs and complete plantlet regeneration through the manipulation of axenic stem-node segments from two tetraploid D. officinale cultivars was reported by Pham et al. (2019), but the regenerated plantlets obtained by Pham et al. (2019) were not tested for ploidy level.
D. officinale has a long juvenile phase that leads to an average time to flowering of three to four years (Wang et al. 1997); therefore, it is difficult to evaluate flower traits in a short time. Wang et al. (1997) found that in D. officinale, in vitro flowers could be induced within three to six months versus the 3 years to flowering in vivo. Compared with in vitro flowering, in vitro flowering allows earlier assessment of flower characteristics ( Teixeira da Silva et al. 2014), although in vitro flower organs are smaller than that of ex vitro flowers (Tee et al. 2008). Thus, flowering in vitro is worth considering. In our study, we used the in vitro flowering ability of D. officinale to evaluate normal flower organ trait in diploid and induced tetraploid plantlets. Generally, changes in ploidy level result in phenotypic variation (Udall and Wendel 2006). In the present study, the tetraploid plantlets had shorter plant heights and leaf and root lengths than the diploid plantlets, which is consistent with a previous study (Pham et al. 2019). Vichiato et al. (2014 ) reported that tetraploidization resulted in wider labella in flowers of D. nobile, similar to our observations in tetraploid plantlets. However, differences in leaf shapes among genetically homogenous genotypes with the same tetraploid level have been found in Pyrus communis (Sun et al. 2011) and D. officinale cultivars (Pham et al. 2019), and variation in flower traits have also been observed in daylily (Podwyszyńska et al. 2015). These observations indicate that chromosome doubling can not only resulted in chromosome and gene redundancy but also chromosome structure variation, gene mutation (Blanc and Wolfe 2004 ), alterations in epigenetic patterns (Marfil et al. 2018) or more complex phenomena. Furthermore, Liu et al. (2019) found that the extent of heterozygosity in genotypes with the same ploidy level affected phenotypic variation. In our study, significant differences were observed between diploid and tetraploid plantlets at 90 days, and variations within tetraploid genotypes under the same growing conditions was also visually apparent. Furthermore, growing years affects phenotypic variation, as observed in daylily (Podwyszyńska et al. 2015), D. nobile (Vichiato et al. 2014 ) and lily (Zhang et al. 2017 ). To compare phenotypic characteristics between diploids and tetraploids and evaluate penotypic variation within tetraploid genotypes, several years of data are necessary. Thus, several cultivation years are required to elucidate the phenotypic variation among multitetraploids induced by oryzalin.
In conclusion, we found that in vitro application of oryzalin to multigenotype protocorms can efficiently induce chromosome doubling in D. officinale. Furthermore, PLBs derived from axenic stem-node segments could be used to establish in vitro plant regeneration of D. officinale. Multiple-tetraploid genotype germplasms not only may be of use in Dendrobium orchid polyploid breeding but also can be used to increase the genetic variation within this species for other various research applications.
References
Anderson NO (2006) Flower breeding and genetics: issues, challenges and opportunities for the 21st century. Springer, New York
Atichart P (2013) Polyploid induction by colchicine treatments and plant regeneration of Dendrobium chrysotoxum. Thai J Agric Sci 46:59–63
Bairu MW, Aremu AO, Van Staden J (2011) Somaclonal variation in plants: causes and detection methods. Plant Growth Regul 63:147–173
Blanc G, Wolfe KH (2004) Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16:1679–1691
Cai X, Kang XY (2011) In vitro tetraploid induction from leaf explants of Populus pseudo-simonii Kitag. Plant Cell Rep 30:1771–1778
Chauvin J, Label A, Kermarrec M (2005) In vitro chromosome-doubling in tulip (Tulipa gesneriana L.). J Hortic Sci Biotechnol 80:693–698
Chen WH, Tang CY (2018) A protocol for the induction of polyploids in Phalaenopsis Orchids by in vitro method without using anti-microtubule agents. In: Lee YI, Yeung EC (eds) Orchid propagation: from laboratories to greenhouses-methods and protocols, 1st edn. Springer, New York, pp 317–330
Chen WH, Tang CY, Lin TY, Weng YC, Kao YL (2011) Changes in the endopolyploidy pattern of different tissues in diploid and tetraploid Phalaenopsis aphrodite subsp. formosana (Orchidaceae). Plant Sci 181:31–38
Choopeng S, Te-chato S, Khawnium T (2019) Effect of colchicine on survival rate and ploidy level of hydrid between Dendrobium santana × D. friedericksianum orchid. Int J Agric Technol 15:249–260
Chugh S, Guha S, Rao IU (2009) Micropropagation of orchids: a review on the potential of different explants. Sci Hortic 122:507–520
Chung MY, Kim CY, Min JS, Lee DJ, Naing AH, Chung JD, Kim CK (2014) In vitro induction of tetraploids in an interspecific hybrid of Calanthe (Calanthe discolor × Calanthe sieboldii) through colchicine and oryzalin treatments. Plant Biotechnol Rep 8:251–257
Dhooghe E, Van Laere K, Eeckhaut T, Leus L, Van Huylenbroeck J (2011) Mitotic chromosome doubling of plant tissues in vitro. Plant Cell Tissue Organ Cult 104:359–373
Fang SC, Chen JC, Wei MJ (2016) Protocorms and protocorm-like bodies are molecularly distinct from zygotic embryonic tissues in Phalaenopsis aphrodite. Plant Physiol 171:2682–2700
Ghanbari MA, Jowkar A, Salehi H, Zarei M (2019) Effects of polyploidization on petal characteristics and optical properties of Impatiens walleriana (Hook.). Plant Cell Tissue Organ Cult 138:299–310
Grosso V, Farina A, Giorgi D, Nardi L, Diretto G, Lucretti S (2018) A high-throughput flow cytometry system for early screening of in vitro made polyploids in Dendrobium hybrids. Plant Cell Tissue Organ Cult 132:57–70
Hajong S, Kumaria S, Tandon P (2013) Effect of plant growth regulators on regeneration potential of axenic nodal segments of Dendrobium chrysanthum Wall. ex Lindl. J Agric Sci Technol 15:1425–1435
Huy NP, Luan VQ, Tung HT, Hien VT, Ngan HTM, Duy PN, Nhut DT (2019) In vitro polyploid induction of Paphiopedilum villosum using colchicine. Sci Hortic 252:283–290
Hwang SH, Kim MS, Park SY (2015) Improvement of chromosome doubling efficiency in Cymbidium hybrids by colchicine and oryzalin treatment. Korean J Hortic Sci Technol 33:900–910
Ishii Y, Takamura T, Goi M, Tanaka M (1998) Callus induction and somatic embryogenesis of Phalaenopsis. Plant Cell Rep 17:446–450
Jiang JL, Ye W, Li YQ, Zhou JJ, Lei FG, Wei DZ (2014) Growth and polysaccharides accumulation in autotetraploid Dendrobium officinale. Plant Physiol J 50:519–526 ((in Chinese with English abstract))
Jones D, Tisserat B (1990) Clonal propagation of orchids. Methods Mol Biol 6:181–191
Kamemoto H, Amore TD, Kuehnle AR (1999) Breeding Dendrobium orchids in Hawaii. University of Hawaii Press, Honolulu
Kron P (2015) Endopolyploidy, genome size, and flow cytometry. Cytom Part A 87:887–889
Larkins BA, Dilkes BP, Dante RA, Coelho CM, Woo YM, Liu Y (2001) Investigating the hows and whys of DNA endoreduplication. J Exp Bot 52:183–192
Leitch I, Dodsworth S (2017). Endopolyploidy in plants eLS.https://doi.org/10.1002/9780470015902.a0020097.pub2
Li DD, Xiang ZX (2017) Comparative differences between autotetraploid and diploid of Dendrobium officinale Kimura et Migo and selection of its cultivation matrixes. Dissertation, Nanjing Agricultural University (in Chinese with English abstract)
Liu WT, Song SY, Li DL, Lu XC, Liu JR, Zhang JW, Wang J (2019) Isolation of diploid and tetraploid cytotypes from mixoploids based on adventitious bud regeneration in Populus. Plant Cell Tissue Organ Cult 140:1–10
Liu WT, Zheng YF, Song SY, Huo BB, Li DL, Wang J (2018) In vitro induction of allohexaploid and resulting phenotypic variation in Populus. Plant Cell Tissue Organ Cult 134:183–192
Luo JP, Wang Y, Zha XQ, Huang L (2008) Micropropagation of Dendrobium densiflorum Lindl. ex Wall. through protocorm-like bodies: effects of plant growth regulators and lanthanoids. Plant Cell Tissue Organ Cult 93:333–340
Marfil CF, Duarte PF, Masuelli RW (2018) Phenotypic and epigenetic variation induced in newly synthesized allopolyploids and autopolyploids of potato. Sci Hortic 234:101–109
Miguel TP, Leonhardt KW (2011) In vitro polyploid induction of orchids using oryzalin. Sci Hortic 130:314–319
Morejohn LC, Bureau TE, Molebajer J, Bajer AS, Fosket DE (1987) Oryzalin, a dinitroaniline herbicide, binds to plant tubulin and inhibits microtubule polymerization in vitro. Planta 172:252–264
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15:473–497
Pham PL, Li YX, Guo HR, Zeng RZ, Xie L, Zhang ZS (2019) Changes in morphological characteristics, regeneration ability, and polysaccharide content in tetraploid Dendrobium officinale. Hortscience 54:1879–1886
Podwyszyńska M, Gabryszewska E, Dyki B, Stępowska AA, Kowalski A, Jasiński A (2015) Phenotypic and genome size changes (variation) in synthetic tetraploids of daylily (Hemerocallis) in relation to their diploid counterparts. Euphytica 203:1–16
Sarathum S, Hegele M, Tantiviwat S, Nanakorn M (2010) Effect of concentration and duration of colchicine treatment on polyploidy induction in Dendrobium scabrilingue L. Europ J Hort Sci 75:123–127
Shiau YJ, Nalawade SM, Hsia CN, Mulabagal V, Tsay HS (2005) In vitro propagation of the Chinese medicinal plant, Dendrobium candidum Wall. Ex Lindl., from axenic nodal segments. In Vitro Cell Dev Biol Plant 41:666–670
Sun QR, Sun HY, Bell RL, Li HF, Xin L (2011) Variation of phenotype, ploidy level, and organogenic potential of in vitro regenerated polyploids of Pyrus communis. Plant Cell Tissue Organ Cult 107:131–140
Tee CS, Maziah M, Tan CS (2008) Induction of in vitro flowering in the orchid Dendrobium Sonia 17. Biol Plant 52:723–726
Teixeira da Silva JA, Cardoso JC, Dobránszki J, Zeng SJ (2015) Dendrobium micropropagation: a review. Plant Cell Rep 34:671–704
Teixeira da Silva JA, Kerbauy GB, Zeng SJ, Chen ZL, Duan J (2014) In vitro flowering of orchids. Crit Rev Biotechnol 34:56–76
Trávníček P, Ponert J, Urfus T, Jersáková J, Vrána J, Hřibová E, Doležel J, Suda J (2015) Challenges of flow-cytometric estimation of nuclear genome size in orchids, a plant group with both whole-genome and progressively partial endoreplication. Cytom Part A 87:958–966
Udall JA, Wendel JF (2006) Polyploidy and crop improvement. Crop Sci 46:3–14
Van Tuyl JM, Meijer B, Van Dien MP (1992) The use of oryzalin as an alternative for colchicine in in-vitro chromosome doubling of Lilium. Lily Yearbook North American Lily Society Inc., 43:19–22
Vendrame WA, Carvalho VS, Dias JM, Maguire I (2008) Pollination of Dendrobium hybrids using cryopreserved pollen. Hortscience 43:264–267
Vichiato MRM, Vichiato M, Pasqual M, Rodrigues FA, Castro DM, Orchidaceae (2014) Morphological effects of induced polyploidy in Dendrobium nobile Lindl. Crop Breed Appl Biot 14:154–159
Wang GY, Xu ZH, Chia TF, Chua NH (1997) In vitro flowering of Dendrobium candidum. Sci China (Ser C) 40:35–42
Wei M, Yang CY, Wei SH (2012) Enhancement of the differentiation of protocorm-like bodies of Dendrobium officinale to shoots by ultrasound treatment. J Plant Physiol 169:770–774
Xu CP, Huang Z, Liao T, Li Y, Kang XY (2016) In vitro tetraploid plants regeneration from leaf explants of multiple genotypes in Populus. Plant Cell Tissue Organ Cult 125:1–9
Yan L, Wang X, Liu H, Tian Y, Lian JM, Yang RJ, Hao SM, Wang XJ, Yang SC, Li QY, Qi S, Kui L, Okpekum M, Ma X, Zhang JJ, Ding ZL, Zhang GJ, Wang W, Dong Y, Sheng J (2015) The Genome of Dendrobium officinale illuminates the biology of the important traditional Chinese orchid herb. Mol Plant 8:922–934
Zahumenická P, Fernández E, Šedivá J, Žiarovská J, Ros-Santaella JL, Martínez-Fernández D, Russo D, Milella L (2018) Morphological, physiological and genomic comparisons between diploids and induced tetraploids in Anemone sylvestris L. Plant Cell Tissue Organ Cult 132:317–327
Zeng QQ, Liu Z, Du K, Kang XY (2019) Oryzalin-induced chromosome doubling in triploid Populus and its effect on plant morphology and anatomy. Plant Cell Tissue Organ Cult 138:571–581
Zhang J, Zhang M, Deng X (2007) Obtaining autotetraploids in vitro at a high frequency in Citrus sinensis. Plant Cell Tissue Organ Cult 89:211–216
Zhang XQ, Cao QZ, Jia GX (2017) A protocol for fertility restoration of F1 hybrid derived from Lilium × formolongi ‘Raizan 3’× Oriental hybrid ‘Sorbonne.’ Plant Cell Tissue Organ Cult 129:375–386
Acknowledgements
This work was supported by the National Natural Science Foundation of China (U1702235) and the Ministry and Province Joint Construction Project of Yunnan University (C176280109).
Author information
Authors and Affiliations
Contributions
JYG and XQZ conceived and designed the experiments; XQZ performed the experiments; XQZ analyzed the data and wrote the manuscript; and XQZ and JYG revised the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Ming-Tsair Chan.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhang, X., Gao, J. In vitro tetraploid induction from multigenotype protocorms and tetraploid regeneration in Dendrobium officinale. Plant Cell Tiss Organ Cult 141, 289–298 (2020). https://doi.org/10.1007/s11240-020-01786-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-020-01786-6