Abstract
In this work, transgenic Salix matsudana expressing the Tamarix hispida ThMT3 gene, which encodes encoding a type 3 metallothionein, showed increased tolerance to copper (Cu) stress. Exposure to 50 μM Cu completely inhibited rooting of wild-type (WT) plants, but induced numerous adventitious roots in the transgenic plants. The nitric oxide (NO) content in the transgenic plants was higher than that in WT plants. The application of an NO inhibitor, 2-phenyl-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide, decreased superoxide dismutase, catalase and ascorbate peroxidase activities under Cu stress. Auxin application-related genes that are known to improve adventitious roots, such as Auxin response factor 8, auxin resistant 1 and pinformed, were highly expressed in transgenic plants under Cu and sodium nitroprusside treatments. These results suggested that the expression of the ThMT3 gene increased Cu tolerance and NO production, and the higher NO release contributed to the induction of adventitious roots under Cu stress.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Phytoremediation of contaminated soils by plants offers an environmentally friendly, cost-effective method to cleanup of toxic pollutants in the environment. There has been an increase in research on improving the ability of plants to remove environmental pollution. For this application, tree species with a large biomass and a tolerance for high levels of toxic metals are needed.
Salix matsudana Koidz. is a large, deciduous, rapidly growing tree that is native to northeastern China, where it experiences a wide range of climatic conditions. The species is one of the most widely distributed and commonly cultivated willow species in China. The species accumulates high level of heavy metals, making it potentially suitable for phytoremediation (Dos Santos Utmazian et al. 2007). However, phytoremediation is limited by the failure of the plants to hyperaccumulate toxins, the unacceptably long time period needed for remediation, and the fact that heavy metal accumulation is restricted mainly to the roots, which complicates their collection for elimination (Dowling and Doty 2009). The efficient genetic engineering of the Salix species is highly desirable to address these limitations. Agrobacterium-mediated transformation is widely to introduce foreign DNA into plant species. Recently, we developed an A. tumefaciens-mediated transformation system for S. matsudana, using the embryonic apical region as an explant (Yang et al. 2013).
Metallothioneins (MTs) are a group of low-molecular-weight Cys-rich metal binding proteins that are present in all eukaryotes, as well as some prokaryotes, and are divided into three classes based on the arrangement of the Cys residues in the N- and C-termini (Cobbett and Goldsbrough 2002). Plant MTs belong to class II and can be further subdivided into four types (MT1, MT2, MT3 and MT4) based on the Cys distribution pattern (Cobbett and Goldsbrough 2002). Numerous studies have observed the expression of plant MTs in response to various heavy metal stresses, such as copper (Cu), zinc and cadmium (Cd). The overexpression of some MTs has led to enhanced heavy metal resistance (Cobbett and Goldsbrough 2002). Arabidopsis MTs 2a and 3 enhanced resistance to Cd in Vicia faba guard cells (Lee et al. 2004), and the overexpression of Paxillus involutus’ PiMT1 in Hebeloma cyclindrosporum resulted in higher tolerance to Cu (Bellion et al. 2007). However, the detailed functions of MTs in plants are still poorly understood.
Currently, it is widely accepted that MTs play an important role in Cu homeostasis during plant growth. MT2 from Brassica juncea (BjMT2) confers a high Cu tolerance to transgenic Escherichia coli and Arabidopsis. The ectopic expression of BjMT2 inhibited root growth in the absence of Cu exposure, whereas root growth of the transgenic plant was identical to the wild-type (WT) plants when exposed to high Cu levels, indicating that the overexpression of the MT2 protein interfered with Cu homeostasis, thus affecting root development (An et al. 2006). MT deficiency (mt1a-2/mt2a-1/mt2b-1/mt3–1) affects Cu accumulation and distribution in Arabidopsis mutants (Benatt et al. 2014). In our previous studies, we found that overexpression of the Tamarix hispida ThMT3 gene, an MT-like gene encoding a type 3 MT, improved the tolerance of transgenic yeast to Zn, Cd and Cu stress (Yang et al. 2011).
Nitric oxide (NO), an important signaling molecule, has roles in the response to Cu. It is a crucial modulator under many stresses, such as pathogen invasion, salt stress, UV-B radiation and drought stress (Shi et al. 2005; Zhang et al. 2007). Recently, NO has been regarded as a molecular signaling response factor in plants against heavy metals, including arsenic (Singh et al. 2009), Cu (Yu et al. 2005), and Cd (Groppa et al. 2008; Laspina et al. 2005). Transgenic antisense-MT2b tomato plants showed an increased sensitivity to Cu stress and the Cu resistance can be efficiently rescued by the addition of sodium nitroprusside (SNP) (Wang et al. 2010). Additionally, NO applications induced MT transcription and accumulation in leaves, indicating the possible roles of NO and MTs in response to heavy metals. However, whether the overexpression of MTs improves Cu tolerance, or whether rooting ability is related to NO, has not been demonstrated.
In the present study, the ThMT3 gene was transformed into S. matsudana. The transgenic plants showed increased tolerance to Cu stress. Additionally, adventitious roots were highly induced in transgenic plants under Cu stress, and the improved induction was shown to be related to NO release under Cu stress. Based on this research, we hope to gain a better understanding of the possible protective mechanism of ThMT3 expression in response to Cu stress in S. matsudana.
Materials and methods
Plant materials and transformation
Open-pollinated mature seeds of S. matsudana Koidz. var. matsudana were collected during late May from 60-year-old trees grown on the campus of Northeast Forestry University, Harbin, China and used for transformation.
A MT-like gene, ThMT3 (GenBank accession number EH057039), isolated from T. hispida was cloned into binary vector PROKII, under the control of the Cauliflower mosaic virus (CaMV) 35S promoter, containing a neomycin phosphotransferase II (npt II) gene under the control of a nopaline synthase (nos) promoter (Chen et al. 2003). The binary vector, PROKII-ThMT3, was introduced into Agrobacterium tumefaciens strain LBA4404.
Transgenic plants were generated by Agrobacterium-mediated transformation of injured S. matsudana seeds, according to our previous method, described by Yang et al. (2013). The regenerated, transformed plantlets were cultured on a selective medium successively at 4-week intervals in flasks. Cultures were placed in a culture room at 24 °C with a 16-h photoperiod and a light intensity of 45 μmol m−2 s−1.
PCR and RT-PCR analysis
DNA was extracted from leaves of WT and transgenic plants using modified cetyltrimethylammonium bromide method (Jaakola et al. 2001). The ThMT3 gene was amplified using primers F: 5′-ATGTCGGGCAAGTGCGGAAACTGCAG-3′ and R: 5′-TCAGTGACTGCCACATGTGCAGTTG-3′ designed to amplify a 210 bp fragment. The PCR conditions were as follows: 94 °C for 3 min; followed by 30 cycles of 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 30 s; and a final 7 min extension at 72 °C. Total RNA was extracted using a Plant RNA Purification Reagent (Invitrogen, Carlsbad, CA, USA). First-strand cDNA synthesized from 0.5 μg of purified RNA was reverse-transcribed using a Reverse Transcriptase kit (Takara Biotech, Dalian, China). The PCR conditions were identical to those described above.
Heavy metal exposure and growth characteristics
In vitro-grown shoots of about 3 cm height were transferred into 1/2 Murashige and Skoog (MS) (1962) medium supplemented with 0, 10, 30 and 50 μM CuSO4, respectively. Growth characteristics, such as plant height, root length and rooting rate (%) were measured after 4 weeks of culture. Five plants were cultured in a flask, with three replicates; three plant lines were tested for each treatment.
Histological observation
Shoots of plants treated with 50 μM CuSO4 for 4 weeks were collected and sections were made, as described previously (Yang et al. 2013). The sections were de-waxed and observed under a light microscope (Olympus SZ, Japan).
Determination of the copper contents
In vitro-grown shoots of about 6 cm height were cultured on 1/2 MS medium containing 50 and 100 μM CuSO4, respectively. After 4 weeks, the roots, stems and leaves of the plants were divided and dried at 80 °C to a constant weight. The samples (0.1–0.2 g) were weighed and dissolved in a mixture of concentrated HNO3–HClO4 (5:1 v/v) and heated at 160 °C for 5 h. After cooling, the beaker was washed with 6 M HCl. Aliquots were analyzed by atomic absorption spectrophotometer (TAS-986), as described previously (Adamis et al. 2003). Absorption was calculated by determining the differences in metal content between non-transgenic and transgenic plant, respectively.
Detection of O2 − and H2O2
The amount of O2 − and H2O2 in the leaves and roots from the WT and three transgenic plant lines (T1, T2, T3) were monitored by incubation with 2 mM nitroblue tetrazolium (NBT; N6876, Sigma-Aldrich) in 20 mM phosphate buffer (pH 6.1) containing 10 mM NaN3 and in 3,3′ diaminobenzidine (DAB) (D5637, Sigma-Aldrich) (pH 3.8). Chlorophyll was destaining by boiling in alcohol (95 %, v/v) for 1 h (Xu et al. 2010).
Transmission electron microscopy (TEM) observation
After treatment with 50 μM CdSO4 for 4 weeks, leaves were collected. Ultrathin sections were produced according to the procedure in Sridhar et al. (2005). Ultrathin sections were obtained using an ultramicrotome (POWTIME-XL), and observed using an H-7650 TEM.
Confocal laser scanning microscopy (CLSM) observation
For NO and H2O2 detection, roots of plants treated with 50 μM CuSO4 (for 5 h) were monitored using the NO-specific fluorescent dye 4,5-diaminofluorescein diacetate (DAF-2DA, Sigma) and H2O2-sensitive fluorescent probe 2′,7′-dichlorofluorescin diacetate (H2DCFDA, Sigma, St Louis, MO, USA). Fluorescence was visualized at 488 nm using a confocal laser scanning microscope (LSM 700, Zeiss, Germany).
Effect of NO on rooting
For rooting investigation, 0, 50 and 100 μM 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazo-line-1-oxyl-3-oxide potassium salt (cPTIO, an NO inhibitor) were added into 1/2 MS medium containing 50 μM CuSO4. The rooting rate (%) was calculated after 4 weeks of culture. Additionally, the phenotype of the WT and transgenic plants (T1 line) that were cultured on 1/2 MS medium containing 50 μM CuSO4, and supplemented with 100 μM cPTIO for 4 weeks was observed.
Measurements of NO and H2O2 contents
NO content was determined as described by Zhou et al. (2005). Powdered leaves (0.6 g) were mixed with 3 mL of 50 mm cool acetic acid buffer (pH 3.6, containing 4 % zinc diacetate), and centrifuged at 10,000×g for 15 min at 4 °C. Activated charcoal (0.1 g) was added to the supernatant, which was then filtrated. Absorbance was determined at 540 nm. The NO content was calculated by comparison with a standard curve of NaNO2.
H2O2 contents were measured according to Zhang et al. (2009). Fresh leaves (0.5 g) were excised and ground to a powder in liquid nitrogen and extracted with 5 mL of 5 % trichloroacetic acid (TCA) and 0.15 g activated charcoal. The mixture was centrifuged at 10,000×g for 20 min at 4 °C. The supernatant was filtered and used for determination of H2O2. Powders of leaves (0.5 g) were extracted in 3 mL of 50 mm cool acetic acid buffer (pH 3.6, containing 4 % zinc diacetate) with 0.1 g of activated charcoal. The homogenates were centrifuged at 10,000×g for 15 min at 4 °C. Absorbance was determined at 540 nm.
Chlorophyll content assay
The fresh leaves (0.5 g) were collected from plants exposed to 50 μM Cu for 4 weeks, and washed with 80 % acetone until the samples turned white. The supernatants were collected and the volume of the samples was fixed to 20 mL using 80 % acetone before being filtered. The chlorophyll content was measured using a 722 s visible spectrophotometer 722 s (Shanghai, China) at the wave-lengths of 663, 645 and 652 nm.
Measurement of SOD, CAT, APX activity and MDA content
Leaf samples (0.5 g) from plants 2 weeks after being treated with 50 μM Cu or Cu + 100 μM cPTIO were ground to a powder with liquid nitrogen respectively. Superoxide dismutase (SOD), catalase (CAT) and ascorbate peroxidase (APX) activities were measured as described previously (Guo et al. 2006). Measurement of malondialdehyde (MDA) content was conducted as described previously (Lei et al. 2007).
Quantitative real-time RT-PCR
Total RNA was extracted from the WT and transgenic plant shoots under conditions of 50 μM CuSO4, 100 μM SNP and 100 μM cPTIO for 5 h, respectively. cDNA was synthesized as described above. PCR was performed with SYBR premix ExTaq (TaKaRa Biotech, Dalian, China), and amplified under the following cycling conditions: 10 s at 95 °C; followed by 40 cycles of 5 s at 95 °C, 30 s at 60 °C; and 1 s at 78 °C for plate reading. The β-tubulin gene was used for normalization of the qRT-PCR analysis. Primers for β-tubulin (GenBank accession number: KM591694) and functional genes NR (KM591683), GR (KM591685), SOD (KM591686), CAT (KM591687), APX (KM591688), ARF8 (KM591689), AUX1 (KM591690), PIN (KM591691), ABCB (KM591692) and PGP (KM591693) are shown in Table S1.
Statistical analysis
All experiments were repeated independently at least three times. Statistical analyses were carried out using SPSS 16.0 for Windows (SPSS Inc., Chicago, IL, USA). Data were compared using Student’s t test. Differences were considered to be significant if P < 0.05.
Results
Overexpression of ThMT3 increases Cu tolerance
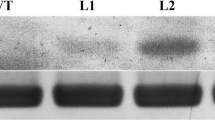
We successfully transformed 13 independent transgenic plant lines, which were confirmed by PCR (Fig. 1a) and RT-PCR analyses (Fig. 1b). Under Cu conditions, plant heights, root growth, and rooting rates of all plants decreased as the Cu concentration increased, however, the plant heights and rooting rates were higher, and the root length was longer, in transgenic plants than in WT plants (Supplementary Fig. S1a–c). In particular, a 50 μM Cu supplement almost completely suppressed the induction of adventitious roots on WT shoots (Fig. 2a, left), while numerous adventitious roots were induced on the transgenic stem within 4 weeks (Fig. 2a right, b). Three plant lines showed the same phenotype. A histological analysis showed that no adventitious roots were induced from the stems of WT plants (Fig. 2c); however, many roots originated from the xylem of the transgenic plant stems and were of multicellular origin (Fig. 2d, e). Thus, we proposed that the ThMT3 gene improves the rooting of transgenic plants under Cu stress.
The overexpression of ThMT3 increase the rooting efficiency under Cu stress. a Wild-type (WT) and transgenic shoots were cultured on 1/2 MS medium containing 50 μM CuSO4 for 4 weeks. Left WT shoots, Right transgenic shoots. Bar = 10 mm. b Transgenic plant with adventitious roots. Bar = 10 mm. Histological observation of transgenic shoots before (c), 2 (d) and 4 weeks (e) after cultured on medium containing 50 μM CuSO4. Bar = 200 μm
The metal contents in the roots, stems and leaves were measured. However, the amounts of Cu in both plants were not significantly different, regardless of lower (Fig. 3a) or higher (Fig. 3b) Cd concentrations.
Cu accumulation in wild-type (WT) and transgenic plants. Both WT and transgenic plants were cultured on 1/2 MS medium containing 50 μM (a) and 100 μM (b) CuSO4 for 4 weeks. T1–T3 transgenic plant lines. Data are mean ± SD from three independent experiments. *Significant (t test, P < 0.05) difference compared with WT plants under each treatment
Oxidative damage caused by Cu
Reactive oxygen species (ROS) are always formed when stress stimulates the photosynthetic electron transport chain (Freeman et al. 2010). The production of ROS in leaves and roots was detected by in situ staining using stains sensitive to O2 − and hydrogen peroxide (H2O2), respectively (Fig. 4). There was no difference between the WT and transgenic plant lines for ROS detection (Fig. 4a, c). The leaves and roots of the WT plant displayed higher O2 − (Fig. 4b) and H2O2 (Fig. 4d) accumulation levels than those measured in three ThMT3-transgenic plant lines (T1, T2 and T3) under Cu stress. This suggested that Cu induced a less serious oxidative stress response in transgenic S. matsudana.
Overexpression of ThMT3 decreases ROS levels under Cu stress. Leaves and roots from wild-type (WT) and transgenic plants were pretreated with 50 μM and 100 μM CuSO4 for 5 h, and were stained with NBT to visualize O2− without Cu (a) or with Cu stress (b), or stained with DAB to visualize H2O2 without Cu (c) or with Cu stress (d). T1–T3 transgenic plant lines. Bar = 3 mm
Overexpression of ThMT3 protects chloroplasts under Cu stress
Chloroplasts of WT plants treated with 50 μM CuSO4 for 4 weeks showed visible damage and contained more starch granules (Fig. 5a), while the chloroplasts of transgenic plants remained intact (Fig. 5b). When the transgenic plants were cultured in medium containing Cu and cPTIO, starch granules also appeared (Fig. 5c). After receiving Cu treatment for 4 weeks, the leaves yellowed and there was a dramatic decrease in the chlorophyll content of WT plants compared with the transgenic plants (Supplementary Fig. S2).
Transmission electron microscopy observation of mesophyll cells. a mesophyll cells of WT plant cultured on 1/2 MS medium containing 50 μM CuSO4 for 4 weeks. Bar = 2 μm. b mesophyll cells of transgenic plants cultured on 1/2 MS medium containing 50 μM CuSO4 for 4 weeks. Bar = 2 μm. c mesophyll cells of transgenic plants cultured on 1/2 MS medium containing 50 μM CuSO4 and supplemented with 100 μM cPTIO for 4 weeks. Bar = 5 μm
Overexpression of ThMT3 increases the NO content and rootings under Cu stress
For rooting investigation, 50 and 100 μM cPTIO were added into 1/2 MS medium containing 50 μM CuSO4. The results showed that, as the level of the NO inhibitor cPTIO increased, the rooting rate dramatically decreased in transgenic plants under 50 μM Cu stress, suggesting that NO was critically important for rooting in the ThMT3-transgenic plants (Supplementary Fig. S3a). No roots showed in the stems of transgenic plants with supplementary of cPTIO in the culture medium containing Cu (Supplementary Fig. S3c), representing the same phenotype as the WT (Supplementary Fig. S3b). Thus, we investigated the potential role of NO in transgenic plants. Only a small amount of NO fluorescence was visible in the roots of the WT (Fig. 6a) and transgenic plants without Cu treatment (Fig. 6b). After Cu treatment, the NO fluorescence increased in the transgenic plant roots compared with that in the WT plant roots (Fig. 6c, d). Without Cu treatment, there was no difference in H2O2 between non- (Fig. 6e) and transgenic plant cells (Fig. 6f), with both having low concentrations. After Cu treatment, higher H2O2 fluorescence levels appeared in non- (Fig. 6g) and transgenic plants (Fig. 6h). However, the H2O2 fluorescence in WT plants (Fig. 6g) was higher than that in transgenic plants (Fig. 6h). Additionally, the NO contents in the roots of non- and transgenic plants were not significantly different (Fig. 7a). However, after Cu treatment, the NO level in WT plants still remained unchanged, while the NO level in transgenic plants was extremely high (Fig. 7a), which was consistent with the NO fluorescence results. We presumed that the increased generation of NO in root cells of the transgenic plants is necessary to protect plants against Cu stress.
Nitric oxide (NO) and hydrogen peroxide (H2O2) fluorescence observation under Cu stress. Confocal laser scanning microscopy (CLSM) observation of NO fluorescence in wild-type (WT) (a) and transgenic plant (b). NO fluorescence in WT (c) and transgenic plants (d) treated with 50 μM CuSO4 for 5 h. H2O2 fluorescence in WT (e) and transgenic plant (f) H2O2 fluorescence in WT (g), and transgenic plants (h) treated with 50 μM CuSO4 for 5 h. Bar = 100 μm
Measurement of nitric oxide (NO) (a) and hydrogen peroxide (H2O2) (b) content in the roots under Cu stress. The roots from wild-type (WT) and transgenic plants were pretreated with 50 μM CuSO4 for 5 h. Data are mean ± SD from three independent experiments. *Significant (t test, P < 0.05) difference compared with WT plants
Overexpression of ThMT3 decreased H2O2 by activating anti-oxidative enzymes under Cu stress
The H2O2 content in the roots of non- and transgenic plants were not significantly different (Fig. 7b). However, the H2O2 content in plant roots under Cu treatment was lower in transgenic plants than in WT plants, although both showed an increased H2O2 content in the roots (Fig. 7b), indicating the H2O2 was scavenged in the transgenic plants.
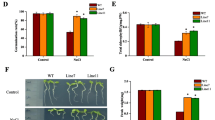
The ThMT3-transgenic plants showed significantly different SOD, CAT and APX activity levels, as demonstrated by three transgenic and WT plants under different conditions (Fig. 8a–c), indicating that there is a relationship between ThMT3 expression and the anti-oxidant enzyme system. The activity levels in transgenic plant increased under Cu stress (Fig. 8a–c), but under the Cu + cPTIO conditions, the SOD, CAT and APX activities were not significantly different between non- and transgenic plants (Fig. 8 a–c). Cu stress also increased the MDA content in non- and transgenic plants (Fig. 8d), which reflected the degree of membrane lipid oxidation. However, the increased level in the transgenic plants was not significantly higher than that in the WT plants (Fig. 8d), indicating membrane lipid oxidation in WT plant similar to that in transgenic plants. Based on our results, we hypothesized that NO plays a central role in adventitious rooting in transgenic plants under Cu stress.
Measurement of anti-oxidative activity in the roots under Cu stress. Both wile type (WT) and transgenic plants were cultured on 1/2 MS medium containing 50 μM CuSO4, 100 μM sodium nitroprusside (SNP), 50 μM Cu + 100 μM SNP, and 50 μM Cu + 100 μM cPTIO for 2 weeks. Activities of Superoxide dismutase (SOD) a catalase (CAT) b ascorbate peroxidase (APX) c and malondialdehyde (MDA). Data are mean ± SD from three independent experiments. *Significant (t test, P < 0.05) difference compared with WT plants under each treatment
Transcript levels of genes involved in NO biosynthesis, anti-oxidation and adventitious root formation under Cu stress
The expression of genes encoding the NO synthesis enzymes nitrate reductase (NR) and Nitric synthase (NOS) were induced in transgenic plants under Cu treatment, while the levels of these enzymes remained unchanged in WT plants after treatment (Fig. 9a). This suggested that NO was indeed induced in ThMT3-transgenic plants by the Cu treatment. Meanwhile, Cu significantly induced genes encoding anti-oxidative enzyme genes, including glutathione reductase (GR), SOD, CAT and APX in ThMT3-transgenic plants.
Expression of NO synthesis, antioxidant and adventitious root formation related genes. (a) The transcript levels of NR (GeneBank accession number: KM591683), GR (KM591685), SOD (KM591686), CAT (KM591687), APX (KM591688) with treatment of 50 μM CuSO4 for 5 h, respectively. b, c, d The transcript levels of ARF8 (KM591689), AUX1 (KM591690), PIN1 (KM591691) under conditions of 50 μM Cu (b) 100 μM SNP (c) and 50 μM Cu + 100 μM cPTIO (d) for 5 h, respectively. The gene expression levels were normalized against the β-tubulin (KM591694) gene expression level. Data are mean ± SD from three independent experiments. *Significant (t test, P < 0.05) difference compared with WT plants under each treatment
Auxin application effectively enhances adventitious root formation (Sukumar et al. 2013), and auxin response factors (ARFs) are transcription factors involved in auxin signaling. There are influx proteins, including auxin resistant 1 (AUX1), which mediate auxin movement (Swarup et al. 2008), and proteins, such as pinformed (PIN1), ATP-binding cassette type B (ABCB), P-glycoprotein (PGP) and multidrug resistant (MDR), which participate in auxin efflux (Zazímalová et al. 2010). Analysis of the expressions of the genes encoding these proteins showed that, Auxin response factor 8 (ARF8), AUX1 and PIN1 were highly expressed in transgenic plants under Cu treatment (Fig. 9b) and SNP (Fig. 9c) treatment compared with WT plants, while the expression levels of ABCB and PGP did not changed significantly (data not shown). After adding cPTIO, the expression levels of the genes were similar in transgenic and WT plants (Fig. 9d).
Discussion
In this study, we introduced a ThMT3 gene from T. hispida into S. matsudana seeds following our previous reported transformation protocol (Yang et al. 2013) to improve the heavy metal tolerance of this important tree species. To the best of our knowledge, this is the first report of the recovery of transgenic Salix plants expressing a functional gene. Previous studies have reported transgenic MT-containing plants that displayed an enhanced tolerance to heavy metals (Bellion et al. 2007; Lee et al. 2004). In the current study, the transgenic plants showed better growth than WT plants when under Cu stress. Moreover, 50 μM Cu treatment damaged the chloroplasts of WT plants and increased the number of starch granules instead of thylakoids in the chloroplasts (Fig. 5a), whereas the chloroplasts of transgenic plants remained intact (Fig. 5b). These results suggested that the transgenic plants are Cu-tolerant. The results were consistent with our previous study that demonstrated that transgenic Saccharomyces cerevisiae overexpressing ThMT3 had increased tolerance to Cu (Yang et al. 2011).
In this study, exposure to 50 μM Cu induced more adventitious roots in transgenic plant stems than in WT plants. Treatment with the NO inhibitor cPTIO decreased the rooting rate of transgenic plants (Supplementary Fig. S3a), suggesting that NO release is related to Cu exposure in ThMT3-transgenic plants. However, NO may be an important element to improve the rooting rate. It was reported that Cu treatments caused NO release in the roots and leaves of tomato and NO can efficiently alleviate the Cu toxicity effect (Wang et al. 2010). NO is a radical molecule that participates in multiple plant physiological processes, such as seed germination, plant growth and response to drought. Exogenous NO protects rice leaves from oxidative stress by increasing the activities of antioxidant enzymes (Hung et al. 2002). Generally, heavy metals cause the rapid release of ROS (Groppa et al. 2008). In the current study, the Cu stress induced a higher anti-oxidative enzyme activity. However, SOD, CAT and APX activities were similar between transgenic and WT plants when exposed to cPTIO, demonstrating that Cu can induce NO, and that NO may play a special role in Cu tolerance. MDA is one of the main products of lipid peroxidation and is highly toxic. An increased MDA level is an indicator of membrane damage, which is closely associated with the accumulation of ROS caused by stress (Cai et al. 2011). The deceased MDA levels in transgenic plants under Cd and Cu stress (Fig. 8d) were probably connected with the decreased H2O2 level that mediated lipid peroxidation. Meanwhile, MTs are thought to function as antioxidants against ROS (Yang et al. 2011). NOS and NR are two major NO synthetic enzymes, and we confirmed that their expression levels increased in transgenic plants under lower Cu conditions, as confirmed by qRT-PCR results (Fig. 9a). This demonstrated that Cu indeed improved NO synthesis, and that NO probably plays a key role in Cu tolerance in transgenic S. matusudana. Additionally, Cu induced GR, SOD, CAT and APX gene expression levels (Fig. 9a), which further indicated that the anti-oxidative enzyme system is induced by Cu to increase tolerance.
Cu promoted adventitious roots growth on the stems of transgenic plants (Fig. 2a, b). The same result was reported by An et al. (2006), low concentrations of Cu in the culture solution enhanced root elongation and increased the lateral root number in Pinus pinaster. The development of the root system is an extremely complex physiological process, which is affected by many internal and environmental factors (Malamy 2005). One factor, the regulation of auxin transport, is essential for normal shoot and root growth. Polar auxin transport (PAT) is the main transport pattern in plants, and interfering with PAT results in the redistribution of endogenous auxins and inhibits gravity responses, which then affect adventitious root development (Geldner et al. 2001; Muday and Murphy 2002). In the current study, NO may have accumulated in transgenic S. matsudana, interfering with PAT to the hypocotyls, and causing accumulation of auxin in stems by an as yet unknown mechanism. Moreover, NO has been reported to regulate adventitious root initiation under the regulation of PAT (Xiong et al. 2009). ARF8, AUX1 and PIN1 in transgenic plants were highly expressed under Cu (Fig. 9b) or SNP (Fig. 9c) treatment, but did not respond to Cu and cPTIO condition (Fig. 9d), indicating that NO is related to the expression of these genes. In Arabidopsis, ARF8 acts as a positive regulator in adventitious root formation (Gutierrez et al. 2009; Wilmoth et al. 2005). Other studies reported that defects in AUX1, PIN1, ABCB19 and PGP1 reduce initiation frequencies and/or the elongation of lateral roots (Lewis et al. 2011; Marchant et al. 2002; Swarup et al. 2008). However, in the current study, ABCB28 and PGP expression levels in transgenic plants were not significantly different from non-treated plants, probably indicating that ARF, AUX and PIN are involved in adventitious root formation in ThMT3-transgenic S. matsudana, but not ABCB and PGP.
In conclusion, overexpression of the T. hispida ThMT3 gene in S. matsudana effectively increased the Cu tolerances and rooting efficiency of the transgenic plants. The NO level in transgenic plants were significantly higher than in WT plants under Cu stress. The application of a NO inhibitor, cPTIO, decreased SOD, CAT and APX activity under Cu stress. Auxin response factors ARF8 and auxin transport proteins, such as AUX1 and PIN1 which are known to improve adventitious roots, were highly expressed in transgenic plants under Cu and SNP treatments. These results suggested that overexpression of the ThMT3 gene increased Cu tolerance and NO production in S. matsudana, and the higher NO release contributed to the induction of adventitious roots under Cu stress.
References
Adamis PDB, Panek AD, Leite SGF, Eleutherio ECA (2003) Factors involved with cadmium absorption by a wild-type strain of Saccharomyces cerevisiae. Braz J Mirobiol 34:55–60
An Z, Li C, Zu Y, Du Y, Wachter A, Gromes R, Rausch T (2006) Expression of BjMT2, a metallothionein 2 from Brassica juncea, increases copper and cadmium tolerance in Escherichia coli and Arabidopsis thaliana, but inhibits root elongation in Arabidopsis thaliana seedlings. J Exp Bot 57:3575–3582
Bellion M, Courbot M, Jacob C, Guinet F, Blaudez D, Chalot M (2007) Metal induction of a Paxillus involutus metallothionein and its heterologous expression in Hebeloma cylindrosporum. New Phytol 174:151–158
Benatt MR, Yookongkaew N, Meetam M, Guo WJ, Punyasuk N, AbuQamar S, Goldsbrough P (2014) Metallothionein deficiency impacts copper accumulation and redistribution in leaves and seeds of Arabidopsis. New Phytol 202:940–951
Cai F, Mei L, An X, Gao S, Tang L, Chen F (2011) Lipid peroxidation and antioxidant responses during seed germination of Jatropha curcas. Int J Agric Biol 13:25–30
Chen PY, Wang CK, Soong SC, To KY (2003) Complete sequence of the binary vector pBI121 and its application in cloning T-DNA insertion from transgenic plants. Mol Breed 11:287–293
Cobbett C, Goldsbrough P (2002) Phytochelatins and metallothioneins: roles in heavy metal detoxification and homeostasis. Annu Rev Plant Biol 53:159–182
Dos Santos Utmazian MN, Wieshammer G, Vega R, Wenzel WW (2007) Hydroponic screening for metal resistance and accumulation of cadmium and zinc in twenty clones of willows and poplars. Environ Pollut 148:155–165
Dowling DN, Doty SL (2009) Improving phytoremediation through biotechnology. Curr Opin Biotechnol 20:204–206
Freeman JL, Tamaoki M, Stushnoff C, Quinn CF, Cappa JJ, Devonshire J, Fakra SC, Marcus MA, McGrath SP, Van Hoewyk D, Pilon-Smits EA (2010) Molecular mechanisms of selenium tolerance and hyperaccumulation in Stanleya pinnata. Plant Physiol 153:1630–1652
Geldner N, Frim J, Stierhof YD, Juergens G, Palme K (2001) Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413:425–428
Groppa MD, Rosales EP, Lannone MF, Benavides MP (2008) Nitric oxide, polyamines and Cd-induced phytotoxicity in wheat roots. Phytochemistry 69:2609–2615
Guo Z, Ou W, Lu S, Zhong Q (2006) Differential responses of antioxidative system to chilling and drought in four rice cultivars differing in sensitivity. Plant Physiol Biochem 44:828–836
Gutierrez L, Bussell JD, Pacurar DI, Schwambach J, Pacurar M, Bellini C (2009) Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant Cell 21:3119–3132
Hung KT, Chang CJ, Kao CH (2002) Paraquat toxicity is reduced by nitric oxide in rice leaves. J Plant Physiol 159:159–166
Jaakola L, Pirttilä AM, Halonen M, Hohtola A (2001) Isolation of high quality RNA from bilberry (Vaccinium myrtillus L.) fruit. Mol Biotechnol 19:201–213
Laspina NV, Groppa MD, Tomaro ML, Benavides MP (2005) Nitric oxide protects sunflower leaves against Cd-induced oxidative stress. Plant Sci 169:323–330
Lee J, Shim D, Song WY, Hwang I, Lee Y (2004) Arabidopsis metallothioneins 2a and 3 enhance resistance to cadmium when expressed in Vicia faba guard cells. Plant Mol Biol 54:805–815
Lei YB, Korpelainen H, Li CY (2007) Physiological and biochemical responses to high Mn concentrations in two contrasting Populus cathayana populations. Chemosphere 68:686–694
Lewis DR, Negi S, Sukumar P, Muday GK (2011) Ethylene inhibits lateral root development, increases IAA transport and expression of PIN3 and PIN7 auxin efflux carriers. Development 138:3485–3495
Malamy JE (2005) Intrinsic and environmental response pathways that regulate root system architecture. Plant, Cell Environ 28:67–77
Marchant A, Bhalerao R, Casimiro I, Eklöf J, Casero PJ, Bennett M, Sandberg G (2002) AUX1 promotes lateral root formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell 14:589–597
Muday GK, Murphy AS (2002) An emerging model of auxin transport regulation. Plant Cell 14:293–299
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plantarum 15:495–497
Shi SY, Wang G, Wang YD, Zhang LG, Zhang LX (2005) Protective effect of nitric oxide against oxidative stress under ultraviolet-B radiation. Nitric Oxide 13:1–9
Singh HP, Kaur S, Batish DR, Sharma VP, Sharma N, Kohli RK (2009) Nitric oxide alleviates arsenic toxicity by reducing oxidative damage in the roots of Oryza sativa (rice). Nitric Oxide 20:289–297
Sridhar BBM, Diehl SV, Han FX, Monts DL, Su Y (2005) Anatomical changes due to uptake and accumulation of Zn and Cd in Indian mustard (Brassica juncea). Environ Exp Bot 54:131–141
Sukumar P, Maloney GS, Muday GK (2013) Localized induction of the ATP-Binding cassette B19 auxin transporter enhances adventitious root formation in Arabidopsis. Plant Physiol 162:1392–1405
Swarup K, Benková E, Swarup R, Casimiro I, Péret B, Yang Y, Parry G, Nielsen E, De Smet I, Vanneste S, Levesque MP, Carrier D, James N, Calvo V, Ljung K, Kramer E, Roberts R, Graham N, Marillonnet S, Patel K, Jones JD, Taylor CG, Schachtman DP, May S, Sandberg G, Benfey P, Friml J, Kerr I, Beeckman T, Laplaze L, Bennett MJ (2008) The auxin influx carrier LAX3 promotes lateral root emergence. Nat Cell Biol 10:946–954
Wang LN, Yang LM, Yang FJ, Li XG, Song YP (2010) Involvements of H2O2 and metallothionein in NO-mediated tomato tolerance to copper toxicity. J Plant Physiol 167:1298–1306
Wilmoth JC, Wang S, Tiwari SB, Joshi AD, Hagen G, Guilfoyle TJ, Alonso JM, Ecker JR, Reed JW (2005) NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J 43:118–130
Xiong J, Lu H, Lu K, Duan Y, An L, Zhu C (2009) Cadmium decreases crown root number by decreasing endogenous nitric oxide, which is indispensable for crown root primordia initiation in rice seedlings. Planta 230:599–610
Xu J, Yin H, Li Y, Liu X (2010) Nitric oxide is associated with long-term zinc tolerance in Solanum nigrum. Plant Physiol 154:1319–1334
Yang JL, Wang YC, Liu GF, Yang CP, Li CH (2011) Tamarix hispida metallothionein-like ThMT3, a reactive oxygen species scavenger, increases tolerance against Cd2+, Zn2+, Cu2+, and NaCl in transgenic yeast. Mol Biol Rep 38:1567–1574
Yang JL, Yi JS, Yang CP, Li CH (2013) Agrobacterium tumefaciens-mediated genetic transformation of Salix matsudana Koidz. using mature seeds. Tree Physiol 33:628–639
Yu CC, Hung KT, Kao CH (2005) Nitric oxide reduces Cu toxicity and Cu-induced NH4+ accumulation in rice leaves. J Plant Physiol 162:1319–1330
Zazímalová E, Murphy AS, Yang HB, Hoyerová K, Hosek P (2010) Auxin transporters–why so many? CSH Perspect Biol 2:a001552
Zhang F, Wang Y, Yang Y, Wu H, Wang D, Liu J (2007) Involvement of hydrogen peroxide and nitric oxide in salt resistance in the calluses from Populus euphratica. Plant, Cell Environ 30:775–785
Zhang Y, Tan J, Guo Z, Lu S, He S, Shu W, Zhou B (2009) Increased abscisic acid levels in transgenic tobacco over-expressing 9 cis-epoxycarotenoid dioxygenase influence H2O2 and NO production and antioxidant defences. Plant, Cell Environ 32:509–519
Zhou B, Guo Z, Xing J, Huang B (2005) Nitric oxide is involved in abscisic acid-induced antioxidant activities in Stylosanthes guianensis. J Exp Bot 56:3223–3228
Acknowledgments
This work was supported by the the Innovation Project of State Key Laboratory of Tree Genetics and Breeding (Northeast Forestry University) (2014B02), National High Technology Research and Development Program of China (863 Program, 2013AA102704), the Fundamental Research Funds for the Central Universities (DL11EA02), and China Postdoctoral Science Foundation (2014M560242).
Conflict of Interest
The authors declare no competing financial interests.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yang, J., Chen, Z., Wu, S. et al. Overexpression of the Tamarix hispida ThMT3 gene increases copper tolerance and adventitious root induction in Salix matsudana Koidz.. Plant Cell Tiss Organ Cult 121, 469–479 (2015). https://doi.org/10.1007/s11240-015-0717-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-015-0717-3