Abstract
A new ancyrocephalid monogenean is described from the gills of wild White-spottedrabbitfish Siganus canaliculatus (Park) based on morphological and molecular analyses. Glyphidohaptor safiensis n. sp. can be distinguished from other members of the genus by the shape of the accessory piece of the male copulatory organ (MCO). Unlike its congeners, the rod-shaped accessory piece of G. safiensis n. sp. is distally broad and flattened. The MCO of G. safiensis n. sp. is curved, enclosed in a heavy sheath with a terminal flap. Partial large subunit (LSU), partial small subunit (SSU) and the partial SSU, entire internal transcribed spacer region 1 (ITS1) and partial 5.8S rDNA of the new species and two species of Tetrancistrum Goto & Kikuchi, 1917 from the same host and locality were sequenced and subjected to phylogenetic analysis. The LSU rDNA analysis grouped G. safiensis n. sp. with Tetrancistrum sp. from the gills of Siganus fuscescens Houttuyn from Australia, indicating a possible misidentification of the latter. Sequences of the SSU rDNA of the new species were most similar to those for Pseudohaliotrema sphincteroporus Yamaguti, 1953, demonstrating the close relatedness of the genera Pseudohaliotrema Yamaguti, 1953 and Glyphidohaptor Kritsky, Galli & Yang, 2007 within the Ancyrocephalidae. The comparison of the partial SSU (424 bp) and entire ITS1 and partial 5.8S rDNA (246 bp) sequences obtained for G. safiensis n. sp. with the only available sequence of another member of Glyphidohaptor Kritsky, Galli & Yang, 2007, G. pletocirra Paperna, 1972 (HE601931-HE601933) yielded on average 1.08% dissimilarity (a difference of 7 bases), with a p-distance of 0.010 ± 0.004%. This is the first record of a species of Glyphidohaptor from S. canaliculatus and from the Persian Gulf, the Gulf of Oman and the Arabian Sea.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Siganidae is a monotypic family of inshore and reef-associated tropical and subtropical fishes consisting of 29 valid species, all from the Indo-Pacific region (Froese & Pauly, 2018). Several authors have investigated the parasite fauna of siganids reporting high parasite diversity (e.g. Paperna, 1972; Diamant & Paperna, 1986; Martens & Moens, 1995; Geets & Ollivier, 1996; Geets et al., 1997; Shih & Jeng, 2002; Aloo et al., 2004; Hassanine & Al Jahdali, 2007). To date, a total of 18 siganid species were investigated for monogenean parasites, resulting in identification of species of the polyopisthocotylean genus Polylabris Euzet & Cauwet, 1967 (see Tingbao et al., 2007; Sailaja & Madhavi, 2010) and four monopisthocotylean genera, including three genera of ancyrocephalids (Goto & Kikuchi, 1917; Young, 1967; Paperna, 1972; Lim, 2002; Kritsky et al., 2007a, b; Kritsky & Galli, 2007) and one viviparous gyrodactylid (Ernst et al., 2001). The ancyrocephalid genus Glyphidohaptor Kritsky, Galli & Yang, 2007 was erected by Kritsky et al. (2007a) to accommodate Glyphidohaptor plectocirra (Paperna, 1972) which was initially described as Pseudohaliotrema plectocirra Paperna, 1972 (syn. Tetrancistrum plectocirra (Paperna, 1972) Lim, 2002) from Siganus luridus (Rüppell) and Siganus rivulatus (Forsskål & Niebuhr) from the Gulf of Aqaba (off Eilat, Red Sea). Two further species were described from the Great Barrier Reef from five siganid hosts, Glyphidohaptor phractophallus Kritsky, Galli & Yang, 2007 from Siganus fuscescens (Houttuyn) and G. sigani Kritsky, Galli & Yang, 2007 from S. doliatus Guérin-Méneville, S. lineatus (Valenciennes), S. punctatus (Schneider & Forster) and S. corallinus (Valenciennes) (see Kritsky et al., 2007a). Other reports include an unidentified Glyphidohaptor sp. from the gills of an unknown Siganus sp. from Macassar, Sulawesi (Indonesia) (Kritsky et al., 2007a).
The class Monogenea has been subject of several major molecular phylogenetic studies (Justine et al., 2002). Molecular-based phylogenetic relationships of the Platyhelminthes, including the Monogenea, were carried out utilising RNA gene sequences (Baverstock et al., 1991; Mollaret et al., 1997; Campos et al., 1998; Littlewood et al., 1999; Olson & Littlewood, 2002). Mollaret et al. (2000) investigated the phylogenetic relationships between the two main subclasses of the Monogenea (Polyopisthocotylea and Monopisthocotylea) using the large subunit (LSU) of the ribosomal RNA gene. Furthermore, a number of authors evaluated the phylogenetic position of the ancyrocephalid genera (e.g. Haliotrema Johnston & Tiegs, 1922; Euryhaliotrema Kritsky & Boeger, 2002; Haliotrematoides Kritsky, Yang & Sun, 2009; and Pseudempleurosoma Yamaguti, 1965) through analyses of the small subunit (SSU) and LSU of the rRNA gene (Plaisance et al., 2005; Wu et al., 2007; Dang et al., 2010; Garcia-Vasquez et al., 2015; Mendoza-Palmero et al., 2015; Theisen et al., 2017, 2018). The internal transcribed spacer region 1 (ITS1) was also used to explore the intraspecific variability (Šimková et al., 2004; Stefani et al., 2012; Šimková et al., 2013; Kmentová et al., 2015).
Although the identification and description of new species of monogeneans is traditionally made through morphological characterisation (Desdevises et al., 2000), the combination of molecular data with morphological analysis for the description of new species is becoming more common (e.g. Freeman & Ogawa, 2010; Bullard et al., 2015; Soo et al., 2015; Theisen et al., 2017, 2018). To date, only a few sequences are available for ancyrocephalids infecting siganids. The first gene sequence from a monogenean infecting siganids was provided by Mollaret et al. (1997). The authors analysed a LSU rDNA sequence of Tetrancistrum sp. obtained from S. fuscescens caught off Heron Island, Queensland, Australia, without providing any morphological information of this species. Later, SSU and LSU sequences of Pseudohaliotrema sphincteroporus Yamaguti, 1953 were obtained by Littlewood & Olson (2001) and used to evaluate the molecular phylogenetic relationships of families within the Monogenea. In addition, SSU sequences for Tetrancistrum sigani Goto & Kikuchi, 1917 (syn. T. nebulosi Young, 1967) were deposited in GenBank by Wang et al. (2010) and Ummey et al. (2015). Furthermore, a comparison of the genetic variation in populations of G. plectocirra was investigated by utilising the cytochrome c oxidase subunit 1 (cox1) gene, and partial SSU and the entire ITS1 region including the 5.8S gene (Stefani et al., 2012).
During a survey of the parasite fauna of Siganus canaliculatus (Park) from Omani waters, a new species of Glyphidohaptor was recovered. The objectives of the present study were to provide a morphological description of the new species and to explore the phylogenetic relationships within the Ancyrocephalidae Bychowsky & Nagibina, 1968 (sensu lato) based on sequence data for the LSU, SSU and partial SSU, entire ITS1 and partial 5.8S rDNA region, focusing on Glyphidohaptor, Pseudohaliotrema Yamaguti, 1953 and Tetrancistrum Goto & Kikuchi, 1917.
Materials and methods
Sample collection and examination
A total of 245 white-spotted rabbitfish, Siganus canaliculatus (Perciformes: Siganidae), were purchased alive from local fish markets from seven locations along the coasts of the Sultanate of Oman (Table 1). Fish were morphologically identified by using the guidelines of Randall (1995) and FAO identification sheets (1984). Fish specimens were transported on ice to the facility of the Laboratory of Aquatic Parasitology at the Fishery Quality Control Center, Sultanate of Oman. The hosts were either immediately subjected for parasitological examination or frozen at -40°C for subsequent investigation. Upon dissection, gills were excised; arches were separated, placed in a Petri dish with filtered seawater and examined under a dissecting microscope. Parasites were detached from the gill filaments by means of fine needles and kept in filtered seawater at 4°C prior to fixation. Monogenean whole mounts were prepared from unflattened worms fixed in AFA (3 parts alcohol: 2 parts formalin: 1 part acetic acid) or 4% neutral buffered formalin, stained with Mayer’s paracarmine, cleared in clove oil and mounted in Canada balsam. Measurements, all in micrometres, are given as the range, followed by the mean and the number (n) of structures measured in parentheses. Body length includes that of the haptor; measurements of the copulatory complex, anchors and hooks and the description of the new species are according to Kritsky et al. (2007a). Additional measurements of anchors were obtained according to Gussev (1985) (see Vignon & Sasal, 2010). An analysis of the available measurements of the known congeners was carried out and statistical comparisons were carried out using Studentʼs t-test for independent samples (two-tailed; α= 95%).
Illustrations were prepared with the aid of a drawing tube attached to an Olympus BX63 motorised light microscope with Nomarski differential interference contrast optics (DIC) and digitalised using Adobe illustrator CC 2018. and the program Inkscape 0.92.2 (Scalable Vector Graphics, 2).
Comparative morphological analysis
Voucher specimens were obtained from the British Natural History Museum (G. phractophallus BMNH 2006.8.8.13-14, G. sigani BMNH 2006.8.8.11-12), Meguro Parasitology Museum (G. phractophallus MPM 18828, G. sigani MPM 18829) and the Queensland Museum (G. phractophallus QMG227443-QMG 227447), G. sigani (QMG227453, QMG 227454, QMG 227457, QMG G227460, QMG G227461) respectively. In addition, micrographs of selected parasites were obtained for comparative purposes (Glyphidohaptor sp. MPM 22839 and MPM 22837 and G. plectocirra USNPC 98587, USNPC 98588, USNPC 98590, USNPC 98591).
Confocal microscopy
Several specimens preserved in 95% ethanol were stained with one step Gomori’s trichrome kit (Morphisto, Frankfurt, Germany), mounted in Histochoice (Amresco, Solon, OH, USA), and imaged using a Carl Zeiss LSM780 confocal fluorescence microscope and a PL APO 63 × 1.4 oil immersion lens following the procedure of Marchiori et al. (2015). Three-dimensional (3D) stacks of the diagnostic features were acquired with a typical voxel size of 66 × 66 × 500 nm (XYZ). The samples were excited using a DP55 561 laser (AOTF 3%) and a main beam splitter 488/561. Spectral emissions (566 to 694 nm window) were detected using the internal PMT detector (grain 800). Maximum projection and three-dimensional images were obtained from a z-stack of 300 planes and the 3D rendering option in ZEN software.
DNA extraction and PCR amplification
Monogeneans infecting S. canaliculatus were preserved in 95% ethanol for molecular analysis. Total genomic DNA was extracted from individual worms using QIAamp mini DNA kit (Qiagen, Hilden, Germany) according to the manufacturerʼs instructions with some modification. Due to the small size of the worms investigated in this study and in order to increase the DNA yield, for the final step in the DNA extraction protocol, the extracted DNA was eluted with 200 μl of elution buffer and incubated for 10 min at room temperature before final spin and collection of genomic DNA. The obtained DNA was then completely dried using a Concentrator plus/Vacufuge®Plus (Eppendorf, Hamburg, Germany), re-suspended with 40 μl elution buffer, then stored in a freezer at − 20°C. This helped increase the yield of extracted DNA. DNA concentration was quantified using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
Polymerase chain reaction (PCR) amplification for partial SSU rDNA were performed in 20 μl reaction volume containing 10 pmol of the forward 390f (5'-AGA GGG AGC CTG AGA AAC G-3') and reverse 870r (5'-GTT GAG TCA AAT TAA GCC GCA-3') primers, and ~10 ng of DNA template using illustra™ puReTaq Ready-To-Go PCR beads (0.2 ml tubes, 96 reactions). After initial denaturation at 98°C for 5 min, samples were subjected to 40 cycles of amplification (denaturation at 95°C for 30 s, primer annealing at 55°C for 45 s, and extension at 72°C for 1 min), followed by a final extension step at 72°C for of 7 min (Freeman et al., 2013).
For amplification of the D1-D2 domain of the LSU rDNA (28S) the primers C1 (5′-ACC CGC TGA ATT TAA GCA T-3′) and D2 (5′-TGG TCC GTG TTT CAA GAC-3′) were used, following Mendlova et al. (2010) with modification of the annealing temperature to 60°C. All PCR reactions were performed in 30 µl reactions containing 5 pmol of each primer and ~30 ng of concentrated genomic DNA using illustra™ puReTaq Ready-To-Go PCR beads. The following amplification conditions were utilised: an initial denaturation at 94°C for 2 min, followed by 40 cycles of amplification (denaturation at 94°C for 20 s, primer annealing at 60°C for 30 s, and extension at 72°C for 90 s), followed by a final extension step at 72°C for 10 min.
Furthermore, partial SSU rDNA and entire ITS1 region were amplified in one round using the primers S1 (5'-ATT CCG ATA ACG AAC GAG ACT-3') and IR8 (5'-GCT AGC TGC GTT CTT CAT CGA-3') that anneal to the SSU and 5.8S rDNA genes, respectively (Šimková et al., 2003). Each amplification reaction was performed in a final volume of 35 µl containing 10 pmol of each primer and ~30 ng of genomic DNA using illustra™ puReTaq Ready-To-Go PCR beads (0.2 ml tubes, 96 reactions). The amplification conditions followed Šimková et al. (2013). All PCR products were viewed on a 0.8% agarose gel stained with ethidium bromide.
Phylogenetic analyses
Contiguous sequences of the investigated worms were obtained manually using BioEdit (Hall, 1999); these were aligned using Clustal W (Thompson et al., 1994) implemented in MEGA v.7 (Kumar et al., 2015). Newly obtained sequences were subjected to a BLAST search in the GenBank database, and sequences for ancyrocephalids showing the highest similarity (88–96%) to the newly acquired data were retrieved from GenBank and used in the phylogenetic analysis (Table 2). Phylogenetic trees were reconstructed utilising three different datasets.
The first analysis used the partial LSU (D1-D2 domain) dataset containing 20 ancyrocephalid ingroup taxa and Thaparocleidus asoti Yamaguti, 1937 (see Wu et al., 2007) as the outgroup. The second analysis used the SSU data from 15 ancyrocephalid species, and Lamellodiscus donatellae Aquaro, Riva & Galli, 2009 as the outgroup. The third dataset consisted of the partial SSU, entire ITS1 and partial 5.8S rDNA sequences of G. safiensis n. sp. and G. plectocirra sequences (GenBank: HE601931-HE601933). Phylogenetic analyses were performed based on best-fit model in MEGA version 7 (Kumar et al., 2015). For the LSU dataset, maximum likelihood method based on the General Time Reversible model was the best model. For the SSU dataset, maximum likelihood method based on the Kimura 2-parameter model was used. The methods chosen for the third dataset was Neighbor-Joining method based on Kimura 2-parameter model. The robustness of the inferred phylogeny was assessed using a bootstrap procedure with 1,000 replications (Wu et al., 2005).
Family Ancyrocephalidae Bychowsky & Nagibina, 1968 ( sensu lato )
Genus Glyphidohaptor Kritsky, Galli & Yang, 2007
Glyphidohaptor safiensis n. sp.
Type-host: Siganus canaliculatus (Park) (Perciformes: Siganidae), white-spotted rabbitfish.
Type-locality: Off Muttrah (23.6249°N, 58.5624°E), Sea of Oman, Sultanate of Oman.
Other localities: Off Khasab fishing harbour (26.1644°N, 56.2426°E), Sea of Oman, Sultanate of Oman; off Dibba Al-Bayya (25.6365°N, 56.2538°E), Sea of Oman, Sultanate of Oman; off Masirah Island (20.4711°N, 58.8153°E), Arabian Sea, Sultanate of Oman; off Al-Lakbi fishing harbour (18.113°N, 56.3255°E), Arabian Sea, Sultanate of Oman; and off Raysut (16.5500°N, 54.0100°E), Arabian Sea, Sultanate of Oman (November and December 2012).
Type-material: The holotype (ZMB Monogenea 7434) and 5 paratypes (ZMB Monogenea 7435) were deposited in the Berlin Natural History Museum, Berlin, Germany; 9 paratypes (MPM. Coll. No. 20959) were deposited in the Meguro Parasitology Museum, Tokyo, Japan.
Site in host: Gills.
Prevalence: Off Khasab fishing harbour: 83% (29 out of 35 fish); off Dibba Al-Bayya: 94% (33 out of 35 fish); off Muttrah: 97% (34 out of 35 fish); off Masirah Island: 97% (34 out of 35 fish); off Al-Lakbi fishing harbour: 97% (34 out of 35 fish); off Raysut: 100% (35 out of 35 fish).
Representative DNA sequences: GenBank MK192832 and MK192898.
ZooBank registration: To comply with the regulations set out in article 8.5 of the amended 2012 version of the International Code of Zoological Nomenclature (ICZN, 2012), details of the new species have been submitted to ZooBank. The Life Science Identifier (LSID) for Glyphidohaptor safiensis n. sp. is urn:lsid:zoobank.org:act:D6911FB7-3E39-4B56-ABF6-4B7B7E0FECAF.
Etymology: The specific name (safiensis) is after the Arabic local name of the host (safi).
Description
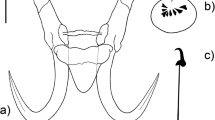
[Based on 25 adult specimens; Figs. 1, 2.] Body fusiform, slightly flattened dorso-ventrally, 926–1,326 (1,113; n = 25) in total length (Fig. 1A); greatest width 217–313 (266; n = 25) at level of gonads; tegument smooth. Cephalic lobes well developed; each head organ consisting of groupings of terminations of cephalic-gland ducts posteriolateral to pharynx. Pharynx a muscular, glandular bulb, spherical to subovate, 43–67 (54; n = 25) long, 37–49 (43; n = 21) wide. Eye-spots absent; chromatic granules scattered throughout cephalic region and posterior trunk. Mouth subterminal, midventral at level of head organs, opening into buccal tube; buccal tube extending posteriorly along body midline to pharynx; intestinal caeca bifurcating posterior to pharynx, confluent, lacking diverticula.
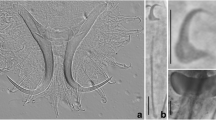
Glyphidohaptor safiensis n. sp. ex Siganus canaliculatus. Male copulatory complex. A, Confocal microscope maximum projection image showing the composition of the MCO and the semi-circular sub-basal flange positioned distally to the fan-shaped basal flange. B, Drawing of the male copulatory organ, dorsal view. Abbreviations: BF, basal flange; PLP, plate-like projection; AP, accessory piece. Scale-bars: 20 µm
Testis fusiform, 173–258 (217; n = 21) long, 35–78 (61; n = 21) wide. Germarium pyriform, 64–132 (96; n = 19) long, 21–43 (32; n = 19) wide. Vaginal vestibule (Fig. 1B) slightly sclerotised, sub-oval; vaginal opening 39–52 (45; n = 16) long, 26–33 (30; n = 6) wide; vaginal tube sinuous or coiled, with frayed opening at the end, 44–70 (54; n = 6) long. Male copulatory organ (MCO), tubular, curved, enclosed in heavy sheath, with semi-circular sub-basal flange (Fig. 2A), 58–76 (64; n = 24) long. Accessory piece rod-shaped, distally flattened, with reniform plate-like projection.
Haptor 41–58 (51; n = 24) long, 84–131 (118; n = 24) wide. Ventral anchor 48–57 (53; n = 17) long, base 32–49 (40; n = 21) long, 29–36 (32; n =17) wide (Fig. 1D), inner roots 12–14 (17; n = 20) long, outer roots 11–25 (17; n= 20) long, length of point 6–10 (8; n = 20). Dorsal anchor 54–63 (58; n = 16) long, base 30–44 (38; n = 22) long, 24–33 (28; n = 16) wide (Fig. 1C), inner roots 13–23 (16; n = 20) long; outer roots 21–32 (25; n = 20) long, length of point 6–10 (8; n = 20). Ventral bar 32–46 (39; n = 16) long (Fig. 1E); dorsal bar 37–48 (41; n = 12) long (Fig. 1F); hooks 11–12 (11; n = 18) long (Fig. 1G).
Remarks
Comparative statistical analysis (see Supplementary Table S1) of the morphometric characteristics of Glyphidohaptor species revealed that Glyphidohaptor safiensis n. sp. can be distinguished from other members of the genus by its significantly larger body length and width. The length of the haptor is significantly shorter in comparison to those of G. phractophallus, G. plectocirra (S. luridus) and G. sigani (S. doliatus and S. lineatus) (Table 3). However, the haptor is significantly wider in comparison to G. phractophallus, G. plectocirra and G. sigani (S. corallinus, S. doliatus and S. lineatus). The size of the pharynx and the male copulatory organ of the new species is significantly larger than all of its congeners. The length of ventral anchor of the new species is larger than that of G. plectocirra and G. sigani (S. doliatus and S. corallinus). The base of the ventral sucker of G. safiensis n. sp. is smaller than those of G. phractophallus and G. sigani (S. doliatus, S. corallinus and S. lineatus), but is significantly wider than that of G. plectocirra (S. luridus). The dorsal anchor length and width are larger in the new species in comparison to its congeners. Also, the ventral and dorsal bars of the new species are significantly larger in comparison to other members of the genus.
Morphologically, the male copulatory complex of all members of Glyphidohaptor is composed of a basally articulated MCO that is equipped with a basal flange and an accessory piece with a plate-like projection (Kritsky et al., 2007a). The shape of the MCO is variable among species of the genus. The MCO tube of the new species is curved, while that of G. phractophallus is arched. In comparison to the new species the MCO tube of G. sigani is recurved and that of G. plectocirra is straight. The accessory piece of Glyphidohaptor spp. appears to be the most suitable structure to differentiate between species of the genus as it is unique to each Glyphidohaptor species. The accessory piece of G. safiensis n. sp. is rod-shaped and distally expanded and flattened. In G. phractophallus the accessory piece is flat and blade-like. Glyphidohaptor sigani displays a rod-shaped accessory piece with a bifid proximal end, while the accessory piece of G. plectocirra is distally pointed. The basal flange of all Glyphidohaptor spp. is fan-shaped, with some species-specific variations among members of the genus. Finally, all Glyphidohaptor spp. possess a plate-like projection that is positioned along the proximal half of the accessory piece whereas the plate-like projection of G. safiensis n. sp. is large and reniform.
Phylogenetic position of Glyphidohaptor safiensis n. sp. using the SSU dataset
The length of the partial SSU sequences of the monogeneans sequenced in the present study was 890 bp for Glyphidohaptor safiensis n. sp. and c.900 bp for both Tetrancistrum labyrinthus Al-Jufaili & Palm, 2017 (GenBank: MN179331 and MN179334) and T. indicum Paperna, 1972 (GenBank: MN179330 and MN179328). The BLAST results showed that G. safiensis n. sp. is 96% similar to P. sphincteroporus, and both sequences of Tetrancistrum spp. investigated in this study showed 98% and 99% similarity to T. sigani (as T. nebulosi; GenBank: KT267177) and 98% similarity to T. sigani (as T. nebulosi; GenBank: HM545910).
The maximum-likelihood tree based on Kimura 2-parameter distance, gamma distributed with invariant sites constructed using the SSU data set revealed that G. safiensis n. sp. and P. sphincteroporus formed a separate clade clustering as sister taxa with a bootstrap value of 69% (Fig. 3). The newly generated SSU rDNA sequences of the two Tetrancistrum spp. and the available SSU rDNA sequence of T. sigani (syn. T. nebulosi) (only one sequence was used, HM545910) formed a well-supported monophyletic clade within the marine ancyrocephalids. The clade containing Glyphidohaptor and Pseudohaliotrema was sister to the clade comprising members of Bravohollisia Bychowsky & Nagibina, 1970 but with low bootstrap support (43%). In the Tetrancistrum clade, T. labyrinthus clustered with T. indicum (Fig. 3).
Phylogenetic position of Glyphidohaptor safiensis n. sp. using the LSU dataset
For the LSU rDNA gene, sequences reaching c.851 bp were successfully generated from G. safiensis n. sp. (accession number: MN176409) and 856 bp and 863 bp sequences were generated for T. indicum (accession numbers: MN179335 and MN179329) and T. labyrinthus (accession numbers: MN179333 and MN179332), respectively. The highest similarity to G. safiensis n. sp. was with the LSU rDNA sequence of Tetrancistrum sp. (GenBank: AF026114) collected from S. fuscescens from Australia (97% similarity), which was deposited by Mollaret et al. (1997). As for the Tetrancistrum spp. investigated in this study, the closest match generated by a BLAST search was with species of Euryhaliotrema Kritsky & Boeger, 2002, especially with Euryhaliotrema perezponcei Garcia-Vargas, Fajer-Ávila & Lamothe-Argumedo, 2008. The resulting neighbor-joining tree that was constructed using the LSU gene data (Fig. 4) showed that G. safiensis n. sp. formed a well-supported sister species relationship with Tetrancistrum sp. (100% bootstrap value), and the two species showed a close phylogenetic relationship clustering with P. sphincteroporus, (bootstrap value of 89%). The species of Tetrancistrum from the Gulf of Oman formed a well-supported monophylytic clade within the Ancyrocephalidae (Fig. 4). The genetic divergence value was the lowest between G. safiensis n. sp. and Tetrancistrum sp. (p-distance of 0.017%) and the highest between G. safiensis n. sp. and Thaparocleidus asoti (p-distance of 0.341%) (see Supplementary Table S2 for details).
Phylogenetic position of Glyphidohaptor safiensis n. sp. using a partial SSU-entire ITS1 region dataset
Approximately 994 bp were generated for the partial SSU, entire ITS1 and partial 5.8S region of rDNA of Glyphidohaptor safiensis n. sp. (accession numbers: MN213150 and MN213152). The two newly obtained sequences were identical. The new sequence was 98.83–98.97% similar to three sequences for another member of Glyphidohaptor available on GenBank, G. plectocirra (GenBank: HE601931-HE601933). The uncorrected p-distance was 0.01% ± 0.004 between G. safiensis n. sp. and G. plectocirra (see Supplementary Table S3) with a difference of seven nucleotides (Supplementary Table S4). The phylogenetic tree shows that G. safiensis n. sp. and G. plectocirra formed separate clades (Fig. 5).
The evolutionary history of Glyphidohaptor spp. using the partial SSU, entire ITS-1 and partial 5.8S rDNA region inferred using the Neighbor-Joining method using the Kimura 2-parameter model. All positions containing gaps and missing data were eliminated. There were a total of 678 positions in the final dataset
Discussion
Diversity of Glyphidohaptor spp.
Prior to this study Glyphidohaptor spp. were only reported from the Great Barrier Reef and the Red Sea. Several micrographs of Glyphidohaptor sp. from an unknown Siganus sp. from Macassar, Celebes, deposited by Yamaguti (1953) were obtained for comparative analysis. It was noted that this species is unlike any of the previously described members of Glyphidohaptor and does not resemble the new species described herein. Thus, it is possible that this undescribed species is another new species of Glyphidohaptor, thereby extending the geographical distribution of these worms.
Geets et al. (1997) reported an unidentified species of Pseudohaliotrema from the gills of Siganus sutor (Valenciennes) off East Africa. It is noteworthy that the species of two of the three ancyrocephalid genera recorded from siganids, Glyphidohaptor and Pseudohaliotrema, are restricted to hosts of the Siganidae (see Kritsky et al., 2007a). However, Glyphidohaptor exhibits a wider geographical range and has been registered from both, reef-associated and colored siganids (e.g. S. corallinus, S. doliatus, S. lineatus and S. punctatus) and drab-coloured off-reef siganids (e.g. S. luridus, S. fuscescens and S. rivulatus). On the other hand, members of Pseudohaliotrema are so far geographically restricted to the Eastern Indian Ocean and Western Pacific and seem to be limited to the deep-bodied reef-associated siganids (Lim, 2002; Kritsky et al., 2007a). Given that the misidentification of Glyphidohaptor and Tetrancistrum species as members of Pseudohaliotrema was a common misconception among researchers (Kritsky et al., 2007b) and based on the geographical restriction of members of Pseudohaliotrema, we believe that the registration of Pseudohaliotrema sp. from S. sutor by Geets et al. (1997) from off East Africa is probably another misidentification of a Glyphidohaptor species. Collection of new material from off East Africa is deemed necessary to confirm this statement.
Molecular characterisation
To the best of our knowledge, the present study provides the first phylogenetic analyses using data for three genera of marine ancyrocephalids known to infect siganids. These three genera show affinities to each other and affinities to other monogeneans within the marine Ancyrocephalidae. The analysis of the SSU rDNA dataset revealed that Glyphidohaptor and Pseudohaliotrema formed a separate clade within the marine Ancyrocephalidae and clustered as sister taxa. Kritsky et al. (2007a) stated that within the order Dactylogyridea, only Glyphidohaptor spp. and Pseudohaliotrema spp. displayed a germarium lying to the right of the anterior portion of the testis and suggested that this feature is apomorphic and that they are phylogenetically related. Future investigations including molecular analysis of all known members of the two genera, in addition to their relationships with their representative hosts, could provide some fascinating insights into the host-parasite phylogenetic relationships and co-evolution.
Species of Tetrancistrum which are found on both siganids and members of the acanthurid genus Naso (Lacépède), formed a distinct monophyletic group within the Ancyrocephalidae (Figs. 4 and 5). All of the Tetrancistrum spp. included in this analysis are collected from siganids; they were divided into two groups, one composed of T. sigani (syn. T. nebulosi) and T. labyrinthus and a separate group containing T. indicum. At the current state of knowledge, it is not clear what main characters are responsible for this phylogenetic division within the members of Tetrancistrum. Obtaining additional molecular data from other species of Tetrancistrum from siganid and nasoid hosts will help clarify the factors influencing the formation of these molecular divisions. The findings of the present study suggest that the SSU rDNA is a reliable marker to understand phylogenetic relationships within the Ancyrocephalidae.
The data obtained from the LSU rDNA region of the worms investigated in the present study revealed that G. safiensis n. sp. disclosed highest similarity to the sequence of Tetrancistrum sp. deposited by Mollaret et al. (1997). Since molecular divergence among species representing different genera is usually higher than that among congeneric species within the family (Wu et al., 2007), it can be suggested that the Tetrancistrum sp. analysed by Mollaret et al. (1997) is actually a species belonging to Glyphidohaptor rather than Tetrancistrum. However, re-evaluation of the species analysed by Mollaret (1997) will require confirmation before drawing any conclusions. Subsequently, the newly generated sequences of Tetrancistrum spp. were always clustering as a separate monophyletic group regardless of the type of dataset used in the molecular analyses. This observation supports the notion that Tetrancistrum sp. analysed by Mollaret et al. (1997) is a member of Glyphidohaptor, and most probably represents G. phractophallus since it is the only species reported from S. fuscescens to date. This suggestion requires further investigation and confirmation by morphological and molecular analysis of Glyphidohaptor and Tetrancistrum-like specimens from S. fuscescens off Australia.
In their study Stefani et al. (2012) observed no intraspecific variability of the partial SSU-ITS1 rDNA marker among individuals of G. plectocirra. Thus, the fact that the p-distance among individuals of G. plectocirra was negligible, supports that G. safiensis n. sp. with a p-distance of 0.010% and a difference of seven nucleotides, is genetically distant from G. plectocirra. The low p-distance value between these two species could be attributed the close geographical localities of these species.
References
Aloo, P. A., Anam, R. O., & Mwangi, J. N. (2004). Metazoan parasites of some commercially important fish along the Kenyan Coast. Western Indian Ocean Journal of Marine Science, 3, 71–78.
Baverstock, P. R., Fielke, R., Johnson, A. M., Bray, R. A., & Beveridge, I. (1991). Conflicting phylogenetic hypotheses for the parasitic platyhelminths tested by partial sequencing of 18S ribosomal RNA. International Journal for Parasitology, 21, 329–339.
Bullard, S. A., Womble, M. R., Maynarda, M. K., Orélis-Ribeiro, R., & Arias, C. R. (2015). Skin lesions on yellowfin tuna Thunnus albacares from Gulf of Mexico outer continental shelf: Morphological, molecular, and histological diagnosis of infection by a capsalid monogenoid. Parasitology International, 64, 609–621.
Campos, A., Cummings, M. P., Reyes, J. L., & Laclette, J. P. (1998). Phylogenetic relationships of Platyhelminthes based on 18S ribosomal gene sequences. Molecular Phylogenetics and Evolution, 10, 1–10.
Dang, B. T., Levsen, A., Schander, C., & Bristow, G. A. (2010). Some Haliotrema (Monogenea: Dactylogyridae) from cultured grouper (Epinephelus spp.) with emphasis on the phylogenetic position of Haliotrema cromileptis. Journal of Parasitology, 96, 30–39.
Desdevises, Y., Jovelin, R., Jousson, O., & Morand, S. (2000). Comparison of ribosomal DNA sequences of Lamellodiscus spp. (Monogenea, Diplectanidae) parasitising Pagellus (Sparidae, Teleostei) in the North Mediterranean Sea: species divergence and coevolutionary interactions. International Journal for Parasitology, 30, 741–746.
Diamant, A., & Paperna, I. (1986). The parasites of wild Red-Sea rabbitfish (Siganus spp.) (Teleostei: Perciformes) as potential pathogens in mariculture. In: Vivares, C. P., Bonami, J.-R. & Jaspers, E. (Eds), Pathology in marine aquaculture. European Aquaculture Society Special Publication 9, pp. 1–83.
Ernst, I., Jones, M. K., & Whittington, I. D. (2010). A new genus of viviparous gyrodactylid (Monogenea) from the Great Barrier Reef, Australia with descriptions of seven new species. Journal of Natural History, 35, 313–340.
Freeman, M. A., Kasper, J. M., & Kristmundsson, A. (2013). Nucleospora cyclopteri n. sp., an intranuclear microsporidian infecting wild lumpfish, Cyclopterus lumpus L., in Icelandic waters. Parasites & Vectors, 6, 49.
Freeman, M. A., & Ogawa, K. (2010). Variation in the small subunit ribosomal DNA confirms that Udonella (Monogenea: Udonellidae) is a species-rich group. International Journal for Parasitology, 40, 255–264.
Froese, R., & Pauly, D. (Eds) (2018). FishBase. World Wide Web electronic publication. www.fishbase.org, version (02/2018).
Geets, A., Coene, H., & Ollevier, F. (1997). Ectoparasites of the whitespotted rabbitfish, Siganus sutor (Valenciennes, 1835) off the Kenyan Coast: distribution within the host population and site selection on the gills. Parasitology, 115, 69–79.
Geets, A., & Ollevier, F. (1996). Endoparasitic helminthes of the whitespotted rabbitfish (Siganus sutor (Valenciennes, 1835) of the Kenyan coast: distribution within the host population and microhabitat use. Belgian Journal of Zoology, 126, 21–36.
Goto, S., & Kikuchi, H. (1917). Two new trematodes of the family Gyrodactylidae. Journal of the College of Science, Imperial University of Tokyo, 39, 1–22.
Gussev, A.V. (1985). Key to parasites of the freshwater fish fauna of the USSR, vol. 2. In: Bauer O. N. (Ed.). Leningrad: Nauka (In Russian).
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Journal of Nucleic Acids Symposium Series, 41, 95–98.
Hassanine, R. M. E., & Al-Jahdali, M. O. (2007). Ecological comments on the intestinal helminths of the rabbitfish Siganus rivulatus (Teleostei, Siganidae) from the northern Red Sea. Acta Parasitologica, 52, 278–285.
ICZN (2012). International Commission on Zoological Nomenclature: Amendment of articles 8, 9, 10, 21 and 78 of the International Code of Zoological Nomenclature to expand and refine methods of publication. Bulletin of Zoological Nomenclature, 69, 161–169.
Justine, J. L., Jovelin, R., Neifar, L., Mollaret, I., Lim, S. L. H., Sherman, S. H., et al. (2002). Phylogenetic positions of the Bothitrematidae and Neocalceostomatidae (Monopisthocotylean monogeneans) inferred from 28S rDNA sequences. Comparative Parasitology, 69, 2–25.
Kmentová, N., Gelnar, M., Koblmüller, S., & Vanhove, M. P. M. (2015). First insights into the diversity of gill monogeneans of Gnathochromis and Limnochromis (Teleostei, Cichlidae) in Burundi: do the parasites mirror host ecology and phylogenetic history? PeerJ, 4, e1629.
Kritsky, D. C., & Galli, P. (2007). Dactylogyrids (Monogenoidea) parasitizing the gills of spinefoots (Teleostei: Siganidae): Revision of Pseudohaliotrema, with redescription of P. sphincteroporus and P. molnari from the Great Barrier Reef, Australia. Comparative Parasitology, 74, 9–22.
Kritsky, D. C., Galli, P., & Tingbao, Y. (2007a). Dactylogyrids (Monogenoidea) parasitizing the gills of spinefoots (Teleostei: Siganidae): Proposal of Glyphidohaptor n. gen., with two new species from the Great Barrier Reef, Australia, and G. plectocirra n. comb. from Ras Mohammed National Park, Egypt. Journal of Parasitology, 93, 39–46.
Kritsky, D. C., Galli, P., & Tingbao, Y. (2007b). Dactylogyrids (Monogenoidea) parasitizing the gills of spinefoots (Teleostei, Siganidae): revision of Tetrancistrum Goto & Kikuchi, with descriptions of two new species from Siganus spp. of the Red Sea and Celebes. Journal of Natural History, 41, 1513–1551.
Kumar, S., Stecher, G., & Tamura, K. (2015). MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33, 1870–1874.
Lim, L. H. S. (2002). Three new species of Pseudohaliotrema Yamaguti, 1953 (Monogenea: Ancyrocephalidae) from Siganus species (Siganidae) and the description of a mechanism for cross-insemination. Journal of Natural History, 36, 1639–1660.
Littlewood, D. T. J., Bray, R. A., & Clough, K. A. (1999). A phylogeny of the Platyhelminthes: towards a total-evidence solution. Hydriobiologia, 383, 155–160.
Littlewood, D. T. J., & Olson, P. D. (2001). Small subunit rDNA and the Platyhelminthes: Signal, noise, conflict and compromise. In: Littlewood, D. T. J. & Bray, R. A. (Eds), Interrelationships of the Platyhelminthes. London: Taylor & Francis, pp. 262–278.
Marchiori, N. C., Pariselle, A., Pereira, J., Agnèse, J. F., Durand, J. D., & Vanhove, M. P. M. (2015). A comparative study of Ligophorus uruguayense and L. saladensis (Monogenea: Ancyrocephalidae) from Mugil liza (Teleostei: Mugilidae) in southern Brazil. Folia Parasitologica, 62, 024.
Martens, E., & Moens, J. (1995). The metazoan ecto- and endoparasites of the rabbitfish, Siganus sutor (Cuvier & Valenciennes, 1835) of the Kenyan coast. I. African Journal of Ecology, 33, 405–416.
Mendlová, M., Pariselle, A., Vyskočilová, M., & Šimková, A. (2010). Molecular phylogeny of monogeneans parasitizing African freshwater Cichlidae inferred from LSU rDNA sequences. Parasitology Research, 107, 1405–1413.
Mendoza-Palmero, C., Blasco-Costa, I., & Scholz, T. (2015). Molecular phylogeny of Neotropical monogeneans (Platyhelminthes: Monogenea) from catfishes (Siluriformes). Parasites & Vectors, 8, 164.
Mollaret, I., Jamieson, B. G. M., Adlard, R. D., Hugall, A., Lecointre, G., Chombard, C., et al. (1997). Phylogenetic analysis of the Monogenea and their relationships with Digenea and Eucestoda inferred from 28S rDNA sequences. Molecular and Biochemical Parasitology, 90, 433–438.
Mollaret, I., Lim, L. H. S., & Justine, J. L. (2000). Phylogenetic position of the monogeneans Sundanonchus, Thaparocleidus, and Cichlidogyrus inferred from 28S rDNA sequences. International Journal for Parasitology, 30, 659–662.
Olson, P. D., & Littlewood, D. T. J. (2002). Phylogenetics of the Monogenea – evidence from a medley of molecules. International Journal for Parasitology, 32, 233–244.
Paperna, I. (1972). Monogenea from Red Sea Fishes. I. Monogenea of fish of the genus Siganus. Proceedings of the Helminthological Society of Washington, 39, 33–39.
Plaisance, L., Littlewood, D. T. J., Olson, P. O., & Morand, S. (2005). Molecular phylogeny of gill monogeneans (Platyhelminthes, Monogenea, Dactylogyridae) and colonization of Indo-West Pacific butterflyfish hosts (Perciformes, Chaetodontidae). Zoologica Scripta, 34, 425–436.
Sailaja, B., & Madhavi, R. (2011). Polylabris bengalensis sp. nov. (Monogenea, Microcotylidae) from siganid fishes of the Visakhapatnam coast, Bay of Bengal. India. Acta Parasitologica, 56, 290–295.
Shih, H., & Jeng, M. (2002). Hysterothylacium aduncum (Nematoda: Anisakidae) infecting an herbivorous fish, Siganus fuscescens, off the Taiwanese coast of the Northwest Pacific. Zoological Studies, 41, 208–215.
Šimková, A., Morand, S., Jobet, E., Gelnar, M., & Verneau, O. (2004). Molecular phylogeny of congeneric monogenean parasites (Dactylogyrus): A case of intrahost speciation. Evolution, 58, 1001–1018.
Šimková, A., Plaisance, L., Matejusova, I., Morand, I., & Verneau, O. (2003). Phylogenetic relationships of the Dactylogyridae Bychowsky, 1933 (Monogenea: Dactylogyridea): the need for the systematic revision of the Ancyrocephalinae Bychowsky, 1937. Systematic Parasitology, 54, 1–11.
Šimková, A., Serbielle, C., Pariselle, A., Vanhove, M. P. M., & Morand, S. (2013). Speciation in Thaparocleidus (Monogenea: Dactylogyridae) parasitizing Asian pangasiid catfishes. BioMed Research International, 2013, 353956.
Soo, O. Y. M., Tan, W. B., & Lim, L. H. S. (2015). Three new species of Ligophorus Euzet & Suriano, 1977 (Monogenea: Ancyrocephalidae) from Moolgarda buchanani (Bleeker) off Johor, Malaysia based on morphological, morphometric and molecular data. Raffles Bulletin of Zoology, 63, 49–65.
Stefani, F., Aquaro, G., Azzurro, E., Colorni, A., & Galli, P. (2012). Patterns of genetic variation of a Lessepsian parasite. Biological Invasions, 14, 1725–1736.
Theisen, S., Palm, H. W., Al-Jufaili, S. H., & Kleinertz, S. (2017). Pseudempleurosoma haywardi sp. nov. (Monogenea: Ancyrocephalidae (sensu lato) Bychowsky & Nagibina, 1968): An endoparasite of croakers (Teleostei: Sciaenidae) from Indonesia. PLoS ONE, 12, e0184376.
Theisen, S., Palm, H. W., Hendrik, S., Al-Jufaili, S. H., & Kleinertz, S. (2018). Endoparasitic Paradiplectanotrema klimpeli sp. nov. (Monogenea: Ancyrocephalidae) from the greater lizardfish Saurida tumbil (Teleostei: Synodontidae) in Indonesia. Parasitology Open, 4, e13.
Thompson, J. D., Higgings, D. G., & Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research, 22, 4673–4680.
Tingbao, Y., Kritsky, D. C., & Jun, P. (2007). Polylabris lingaoensis sp. n. and Polylabris cf. mamaevi Ogawa et Egusa, 1980 (Monogenoidea: Microcotylidae) from perciform fishes in the Gulf of Tonkin, South China Sea. Folia Parasitologica, 54, 27–33.
Vignon, M., & Sasal, P. (2010). The use of geometric morphometrics in understanding shape variability of sclerotized haptoral structures of monogeneans (Platyhelminthes) with insights into biogeographic variability. Parasitology International, 59, 183–191.
Wu, X. Y., Li, A. X., Zhu, X. Q., & Xie, M. Q. (2005). Description of Pseudorhabdosynochus seabassi sp. n. (Monogenea: Diplectanidae) from Lates calcarifer and revision of the phylogenetic position of Diplectanum grouperi (Monogenea: Diplectanidae) based on rDNA sequence data. Folia Parasitologica, 52, 231–240.
Wu, X. Y., Zhu, X. Q., Xie, M. Q., & Li, A. X. (2007). The evaluation for generic-level monophyly of Ancyrocephalinae (Monogenea, Dactylogyridae) using ribosomal DNA sequence data. Molecular Phylogenetics and Evolution, 44, 530–544.
Young, P. C. (1967). Some species of the genus Tetrancistrum Goto and Kikuchi, 1917 (Monogenoidea: Dactylogyridae). Journal of Parasitology, 53, 1016–1022.
Acknowledgements
We are grateful to the Ministry of Agriculture and Fisheries Wealth, Sultanate of Oman, for the TRC Project ORG-EBR_11-TRC grants, to Laura Machkevska for her help with the illustrations for this paper. Also, we would like to extend our thanks to Professor Kazuo Ogawa (Meguro Parasitological Museum, Tokyo, Japan), Mrs Eileen Harris (Natural History Museum, London, UK), Mrs Patricia Pilitt, (US National Parasite Collection, Beltsville, USA), Dr Mal Bryant (Queensland Museum, Australia) for the loan of voucher specimens and worm micrographs. In addition, we would like to thank Dr Juan Luis Ribas of the Servicio de Microscopía Centro de Investigación, Tecnología e Innovación at the Universidad de Sevilla, Spain, for his outmost help and kindness with obtaining the confocal microscopy images.
Funding
This project was supported by The Research Council of Sultanate of Oman (ORG-EBR_11-TRC).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All applicable institutional, national and international guidelines for the care and use of animals were followed.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article was registered in the Official Register of Zoological Nomenclature (ZooBank) as E2CC7FE8-BEE0-482E-AB48-7590C25E1B6F. This article was published as an Online First article on the online publication date shown on this page. The article should be cited by using the doi number. This is the Version of Record.
This article is part of the Topical Collection Monogenea.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Al Jufaili, S.H., Machkevsky, V.K., Al Kindi, U.H. et al. Glyphidohaptor safiensis n. sp. (Monogenea: Ancyrocephalidae) from the white-spotted rabbitfish Siganus canaliculatus (Park) (Perciformes: Siganidae) off Oman, with notes on its phylogenetic position within the Ancyrocephalidae Bychowsky & Nagibina, 1968 (sensu lato). Syst Parasitol 97, 727–741 (2020). https://doi.org/10.1007/s11230-020-09949-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11230-020-09949-x