Abstract
Members of the genus Clinostomum Leidy, 1856, colloquially known as yellow grubs, are cosmopolitan parasites of piscivorous birds, freshwater snails, fish and amphibians. In the southeastern United States, piscivorous birds present a continuous challenge for producers of farm-raised catfish. Ciconiiform birds are common hosts of Clinostomum spp. in North America and are endemic on most commercial catfish operations. The great egret Ardea alba L. is an avian predator often found foraging on commercial catfish operations, but to date the trematode fauna of great egrets preying on catfish ponds remains mostly understudied. Thirteen great egrets were captured from commercial catfish ponds in northeast Mississippi, and examined for trematode infections. Two morphologically distinct Clinostomum spp. were observed in the great egrets sampled, one morphologically consistent with Clinostomum marginatum (Rudolphi, 1819) and one morphologically unique species. These morphological descriptions were supplemented with molecular sequence data (c.4,800 bp of ribosomal DNA and c.600 bp of mitochondrial DNA). Gene sequences confirmed the identification of C. marginatum. However, the second species differed significantly from its congeners in both morphology and DNA sequence. Given these distinct morphological and molecular characters we propose this second species as Clinostomum album n. sp.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Digenetic trematodes of the genus Clinostomum Leidy, 1856 are widely distributed parasites of piscivorous birds, molluscs, fishes and amphibians (Kanev et al., 2002). Adults are found in the oral cavity or oesophagus of the bird hosts, the cercariae develop in freshwater snails and metacercariae encyst in freshwater fish or amphibians (Dias et al., 2003). The debate on the number of species is ongoing, but contemporary studies investigating the genetic variation between and among species have revealed new insights into the taxonomy of the genus (Locke et al., 2015). As with other groups of digeneans, discriminatory morphological characters that differentiate closely related species are lacking. This has led to an underestimation of true species richness, which was revealed primarily through molecular DNA sequencing (Caffara et al., 2011; Locke et al., 2015; Rosser et al., 2016a). Supplemental molecular data coupled with detailed morphological descriptions have facilitated the identification of new species and offer more sound support of amended descriptions of established species. Genetic markers typically used to distinguish between species include ribosomal (e.g. internal transcribed spacer regions) and mitochondrial (e.g. cytochrome c oxidase subunit 1) genes (Caffara et al., 2011; Gustinelli et al., 2010; Sereno-Uribe et al., 2013; Locke et al., 2015).
The great egret Ardea alba L. (Pelecaniformes: Ardeidae) is a species of piscivorous bird ranging throughout the USA, southern Canada, Central America and South America. Given the frequent occurrence of the great egret on commercial catfish operations in the southeastern USA, the great egret is widely considered a nuisance species in catfish aquaculture (Glahn & King, 2004). While the effect of predatory foraging by great egrets on catfish aquaculture has been documented (Glahn et al., 1999; Werner et al., 2001), the impact of the trematodes they introduce to catfish production systems is largely understudied.
In North America the number of Clinostomum species continues to expand. Clinostomum heluans Braun, 1899 was reported in the great blue heron Area herodias L. and Clinostomum intermedialis Lamont, 1920 in the Brandt’s cormorant Phalacrocorax penicillatus Brandt from Mexico (Bravo-Hollis, 1947). In a survey of the helminth parasites of great egrets in Florida, USA, two species of Clinostomum were reported, Clinostomum attenuatum Cort, 1913 and Clinostomum complanatum (Rudolphi, 1814) (see Sepúlveda et al., 1999). Overstreet & Curran (2004) reported Clinostomum marginatum (Rudolphi, 1819) from herons, egrets and catfish obtained from production ponds in Louisiana and Mississippi, USA. In Mexico, C. complanatum has been found in great egrets (Violante-González et al., 2012); however recently Sereno-Uribe et al. (2013) suggested that previous records of C. complanatum in Mexico are likely C. marginatum or the more recently recognized Clinostomum tataxumui Sereno-Uribe, Pinacho-Pinacho, García-Varela & Pérez-Ponce de León, 2013 from great egret, great blue heron, and bare-throated tiger heron Tigrisoma mexicanum Swainson. Furthermore, Caffara et al. (2011) combined morphological and molecular data to differentiate adult and metacercaria stages of C. complanatum and C. marginatum and concluded that C. complanatum is the “European” species and is not present in the Americas.
A recent collection of great egrets from catfish production operations in the northeastern area of Mississippi was evaluated for Clinostomum spp. infection and a novel species is described herein.
Materials and methods
Trematode collection and morphological characterisation
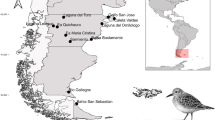
Thirteen great egrets were collected from commercial catfish operations in Noxubee County, Mississippi using soft catch leg hold traps and euthanized using CO2. Immediately following euthanasia, the oral cavity and sublingual area were inspected for adult Clinostomum spp. These were removed manually with sterile featherweight forceps (BioQuip Products, Rancho Dominguez, California) and placed in 0.09% sterile saline. The oesophagus and trachea were separated and opened longitudinally, the contents emptied into a 38-µm aperture brass sieve and washed with dechlorinated water. The intestinal lining was scraped manually and the contents rinsed onto the screen. The entire screen contents were then examined in a lined Petri dish under a dissecting stereomicroscope (Olympus SZ60, Olympus Optical Co. Ltd., Tokyo, Japan). Remaining intestinal contents were removed and examined for additional trematodes. All Clinostomum spp. adults were washed into containers with 0.09% saline.
Adult trematodes were relaxed in slightly boiling saline and fixed in 70% ethanol. A subsample of each suspected species was stained with acetocarmine for at least 5 h, destained in 1% acidic ethanol, and rinsed in increasing concentrations of ethanol (70–100%) for at least 1 h each. Specimens were cleared in Hemo-De (Fisher Scientific, Pittsburgh, Pennsylvania, USA) and mounted on clean glass microscope slides using Permount™ Mounting Medium (Fisher Scientific, Pittsburgh, Pennsylvania, USA). Representative specimens were submitted to the Smithsonian Institution, National Museum of Natural History, Washington, DC, USA under accession numbers: USNM 1422013–1422018. Line drawings of each species were made with the aid of a camera lucida and digitized using Adobe Illustrator CC 2014 (Adobe, San Jose, California). Photomicrographs of adult specimens were captured using an Olympus DP72 digital camera and DP-2-Twain/cellSens software (Olympus Optical Co. Ltd., Tokyo, Japan). Morphological characteristics of the collected Clinostomum spp. were compared to other species within the genus (Caffara et al., 2011). Measurements are presented as the range followed by the mean in parentheses and are reported in micrometres.
DNA extraction and molecular characterisation
Genomic DNA was extracted from three adult specimens of each species with the DNeasy Blood and Tissue Kit (QIAGEN, Valencia, California, USA). Approximately 4,800 bp of ribosomal DNA, including the 18S rRNA gene, ITS1 region, 5.8S rRNA gene, ITS2 region, and partial 28S rRNA gene, was amplified by polymerase chain reaction (PCR) from one representative adult for each species. To identify the isolate to a lower taxonomic level, an approximately 600 bp sequence of the cytochrome c oxidase subunit 1 (cox1) gene was amplified for all adult specimens from both species. Primers used for each region are listed in Table 1. Briefly, each PCR reaction contained 22 µl of Platinum® PCR Supermix (Invitrogen, Carlsbad, California, USA), 10 pmol of forward and reverse primer, and 1 µl of gDNA (c.15 ng/µl) as template. For Barker3/Barker4 primers the thermal cycling program consisted of 94°C for 3 min, 35 cycles of 94°C for 30 s, 50°C for 30 s, followed by 72°C for 1 min. Parameters for the 1F/5R, BD1/BD2, and LSU5/1500R primer combinations were similar, but employed an annealing temperature of 45°C and an elongation step of 1 min 30 s. Likewise, the PCR thermal cycling program for Diplo1795F/Diplo2549R, Diplo2617F/Diplo3170R, 28S 3431F/28S 4779R, and 28S 4759F/28S 5699R primer combinations used previous parameters with an annealing temperature of 55°C. Finally, the thermal cycling protocol for the cox1_schist 5′/acox650r primer set was the same as above, but used a 45°C annealing temperature. Amplification products were electrophoresed through 0.8% agarose gels stained with ethidium bromide (0.5 µg/ml) and visualized under ultraviolet fluorescent light. Each gel was run concurrently with a molecular weight ladder (HyperLadder™ 50 bp, Bioline, London, UK) to confirm the presence of appropriate sized bands.
Amplicons were excised and purified using the QIAquick Gel Extraction Kit (QIAGEN Inc., Valencia, California, USA) and sequenced commercially (Eurofins MWG Operon LLC, Huntsville, Alabama, USA) using the same forward and reverse primers used to generate the amplicons. Ambiguous base calls were annotated manually from respective chromatograms in SeqMan™ (DNAStar, Madison, Wisconsin, USA). The contiguous rRNA and cox1 gene sequences for each species were compared to other sequenced Clinostomum species by a Blastn search of the National Center for Biotechnology Information non-redundant nucleotide database (NCBI nr/nt) (Altschul et al., 1990).
Published cox1 gene sequences from the genus Clinostomum available in the NCBI nr/nt database were downloaded and ClustalW aligned and trimmed in MEGA6 (Tamura et al., 2013). The final dataset contained a total of 427 positions across 88 sequences. Accession numbers for sequences used in phylogenetic analysis are provided in Supplementary Table S1. The best-fit nucleotide substitution model for phylogenetic analysis was determined using the Bayesian Information Criterion as the Hasegawa Kishino-Yano (HKY) model including gamma distribution site variation (Nei & Kumar, 2000). Bayesian inference analysis was performed in MrBayes 3.2.6 with Markov chain Monte Carlo searches of two simultaneous runs of four chains. Chain sampling occurred every 100th tree over 10,000,000 generations (Ronquist & Huelsenbeck, 2003) and the first 25% were discarded as ‘burn-in’ with the posterior probabilities calculated from the remaining trees. The consensus tree was visualized in FigTree 1.4.2 (Rambaut, 2014) and annotated in Adobe Illustrator (Adobe, San Jose, California, USA). Pairwise distances were calculated in MEGA6 based on the alignment of the two Clinostomum spp. encountered in this study with those used to construct the phylogenetic tree.
Results
Two distinct species of Clinostomum, characterised morphologically and molecularly, were observed in the oral cavity and occasionally the oesophagus of 11/13 (overall prevalence of 85%) great egrets. Clinostomum marginatum was identified in 10/13 (prevalence of 77%) and a second morphologically and molecularly distinct species in 4/13 (prevalence of 31%) great egrets. No Clinostomum spp. were observed in the lower intestinal tracts of any bird.
Family Clinostomidae Lühe, 1901
Genus Clinostomum Leidy, 1856
Clinostomum album n. sp.
Type host: Great egret Ardea alba Linnaeus (Pelecaniformes: Ardeidae).
Type-locality: Noxubee County, Mississippi, USA.
Type-material: Holotype USNM 1422013 and 2 paratypes USNM 1422014–1422015 are deposited in the Smithsonian Institution, National Museum of Natural History, Washington, D.C., USA
Site in host: Oral cavity (sublingual) and oesophagus.
Infection parameters: Prevalence: 31% (4 out of 13 birds); abundance: range 0–6, mean 0.9 worms per bird; mean intensity 3.0 worms per infected bird.
Representative DNA sequences: GenBank KU708008 (ribosomal genes) and KU708010 (cox1).
Etymology: The specific epithet is in reference to the host specific name.
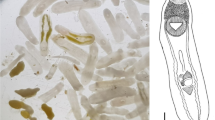
Description (Figs. 1A, 2A)
[Based on the holotype and 5 paratypes. All measurements were taken from stained and mounted gravid adult worms.] Body, linguiform, slender anterior region, widest at level of gonads, 4,402–5,929 × 969–1,108 (5,269 × 1,047). Anterior extremity with distinct oral collar-like fold typical of the genus, 357–507 × 526–690 (428 × 617). Oral sucker small, 207–307 × 234–344 (256 × 263). Pharynx present, intestine bifurcates just posterior to oral sucker and intestinal caeca laterally extend almost to posterior of body. Ventral sucker, large, located in lower anterior third of the body, 480–648 × 485–610 (578 × 560). Distance between oral and ventral sucker 360–827 (656).
Testes, tandem, located in upper region of posterior third of body. Anterior testis triangular, lobed, laterally compressed by cirrus-sac on right margin, 266–469 × 449–535 (375 × 488). Posterior testis larger, triangular, lobed, 313–473 × 416–571 (425 × 501). Distance between testes 264–354 (298). Cirrus-sac, laterodorsally surrounds right margin of anterior testis, 140–307 × 117–162 (222 × 146).
Ovary small, intertesticular, dextral, ovoid, 201–281 × 180–254 (235 × 199). Uterine duct intracaecal, extending anteriorly along left margin of anterior testis before opening into uterine sac. Uterine sac filled with eggs, occupies lower field between ventral sucker and anterior testis. Metraterm muscular, leads into uterus close to genital atrium. Genital pore pretesticular. Vitelline fields restricted mostly to lateral margins of body, begin just posterior to ventral sucker and extend to posterior extremity of body. Eggs yellowish, 90–108 × 53–67 (100 × 61), on average 70–82 (75) in number, often located within uterine sac, uterine duct, and oötype region.
Molecular data
Molecular analysis of c.4,800 bp of ribosomal DNA from a single adult showed a high level of conservation between the two species collected in this study, as they were 99.4% (4,801/4,832 bp) similar across the five ribosomal targets examined. When compared to other sequences deposited in the GenBank database, C. album n. sp. shared 99.6% (1,952/1,960) sequence similarity at the 18S rRNA gene with an unpublished sequence of C. marginatum from North America (AY245760). Additionally, C. album n. sp. shared 96.5–97.2% sequence similarity with C. complanatum (AY245701 and FJ609420; Dzikowski et al., 2004; Gustinelli et al., 2010), Clinostomum cutaneum Paperna, 1964 (GQ339114 and FJ609421; Gustinelli et al., 2010), and Clinostomum phalacrocoracis Dubois, 1931 (FJ609422–FJ609423; Gustinelli et al., 2010) across partial 18S rDNA, complete ITS1, 5.8S rDNA, ITS2, and partial 28S rDNA sequences.
A 604 bp sequence of the mitochondrial cox1 gene from three individual adults was identical for all three specimens and revealed that C. album n. sp. shared limited sequence similarity with any Clinostomum spp. in the GenBank database. The highest sequence similarity was with C. detruncatum at 85.3–85.4% (KP110517–KP110519; Locke et al., 2015). Clinostomum album n. sp. shared 85.1% sequence similarity with C. attenuatum (KP150305–KP150306; Locke et al., 2015), and <85% sequence similarity with two unidentified species of Clinostomum, designated as Clinostomum sp. 4 from Apistogramma sp. Regan in Peru (KP110531; Locke et al., 2015) and Clinostomum sp. from Rana clamitans Latreille and Rana pipiens Schreber in Canada (JF718587 & JF718585; Caffara et al., 2011). Similarly, C. album n. sp. shared only 82.9–84.9% sequence similarity with isolates of C. marginatum available in the GenBank database and from great egrets in this study.
Remarks
Morphologically C. album n. sp. was distinct from C. marginatum infecting the great egrets examined in this study. Although similar in mean length, C. album n. sp. was considerably narrower than C. marginatum (1,047 vs 1,562 µm). Furthermore the oral sucker, ventral sucker, testes, and cirrus-sac of C. album n. sp. were all smaller on average when compared to those of C. marginatum observed in this study and in previous records of this species from North American Ardeidae (Caffara et al., 2011). Clinostomum album n. sp. also tended to have less diffuse vitelline follicles, usually confined to the lateral margins of the body compared to the more expansive vitelline follicles of C. marginatum. Eggs of C. album n. sp. are roughly the same size as C. marginatum, although overall body length and width are considerably smaller. Morphological data of the Clinostomum spp. of North America are presented in Table 2.
Clinostomum marginatum (Rudolphi, 1819)
Host: Great egret Ardea alba (Linnaeus) (Pelecaniformes: Ardeidae).
Locality: Noxubee County, Mississippi, USA.
Site in host: Oral cavity (sublingual) and oesophagus.
Infection parameters: Prevalence: 77% (in 10 out of 13 birds); abundance: range 0–29, mean 5 worms per bird; mean intensity 6.5 worms per infected bird.
Voucher material: Vouchers USNM 1422016–1422018 are deposited in the Smithsonian Institution, National Museum of Natural History, Washington, D.C., USA
Representative DNA sequences: GenBank KU708007 (ribosomal genes) and KU708009 (cox1).
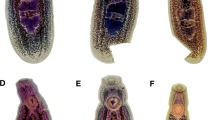
Description (Figs. 1B, 2B)
[Based on 7 stained and mounted gravid adult worms.] Body, stout, linguiform, 5,132–6,210 × 1,350–1,820 (5,697 × 1,562). Anterior end possessing oral collar-like fold typical of the genus, 432–793 × 742–963 (561 × 823), surrounding oral sucker. Oral sucker, small, 246–299 × 237–318 (268 × 267). Ventral sucker, large, located near anterior of the body, 550–694 × 589–677 (612 × 611). Intestinal caeca bifurcate immediately behind oral sucker and extend to the terminal end of the body. Distance between the oral and ventral sucker, 640–928 (770).
Testes, tandem, located toward the middle or upper portion of the posterior of the body. Anterior testis, triangular, lobed, 373–526 × 534–737 (453 × 667) and laterally compressed on the right margin by cirrus sac. Posterior testis, triangular, lobed, 319–589 × 569–826 (424 × 700). Distance between testes 252–378 (312). Cirrus sac, laterodorsally surrounds the right margin of the anterior testis, 316–544 × 154–298 (418 × 220).
Ovary, ovoid, intertesticular, dextral, 187–261 × 176–256 (236 × 217). Uterine duct lies at the level of the caecae and extending anteriorly along the left margin of the anterior testis before opening into the uterine sac. Uterine sac occupies almost the entire area of the body between the ventral sucker and anterior testis. Metraterm, muscular, joins the uterus close to the genital atrium. Genital pore, when observed, pretesticular. Vitelline follicles diffuse, concentrated in the lateral regions of the body and beginning at the level of the ventral sucker and extending to the end of the body. Eggs, yellow in color, 94–105 × 63–72 (101 × 68), 37–94 (68) eggs throughout the uterine ducts and sac, and also often obscuring the oötype region.
Remarks
Morphologically C. marginatum specimens identified in this study were consistent with those previously described for the species (Cort, 1913; Caffara et al., 2011). Clinostomum marginatum collected from great egrets in this study shared 98.9–100% sequence similarity at ribosomal genes available for C. marginatum in the GenBank database, with most archived sequences covering the ITS1, 5.8S rRNA, and ITS2 region. Additionally, C. marginatum shared 97.5–96.9% sequence similarity with > 4,500-bp of ribosomal DNA from C. cutaneum (GQ339114 & FJ609421; Gustinelli et al., 2010), C. complanatum (AY245701 & FJ609420; Dzikowski et al., 2004; Gustinelli et al., 2010), and C. phalacrocoracis (FJ609422–FJ609423; Gustinelli et al., 2010).
Moreover, the 612-bp cox1 sequence of three C. marginatum specimens was > 99% similar to isolates of C. marginatum in the GenBank database. The morphological description and limited interspecific variation at the cox1 gene support the identification as C. marginatum.
Cox 1 phylogeny of Clinostomum spp.
Genetic divergence of cox1 sequences (Table 3) of C. album n. sp. with other species of the genus ranged on average 15.29–19.86% (14.12–20.0%). Whereas, C. marginatum had minimal intraspecific genetic divergence at the cox1 sequence when compared to other isolates of C. marginatum. The genetic divergence of C. marginatum to other isolates of C. marginatum was 1.49% (0.47–8.71%). Bayesian inference based on cox1 sequences demonstrated distinct clustering of the Clinostomum marginatum from this study with other isolates obtained throughout North America (Fig. 3) and as sister taxa to C. attenuatum. Clinostomum album n. sp. was basally located within a clade containing C. tataxumui, Clinostomum sp. 5, Clinostomum sp. 2, Clinostomum sp. 1, Clinostomum sp. 3, and Clinostomum sp. 4. Topology of the tree was similar to previously published phylogenetic trees of cox1 sequences of clinostomes (Locke et al., 2015) and well supported. For an uncollapsed tree, see Supplementary Figure S1.
Discussion
Clinostomum species are cosmopolitan parasites of avian, mollusc and amphibian or fish hosts. These digeneans are of significant commercial and ecological importance as parasites of ecologically threatened species of amphibians, as well as wild and farm-raised fish (Paperna, 1991). In North America, six named species have been reported (C. attenuatum, C. complanatum, C. heluans, C. intermedialis, C. marginatum and C. tataxumui) from avian, mollusc, fish and amphibian hosts (Bravo-Hollis, 1947; Stuart et al., 1972; McAllister, 1990; Sepúlveda et al., 1994, 1996, 1999; Kinsella et al., 2004; Caffara et al., 2011; Sereno-Uribe et al., 2013). The larval stages of C. marginatum have been reported from hosts in commercial catfish ponds, marsh ramshorn snails Planorbella trivolvis (Say) and channel catfish Ictalurus punctatus (Rafinesque) (Lorio, 1989) collected from commercial catfish operations. Moreover, Overstreet & Curran (2004) reported C. marginatum in egrets and herons in the southeastern United States. Herein we report two species, C. marginatum and C. album n. sp., from great egrets foraging on catfish aquaculture operations in Mississippi.
Morphologically C. album n. sp. was distinct from other Clinostomum spp. reported from North American avian hosts. The isolate was consistently smaller across numerous features. Gonads were diminutive and placement of vitelline follicles was restricted to the lateral margins of the body rather than the more diffuse vitelline follicles of C. marginatum. Clinostomum spp. from South America and Mexico, specifically C. detruncatum and C. heluans differ considerably, not only in their much larger size, but also placement of the gonads at the posterior of the body (Bravo-Hollis, 1947; Travassos et al., 1969). In addition, eggs of C. album n. sp. were similar in size to C. marginatum, even though the body and other morphological features of C. album n. sp. are markedly smaller.
While morphological descriptions have been the basis of identification of digeneans over the past two centuries, molecular identification has afforded more precise differentiation of morphologically similar species (Caffara et al., 2011). While unremarkable at the c.4,800 bp rDNA region, C. album n. sp. was markedly divergent at the cox1 gene from all other species of Clinostomum available in the GenBank database. That said, the interspecific variability between C. album n. sp. and C. marginatum was consistent with intrageneric variability described for the genus (Caffara et al., 2011; Locke et al., 2015). Additionally, the limitation of ribosomal genes as the only molecular marker for species delimitation was exemplified in this study. The c.4,800-bp of C. album n. sp. ribosomal DNA demonstrated less than 0.7% divergence from C. marginatum, while demonstrating less than 4% divergence from other Clinostomum species in the GenBank database. These results are consistent with those reported by Gustinelli et al. (2010), where ribosomal DNA sequences of C. cutaneum were less than 3% divergent from other closely related species of Clinostomum. In order to fully appreciate the species richness of the Clinostomum, further sampling from avian, fish and mollusc hosts from different continents is needed, coupling detailed morphological descriptions with sequences from both conserved (ribosomal) and fast evolving genes (mitochondrial).
At present, the importance of C. album n. sp. as a pathogen of amphibians or fish is unclear as the intermediate hosts involved in the life-cycle are unknown. Clinostomum marginatum has been reported from farm-raised catfish in the southeastern United States, where infections may lead to unmarketable fish at processing. However, C. marginatum infections are rare and generally of little consequence to catfish aquaculture compared to other more damaging digeneans (Lorio, 1989; Wise et al., 2008; Griffin et al., 2012).
In catfish production ponds, marsh ramshorn snails serve as intermediate hosts for C. marginatum, and possibly other Clinostomum spp. (Hunter & Hunter, 1934; Lorio, 1989; Overstreet & Curran, 2004). In Brazil, planorbid snails in the genus Biomphalaria Preston serve as the first intermediate host for a Clinostomum sp., suggesting that other planorbid snails may be suitable hosts in the life-cycle of Clinostomum spp. (Pinto et al., 2015). In catfish production ponds in Mississippi, USA, there are at least two species of planorbid snail, namely P. trivolvis and Biomphalaria havanensis (L. Pfeiffer), that host digeneans infecting farmed catfish (Yost et al., 2009; Rosser et al., 2016a, b). The importance of B. havanensis as a first intermediate host in the life-cycle of Clinostomum spp. is currently unknown, but B. havanensis has been shown to host several genera of diplostomids including, Austrodiplostomum ostrowskiae Dronen, 2009, an uncharacterised Austrodiplostomum sp., Bolbophorus damnificus Overstreet, Curran, Pote, King, Blend & Grater 2002, Drepanocephalus auritus Kudlai, Kostadinova, Pulis & Tkach 2015, and an unidentified Tylodelphys sp. (Alberson et al. unpublished data; Rosser et al., 2016a, b).
Herein we report the clinostomid trematodes of great egrets collected from commercial catfish operations in the northeastern part of Mississippi, USA. Clinostomum album n. sp. represents the fourth named species of Clinostomum described in North America. Molecular sequencing data will allow further elucidation of life-cycle stages of C. album n. sp. as they are discovered. Additionally C. marginatum was observed and molecularly confirmed as a parasite of the great egret. The Clinostomum species of other piscivorous birds foraging from commercial catfish ponds and their effects on catfish production warrant further study.
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., & Lipman, D. J. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215, 403–410.
Barker, S. C., Blair, D., Cribb, T. H., & Tonion, K. (1993). Phylogenetic position of Heronimus mollis (Digenea): Evidence from 18S ribosomal RNA. International Journal for Parasitology, 23, 533–536.
Bonett, R. M., Steffen, M. A., Trujano-Alvarez, A. L., Martin, S. D., Bursey, C. R., & McAllister, C. T. (2011). Distribution, abundance, and genetic diversity of Clinostomum spp. metacercariae (Trematoda: Digenea) in a modified Ozark stream system. Journal of Parasitology, 97, 177–184.
Bravo-Hollis, M. (1947). Dos especies de Clinostomum (Trematoda), de aves procedentes del estado de Nuevo Leon, Mexico. Anales del Instituto de Biología, México, 18, 489–498.
Caffara, M., Bruni, G., Paoletti, C., Gustinelli, A., & Fioravanti, M. L. (2014a). Metacercariae of Clinostomum complanatum (Trematoda: Digenea) in European newts Triturus carnifex and Lissotriton vulgaris (Caudata: Salamandridae). Journal of Helminthology, 88, 278–285.
Caffara, M., Davidovich, N., Falk, R., Smirnov, M., Ofek, T., Cummings, D., Gustinellis, A., & Fioravanti, M. L. (2014b). Redescription of Clinostomum phalacrocoracis metacercariae (Digenea: Clinostomidae) in cichlids from Lake Kinneret, Israel. Parasite, 21, 32.
Caffara, M., Locke, S. A., Gustinelli, A., Marcogliese, D. J., & Fioravanti, M. L. (2011). Morphological and molecular differentiation of Clinostomum complanatum and Clinostomum marginatum (Digenea: Clinostomidae) metacercariae and adults. Journal of Parasitology, 97, 884–891.
Carranza, S., Baguña, J., & Riutort, M. (1997). Are the Platyhelminthes a monophyletic primitive group? An assessment using 18S rDNA sequences. Molecular Biology and Evolution, 14, 485–497.
Cort, W. W. (1913). Notes on the trematode genus Clinostomum. Transactions of the American Microscopical Society, 32, 169–182.
Dias, M. L. G. G., Eiras, J. C., Machado, M. H., Souza, G. T. R., & Pavanelli, G. C. (2003). The life cycle of Clinostomum complanatum Rudolphi, 1814 (Digenea, Clinostomidae) on the floodplain of the high Paraná River, Brazil. Parasitology Research, 89, 506–508.
Dzikowski, R., Levy, M. G., Poore, M. F., Flowers, J. R., & Paperna, I. (2004). Clinostomum complanatum and Clinostomum marginatum (Rudolphi, 1819) (Digenea: Clinostomidae) are separate species based on differences in ribosomal DNA. Journal of Parasitology, 90, 413–414.
Glahn, J. F., & King, D. T. (2004). Bird depredation. In: C. S. Tucker, C. S & Hargreaves, J. A. (Eds), Biology and culture of the channel catfish. Amsterdam, The Netherlands: Elsevier, pp. 503–529.
Glahn, J. F., Reinhold, D. S., & Smith, P. (1999). Wading bird depredations on channel catfish Ictalurus punctatus in northwest Mississippi. Journal of the World Aquaculture Society, 30, 107–114.
Griffin, M. J., Khoo, L. H., Quiniou, S. M., O’Hear, M. M., Pote, L. M., Greenway, T. E., & Wise, D. J. (2012). Genetic sequence data identifies the cercaria of Drepanocephalus spathans (Digenea: Echinostomatidae), a parasite of the double-crested cormorant (Phalacrocorax auritus), with notes on its pathology in juvenile channel catfish (Ictalurus punctatus). Journal of Parasitology, 98, 967–972.
Gustinelli, A., Caffara, M., Florio, D., Otachi, E. O., Wathuta, E. M., & Fioravanti, M. L. (2010). First description of the adult stage of Clinostomum cutaneum Paperna, 1964 (Digenea: Clinostomidae) from grey herons Ardea cinerea L. and a redescription of the metacercaria from the Nile tilapia Oreochromis niloticus niloticus (L.) in Kenya. Systematic Parasitology, 76, 39–51.
Hunter, G. W., & Hunter, W. S. (1934). The life cycle of the yellow grub of fish. Journal of Parasitology, 20, 325.
Hutton, R. F., & Sogandares-Bernal, F. (1960). Studies on the helminth parasites from the coast of Florida. II. Digenetic trematodes from shore birds of the west coast of Florida. I. Bulletin of Marine Science, 10, 40–54.
Kanev, I., Radev, V., & Fried, B. (2002). Family Clinostomidae Lühe, 1901. In: Gibson, D. I., Jones, A. & Bray, R. A. (Eds), Keys to the Trematoda, Vol. 1. Wallingford, UK: CAB International, pp. 113–120.
Kinsella, J. M., Spalding, M. G., & Forrester, D. J. (2004). Parasitic helminths of the American White Pelican, Pelecanus erythrorhynchos, from Florida, U.S.A. Comparative Parasitology, 71, 29–36.
Kudlai, O., Kostadinova, A., Pulis, E. E., & Tkach, V. V. (2015). A new species of Drepanocephalus Dietz, 1909 (Digenea: Echinostomatidae) from the double-crested cormorant Phalacrocorax auritus (Lesson) (Aves: Phalacrocoracidae) in North America. Systematic Parasitology, 90, 221–230.
Littlewood, D. T. J., Curini-Galletti, M., & Herniou, E. A. (2000). The interrelationships of Proseriata (Platyhelminthes: Seratia) flatworms tested with molecules and morphology. Molecular Phylogenetics and Evolution, 16, 449–466.
Locke, S. A., Caffara, M., Marcogliese, D. J., & Fioravanti, M. L. (2015). A large-scale molecular survey of Clinostomum (Digenea, Clinostomidae). Zoologica Scripta, 44, 203–217.
Lockyer, A. E., Olson, P. D., Ostergaard, P., Rollinson, D., Johnston, D. A., Attwood, S. W., et al. (2003). The phylogeny of the Schistosomatidae based on three genes with emphasis on the interrelationships of Schistosoma Weinland, 1858. Parasitology, 126, 203–224.
Lorio, W. J. (1989). Experimental control of metacercariae of the yellow grub Clinostomum marginatum in channel catfish. Journal of Aquatic Animal Health, 1, 269–271.
McAllister, C. T. (1990). Metacercaria of Clinostomum complanatum (Rudolphi, 1814) (Trematoda: Digenea) in a Texas Salamander, Eurycea neotenes (Amphibian: Caudata), with comments on C. marginatum (Rudolphi, 1819). Journal of the Helminthological Society of Washington, 57, 69–71.
Morgan, J. A. T., & Blair, D. (1995). Nuclear rDNA ITS sequence variation in the trematode genus Echinostoma: An aid to establishing relationships within the 37-collar-spine group. Parasitology, 111, 609–615.
Moszczynska, A., Locke, S. A., McLaughlin, D., Marcogliese, D. J., & Crease, T. J. (2009). Development of primers for the mitochondrial cytochrome c oxidase I gene in digenetic trematodes (Platyhelminthes) illustrates the challenge of barcoding parasitic helminths. Molecular Ecology Resources, 9, 75–82.
Nei, M., & Kumar, S. (2000). Molecular Evolution and Phylogenetics. New York: Oxford University Press.
Overstreet, R. M., & Curran, S. S. (2004). Defeating diplostomoid dangers in USA catfish aquaculture. Folia Parasitologica, 51, 153–165.
Paperna, I. (1991). Diseases caused by parasites in the aquaculture of warm water fish. Annual Review of Fish Diseases, 1, 155–194.
Pinto, H. A., Caffara, M., Fioravanti, M. L., & Melo, A. L. (2015). Experimental and molecular study of cercariae of Clinostomum sp. (Trematoda: Clinostomidae) from Biomphalaria spp. (Mollusca: Planorbidae) in Brazil. Journal of Parasitology, 101, 108–113.
Rambaut, A. (2014). FigTree: Tree Figure Drawing Tool v. 1.4.2. Institute of Evolutionary Biology, University of Edinburgh, http://tree.bio.ed.ac.uk/
Ronquist, F., & Huelsenbeck, J. P. (2003). MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19, 1571–1574.
Rosser, T. G., Alberson, N. R., Khoo, L. H., Woodyard, E. T., Pote, L. M., & Griffin, M. J. (2016a). Characterization of the life cycle of a fish eye fluke, Austrodiplostomum ostrowskiae (Digenea: Diplostomidae), with notes on two other diplostomids infecting Biomphalaria obstructa (Mollusca: Planorbidae) from catfish aquaculture ponds in Mississippi, USA. Journal of Parasitology, 102, 260–274.
Rosser, T. G., Alberson, N. R., Khoo, L. H., Woodyard, E. T., Wise, D. J., Pote, L. M., & Griffin, M. J. (2016b). Biomphalaria havanensis is a natural first intermediate host for the trematode Bolbophorus damnificus in commercial catfish production in Mississippi. North American Journal of Aquaculture, 78, 189–192.
Sepúlveda, M. S., Spalding, M. G., Kinsella, J. M., Bjork, R. D., & McLaughlin, G. S. (1994). Helminths of the roseate spoonbill, Ajaia ajaja, in southern Florida. Journal of the Helminthological Society of Washington, 61, 179–189.
Sepúlveda, M. S., Spalding, M. G., Kinsella, J. M., & Forrester, D. J. (1996). Parasitic helminths of the little blue heron, Egretta caerulea, in southern Florida. Journal of the Helminthological Society of Washington, 63, 136–140.
Sepúlveda, M. S., Spalding, M. G., Kinsella, J. M., & Forrester, D. J. (1999). Parasites of the great egret (Ardea albus) in Florida and a review of the helminthes reported for the species. Journal of the Helminthological Society of Washington, 66, 7–13.
Sereno-Uribe, A. L., Pinacho-Pinacho, C. D., García-Varela, M., & Pérez-Ponce de León, G. (2013). Using mitochondrial and ribosomal DNA sequences to test the taxonomic validity of Clinostomum complanatum Rudolphi, 1814 in fish-eating birds and freshwater fishes in Mexico, with the description of a new species. Parasitology Research, 112, 2855–2870.
Stuart, J. J., Dismukes, J. F., & Dixon, C. F. (1972). Endoparasites of the cattle egret (Bubulcus ibis) in Alabama. Journal of Parasitology, 58, 518.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., & Kumar, S. (2013). MEGA: Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biology and Evolution, 30, 2725–2729.
Travassos, L., Freitas, J. T., & Kohn, A. (1969). Trematódeos do Brasil. Memórias do Instituto Oswaldo Cruz, 67, 1–886.
Van Steenkiste, N., Locke, S. A., Castelin, M., Marcogliese, D. J., & Abbott, C. L. (2015). New primers for DNA barcoding of digeneans and cestodes (Platyhelminthes). Molecular Ecology Resources, 15, 945–952.
Violante-González, J., Monks, S., Gil-Guerrero, S., Rojas-Herrera, A. A., & Flores-Rodriguez, P. (2012). Helminth communities of two species of piscivorous birds, Ardea alba (Linnaeus) and Nyctanassa violacea (Gmelin) (Ciconiiformes: Ardeidae), in two coastal lagoons from Guerrero state, Mexico. Parasitology Research, 111, 309–315.
Werner, S. J., Tobin, M. E., & Fioranelli, P. B. (2001). Great egret preference for catfish size classes. Waterbirds: The International Journal of Waterbird Biology, 24, 381–385.
Wise, D. J., Hanson, T. R., & Tucker, C. S. (2008). Farm-level economic impacts of Bolbophorus infections of channel catfish. North American Journal of Aquaculture, 70, 382–387.
Yost, M. C., Pote, L. M., Wise, D. J., Dorr, B. S., & Richardson, T. D. (2009). Biomphalaria havanensis identified as a potential intermediate host for the digenetic trematode Bolbophorus damnificus. North American Journal of Aquaculture, 71, 10–15.
Acknowledgements
We would like to thank Katie Hanson-Dorr, Lanna Durst, Alex Crain, Lorelei Ford, and Raleigh Middleton for their assistance in capturing the egrets and during egret necropsies.
Funding
This work was supported by the National Institute of Food and Agriculture, U.S. Department of Agriculture, Project Number MIS-371530, the U.S. Department of Agriculture, Agricultural Research Service-Catfish Health Initiative, the Mississippi State University College of Veterinary Medicine, and the Mississippi Agriculture and Forestry Experiment Station (MAFES).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All applicable institutional, national and international guidelines for the care and use of animals were followed (IACUC QA 2458).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rosser, T.G., Alberson, N.R., Woodyard, E.T. et al. Clinostomum album n. sp. and Clinostomum marginatum (Rudolphi, 1819), parasites of the great egret Ardea alba L. from Mississippi, USA. Syst Parasitol 94, 35–49 (2017). https://doi.org/10.1007/s11230-016-9686-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11230-016-9686-0