Abstract
Ontology is a significant data model for identifying semantic information for Coronavirus disease (COVID-19) discovery. Large-scale biological and general datasets have recently been more readily available for study and to support the development of COVID-19 repositioning, but efficiently utilizing and standard metadata modeling of these datasets still remains challenging. Coronavirus disease is a deadly disease. Researchers have focused on the different aspects of COVID-19, such as studies about COVID-19, symptoms, treatment, prevention, resources, infected cases, patient information, global impact, and related research. Compilation and analysis of metadata by knowledge gathered in various field studies can help in efficient data sharing and better decision-making of data related to COVID-19. However, considering the heterogeneous nature of data sources, it is challenging and innovative to make the COVID-19 information human and machine understandable and to provide answers to user queries at all possible aspects of the COVID-19 pandemic. This paper propose a COVID-19 ontology named as CovidO metadata model that provides a common conceptual model to facilitate interoperability between metadata ontologies from various heterogeneous data sources. The CovidO has the following objectives: (1) The ontology serves as a reference schema for reporting COVID-19 data. (2) It also offer a customized schema of the existing ontologies to build a standard global data model. (3) It covers all the possible aspects/dimensions of COVID-19. (4) the proposed model aims to utilize findability, accessibility, interoperability, and reusability (FAIR) principles by providing common conceptualization metadata and thereby abstracting from heterogeneous structures of existing sources of (static) data. The experiments are conducted over CovidO using OOPS, OntoMetric, and RDF/SPARQL query to evaluate the efficiency of the proposed model. The result shows that the proposed model has a broad scope with fewer pitfalls than existing COVID-19 ontologies.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Novel coronavirus disease (COVID-19) is a global pandemic declared by the World Health Organization (WHO) as an outbreak of a Public Health Emergency of International Concern on 30 January 2020 [1]. Since March 11, 2020, various data sources like WHO (https://covid19.who.int), BNO News (https://bnonews.com), DXY.CN (https://dxy.cn), National Health Commission (NHC) (http://en.nhc.gov.cn), China CDC (CCDC) (https://chinacdc.cn/en/), Hong Kong Health Department (https://www.dh.gov.hk), Macau Government (https://www.gov.mo/en/), Government of India (https://www.mygov.in/covid-19), Indian Statistical Institute Bangalore (https://www.isibang.ac.in/~athreya/incovid19/), and Maryland department of health (https://coronavirus.maryland.gov/) described in literature have collected COVID-19 information. But data have been stored in such a way that interoperability and interlinking are difficult between the key data terms [2]. All data sources, however, provide a static representation of COVID-19 data. This is posing a significant problem for users to analyze and access semantic information [3]. The lack of consistent metadata makes it difficult to understand and process the correct information for both humans and machines [4].
COVID-19 includes studies about the biology and evolution of the virus SARS-CoV-2, pathogenesis, and epidemiology of the disease [5], but has created a global health crisis that has a profound impact on human life every day. It has an impact on slowing down the global economy, increasing the risk, creating new resources, etc. Therefore, it becomes necessary to examine and represent all possible aspects of information related to COVID-19 and take initiatives at both strategic and tactical levels. Ontology is a knowledge representation model that uses classes, relationships, properties, instances, and axioms to encode knowledge [6, 7]. With this information, we introduced a coronavirus disease ontology, “CovidO,” that provides semantic information about COVID-19 in all possible aspects. The ontology as a metadata model provides the solution to consistently access numerous heterogeneous data sources on the Web and accurately answer millions of user questions. So CovidO, like humans, has a reasoning capacity that allows it to infer hidden knowledge, data integration, sharing, re-indexing, and computer-assisted COVID-19 data analysis. As a result, CovidO provides the metadata to COVID-19 real-world data in various applications such as knowledge management, information extraction, decision making, recommender, disease analysis, treatment, searching, etc. The major contributions of the paper are listed below:
-
We provide dimensions for the COVID-19 pandemic which will highlight all the different aspects of COVID-19 knowledge.
-
An ontology development approach (ODA) has been proposed and discussed.
-
Development of a reference schema (CovidO) for use in reporting COVID-19-related data.
-
Evaluate the pitfall and schema metrics of CovidO ontology and compare it with existing ontologies to determine the efficiency, scope, and usability of ontology in the field of Covid-19 information.
We have found thirteen COVID-19-related ontologies or taxonomy that have been explained in a further section. All of these ontologies share a common objective to address COVID-19. But, they each differ because each discusses the different aspects of COVID-19 and has a different scope/dimension. For example, the diseases and treatment aspects including clinical test tracking are covered in infection disease ontology (IDO) [8], test report history, illness, symptoms, and medication are covered in virus infectious disease ontology (VIDO) [9], and clinical measurement and diagnosis are covered by Covid infectious disease ontology (CIDO) [10]. Similarly, CovidO traces other dimensions as well. To cover all aspects of COVID-19, we have grouped them and created seven dimensions D1 to D7 which cover all essential aspects of COVID-19 explained in Sect. 2.
This paper presents the extension of work [11] on CovidO version 1.0.0, https://w3id.org/CovidO consolidating Covid-19 concepts. To be independent of external ontologies, we defined a new namespace https://w3id.org/covido with the prefix Covido (registered entry at http://prex.cc for all classes used in the ontology. As a permanent URL service, we use w3id.org. The relevant conceptual design, competency questions, ontology, row data, and the results are publicly available to the community through a GitHub repository at https://github.com/sumitsnit/CovidO.
The paper is organized into the following sections: Section 2 describes the dimensions to determine the scope of the coronavirus in various aspects. The literature review on the existing ontologies related to Covid-19 is discussed in Sect. 3. Section 4 addresses the existing step-based ontologies development methodologies and based on that we categorize these steps into four general steps as a proposed ontology development approach to create any domain ontology. Based on the proposed approach, we are creating CovidO in Sect. 4. In Sect. 5, we have used verification and validation methods to evaluate the proposed CovidO ontology. Section 6 provides a summary of the work.
2 Dimension for COVID-19 scope
To allow stakeholders in the biological research community and application developers to reach out and benefit, CovidO has created a platform to link biological domains with real-word through specific dimensions. COVID-19 has many domains and subdomains; we group these domains into seven significant divisions that cover all the aspects of COVID-19-related knowledge. We created dimensions as a platform to allow stakeholders in the research community, including academic bibliographers, health workers, public authorities, and businesses, to more easily create and understand the context of different types of research objects through the relationships between these objects. These seven dimensions are shown and explained in Table 1. The CovidO ontology is developed based on these dimensions representing COVID-19 information in OWL format and other W3C standards utilized by other ontologies and software systems. The last column of Table 1 describes the core CovidO classes associated with a particular dimension.
Dimension:- D1: COVID-19 Disease, Symptom, and Treatment, represents all the information related to the COVID-19 disease. D2: Cases Information, provide statistics about the COVID-19 cases such as active, recovered, and deceased across the Geo-location (district, state, and country). D3: Research Domain, store information regarding the research, news, and social media post related to COVID-19. D4: Resources Utilized for COVID-19 monitor the COVID-19-related resources used and required by the patients. D5: Patient Information, track the medical history of patients, including their travel history, patient symptoms, and interpersonal relationship with other persons. D6: COVID-19 Related Events and Decisions represent the various exposure to COVID-19 and the order and advisory issued by the public authority. D7: The Impact of Coronavirus covers the implications of COVID-19 on the person and organization.
3 Review on COVID-19 ontologies
This section highlights the relevant literature that we undertook before creating CovidO. We have discussed the existing ontologies and their approach to creating the ontology. We have found 13 ontologies related to COVID -19 which represent the different scopes of COVID-19. COVID-19 ontologies have been explained as follows:
O1: Infectious Disease Ontology (IDO) IDO [8] Core is designed to be a disease and pathogen-neutral ontology, covering just those types of entities and relations that are relevant to infectious diseases generally. IDO is an inter-operable ontology that contains the domain information about an infectious disease where entities are related to the clinical and biomedical aspects of the disease.
O2: Coronavirus Infectious Disease Ontology (CIDO) CIDO [10] extends the IDO to develop a coronavirus-specific ontology, which makes it a more generalized ontology as compared to VIDO. CIDO familiarizes eight areas (coronavirus diseases, including etiology, transmission, epidemiology, pathogenesis, diagnosis, prevention, and treatment) that basically belong to the coronavirus infectious disease domain.
O3: Virus Infectious Disease Ontology (VIDO) VIDO [9] solves some of the most prevalent ontological issues seen in virus ontologies. In particular, VIDO inherits the entities from the IDO by adding the term ’virus’ to create a subclass, and the logical and textual information about the classes are adjusted accordingly. VIDO also uses entities that follow the OBO Foundry standards starting with the Basic Formal Ontology and encompassing epidemiology, classification, pathophysiology, and treatment of terminology used by virologists such as virus, prion, satellite, viroid, and so on.
O4: Covid-19 Infectious Disease Ontology (IDO-Covid-19) In IDO-Covid-19 [12], the acellular structure was added to IDO core to cover viruses and other acellular entities investigated by virologists. IDO-Covid-19 extends VIDO and CIDO, and IDO-core. It follows OBO Foundry criteria, start with the Basic Formal Ontology, and covers epidemiology, categorization, pathophysiology, and treatment of terminology used to describe SARS-CoV-2 virus infection and the related COVID-19 disease.
O5: CIDO-COVID-19 CIDO-COVID-19 [13] is a COVID-19 ontology that includes disease, diagnosis, etiology, virus, transmission, symptoms, treatment, medications, and prevention. CIDO-COVID-19 expands and improves on previous CIDO concepts, particularly regarding COVID-19 diagnosis and treatment. CIDO-COVID-19 intends to provide a more comprehensive knowledge base on COVID-19 for medical practitioners and researchers and lay the framework for future CIDO-COVID-19 applications. CIDO-COVID-19 follows the ontology creation principles specified by OBO and BFO.
O6: COVID-19 Ontology The new major entity of the coronavirus (SARS-CoV-2) is represented by the COVID-19 ontology [14]. Ontology has a healthful range of chemicals suitable for drug reuse as it is an important target for the ongoing therapeutic development of COVID-19 ontology. The ontology covers the role of molecular and cellular entities in virus-host interactions in the virus life cycle and a wide spectrum of medical and epidemiological concepts linked to COVID-19.
O7:COviD-19 Ontology for the Case and Patient Information (called CODO) The CODO [15] is a knowledge graph that provides information about the Covid-19 cases and patient-related history. The CODO knowledge model is used to collect and analyze the data about the COVID-19 pandemic in statistics and patient information domains only. CODO tracks clinical tests, travel history, available resources, actual needs (e.g., ICU beds, invasive ventilators), trend studies, and growth projections. The CODO model follows the FAIR principles to publish the data.
O8: COVID-19 Surveillance Ontology (COVID19) The goal of surveillance [16] application ontology is to offer COVID-19 cases and their related respiratory information by accessing data from several computerized medical records systems. However, this ontology is designed as a taxonomy having 32 classes only and based on the use case of the Royal College of General Practitioners (RCGP) Research and Surveillance Centre (RSC). The foundational concepts of ontology are definite COVID-19, COVID-19 confirmed by a laboratory test, SARS-CoV-2 detected, Probable COVID-19, clinical codes, possible COVID-19, suspected COVID-19, virology, exposure, COVID-19 excluded. The COVID-19 ontology is developed by the protégé tool, whereas its format is based on OWL language.
O9: The Covid19 Impact on Banking Ontology (Covid19-IBO) Covid19-IBO [17] contains structured knowledge about the impact of Covid-19 on the banking sector of India. To avoid the problem of overlapping information, the authors propose a schema-matching strategy for linking available Covid-19 ontologies.
O10: COVIDCRFRAPID The World Health Organization (WHO) COVIDCRFRAPID [18] ontology is a semantic data model for the WHO’s COVID-19 RAPID case record form from 23 March 2020. This model provides semantic references to the questions and answers of the form. It demonstrates several use cases including graph-based machine learning.
O11: KG-COVID-19 KG-COVID-19 [19] is a flexible framework for collecting and integrating heterogeneous biomedical data to create a knowledge graph (KG) used to create a KG for COVID-19 responses. It presents a comprehensive COVID-19 KG derived from 13 knowledge sources containing 377,482 nodes and 21,433,063 edges. The knowledge graph was constructed using modern ontology best practices whereby different data sources were normalized and merged.
O12: DRUGS4COVID19 DRUGS4COVID-19 [20] defines medications and their relationships related to COVID-19. Some of the key classes of ontology are drug, effect, disease, symptoms, disorder, chemical substance, etc. Drug4covid-19 is an ontology that consists of classes to enable the description of COVID-19 datasets in RDF. Some of the classes of this ontology are extracted from the Dataset of Johns Hopkins University.
O13: ROC: Ontology (Country Responses toward COVID-19) ROC [21] enables data integration from heterogeneous data sources and answers interesting questions. ROC intended to facilitate statistical analysis to investigate and evaluate such responses’ effectiveness and side effects of government responses to COVID-19 in different countries.
We also discuss the FAIR (Findable, Accessible, Interoperable, and Reusable) principle [22] and Open Biological and Biomedical Ontologies (OBO) Foundry [23], a set of overarching principles used as guidelines to develop, data harmonization, application, and sharing of ontologies. OBO aims to create and maintain a set of interoperable, well-formed ontologies representing biomedical knowledge. The initiative of OBO began in the 2000s, producing a set of principles considered good practices in the construction of ontologies [24]. The OBO principles have not been initially encoded in a precise fashion, and interpretation was subjective. OBO’s interoperability with other ontologies is not consistent, and the lack of a standard for biomedical ontologies hampers their applicability and subsequent adoption in real-world applications. Researchers in the ontology community also recognize the need for ontologies to follow the FAIR principles[22]. Thus, the goals of OBO and FAIR are highly compatible, and there is no conflict between these principles. Furthermore, the current pandemic has necessitated the development of a COVID-19-specific ontology that is semantically linked to ontologies reflecting geography, healthcare policy, and biomedical domains.
We investigate these existing taxonomies in Table 2, to ascertain the scope, development guidelines, availability on the linked open data (LoD) cloud, openness as a license to reuse, covering dimension, and ontology limitations. Despite sharing a similar objective, they are all different from one another due to their varied scopes. We have found that ontologies referring to the Covid-19 pandemic are mainly focused on a biological scale (gene, cell, organ, organism, population, disease, disorder, and infectious). Some ontologies refer to clinical treatment (drug product, drug substances, vaccine, drug pathway, diagnosis), primary health care of the patient (age, gender, ethnicity, deprivation, rurality, test), and response (government agencies, hospitals, academic researchers, publishers, news agencies, etc.), monitoring condition (symptoms, signs, key clinical features, virology, clinical research, risk factor), health measures (hospitalization, oxygen, therapy, intensive care admission, and mortality) domain. We have conglomerated all these domains into seven distinct dimensions in Table 1. We found that no single ontology covers all defined dimensions. All ontologies may have further scope to expand in this direction.
4 Development of coronavirus disease ontology (CovidO)
Many ontology engineering methodologies have been proposed to develop ontologies. Still, the field lacks widely accepted and mature methods. Most procedures lack sufficient details of techniques and activities employed in them [25]. However, some methodologies provide fine details, including METHONTOLOGY [26].
-
TOVE Based on their experience developing the TOVE (Toronto Virtual Enterprise)[27] project, GrRuninger, and Fox produced some steps toward a methodological construction and evaluation of ontologies in 1995.
-
Enterprise Model Based on knowledge gained through the creation of enterprise ontology, an Enterprise Model [28] approach is suggested. An Enterprise Model is a computational representation of the structure, knowledge, processes, resources, people, behavior, goals, and constraints of a business, government, or other enterprise information.
-
METHONTOLOGY METHONTOLOGY [26] was considerably influenced by software engineering methodologies, therefore, its terminology is related to the development and support of an evolving prototyping life cycle and has been continuously updated. Ontology reuse was proposed as an activity in the development process.
-
Systematic Approach Systematic approach [29] is composed of a set of directives, design patterns, and transformation rules. The directives are used to guide the mapping from the epistemological structures of the domain ontology (concepts, relations, properties, and roles) to their counterparts in the object-oriented paradigm (classes, associations, attributes, and roles).
-
KBSI IDEF5 KBSI IDEF5 [30] has some similarities to the methodologies discussed in that paper. Their approach involves: 1. Organizing and scoping, 2. Data collection, 3. Data analysis and initial ontology development and 4. Ontology refinement and validation step.
-
On-To-Knowledge A collaborative ontology engineering technique, On-To-Knowledge [31], demonstrates several KM project process drivers and offers tips for pragmatic domain experts working in industrial settings. Still a lot of unresolved problems, such as how to manage the scattered process of emerging and aligned ontologies that the semantic web is anticipated to experience.
-
DILIGENT With the use of a sophisticated methodology based on rhetorical structure theory [32], a DILIGENT [33] approach is taken to support the domain experts in a distributed situation as they construct and build ontologies. It recognizes ontology engineering methodologies like On-To-Knowledge or METHONTOLOGY as proven useful for the initial design.
We have examined strategies that use a step- or process-based approach to the ontology design and development methodology to capture shared knowledge, as shown in Fig. 1. The process used in these ontologies development approaches may be categorized into four primary phases, represented by four different colors. This stage-based approach is preferable if the goals and specifications for creating the ontology are apparent from the start. Otherwise, use an evolving prototype approach for creating ontology. Creating an ontology is a creative process, and no two ontologies created by different individuals would be alike [34]. Creating ontologies requires craft expertise rather than following a well-known technical method [35]. As a result, no one ideal ontology design process can be used to develop all domain ontologies.
After reviewing the available methodologies, we proposed an ontology development approach (ODA). ODA is an ontology development approach that can be applied to develop any kind of domain ontology from the beginning. ODA has four phases: (1) purpose identification and requirement specification, (2) ontology development phase, (3) evaluation and validation, and (4) post-development phase as shown in Fig. 2. These phases are related to the phases used in existing development approaches which are depicted in the four different colors in Fig. 1. We have derived these steps from the current ontology development approach. We consider the ODA approach to developing CovidO. ODA phases may contain several activities to achieve the goal. We demonstrate these phases considering the case of our CovidO Ontology in detail.
4.1 Purpose identification and requirement specification for CovidO ontology
In the first phase, draw some inspirational scenarios for the objective of better understanding the ontology’s scope, goals and help to create a suitable ontology as per user requirements. This phase contains the following sub-phases:
-
State the purpose This step describes the purpose and application of the ontology. As previously stated, the CovidO ontology’s goal is to make it easier to publish COVID-19 data as a knowledge graph and to develop semantic services and applications (e.g., decision support system, advanced analytics, or all the application related to COVID-19).
-
Identify the scope To define the scope, we recommend formalizing the requirements through user stories, competency questions (CQ:s), contextual statements, and reasoning requirements to help structure and delimit the modeling problems. Some examples of CQ:s are given in Table 3 in the domain of COVID-19; The purpose of this CQ is to: answer all of the mentioned constraints.
-
Ontology requirements specification document (ORSD) The ORSD contains the following key concept: (1) Define the goal of the ontology to be developed that has a specified purpose, (2) The intended uses/applications of ontology and end user who may use it [36]. For example, CovidO has several uses, such as annotation, recommendation, and semantic representation of COVID-19 information. The users of CovidO are the developer, domain experts, and end users who require information regarding COVID-19. (3) Identify the data sources which motivate the construction of Ontology. For example, CovidO, as discussed in the introduction has various data sources. We used the WHO and voltmeter as the necessary data sources for the case study to demonstrate CovidO. (4) Find similar existing knowledge resources for re-engineering them into ontologies. Ontologies O1 to O12 are the existing ontologies available for COVID-19. (5) Develop ORSD, which contains the ontology requirements such that the ontology should satisfy after being formally implemented as CQ. We upload ORSD to the GitHub repository.
4.2 Ontology development phase (CovidO construction)
In this phase, we discuss the list of the subtasks of ODA utilized to create the ontology. Keeping the scope of ontology captured, we identify the essential terms and relationships in the relevant field. After this creating a clear, concise, and unambiguous textual description of each essential term and relationship and then a hierarchy of their usage is created. In ODA, the capture of specific ontology design processes has been improved from the early phases of non-systematic work to the current activities performed in the framework of ontological engineering [37]. In ODA, we are not lacking any explicit explanation of how ontology is created or no capture of the rationale behind the process (such as ODRS, CQ:s, Class hierarchy, Conceptual design, implementations in particular languages, linking and reuse, etc.). More specifically, in the existing methodologies, there is no trace of the activities that have originated any of the products. The requirements imposed, the actors that have performed each activity, and the underlying rationale behind each decision made [38].
4.2.1 Concept extraction
In this phase, we produce clear, unambiguous text and extract the important terms or concepts for the ontology. It offers a wide range of representations that reflect various necessary concepts required for ontology within scope. In developing the CovidO ontology, we extract the concepts from sources (as mentioned below) according to the dimensions and scope of the domain.
-
1.
The references list includes studies on the COVID-19 epidemic that can be found in conferences, journals, and book chapters.
-
2.
The ontology repositories/portals already in existence, such as the Bio portal, EMBL-EBI, and Agro portal.
-
3.
COVID-19-related articles published on websites (Like Wikipedia, blogs, different sites, and so on).
-
4.
COVID-19-related databases (https://dea.gov.in, https://who.int, https://www.mygov.in/covid-19, etc.).
-
5.
COVID-19-related ontologies O1 to O12 (CODO, CIDO, VIDO, IDO, COVID-19, etc.).
-
6.
Conducting interviews with an expert like doctors, Auditors.
-
7.
Various COVID-19 reports, GoI press releases about COVID-19 impact on the Indian economy, WHO report of COVID-19 cases.
These sources generate a list of potential entities (like classes, properties, and instances). An example of CQ:s and extracted entities from this CQ:s are kept in the GitHub repository and likely are chosen after considering all of their potential synonyms. We determined the concept coverage of COVID-19 and then retrieved, filtered, and reused terms from existing ontologies. Based on a literature review of all relevant dimensions, recognized the scope of the core concepts for CovidO. Some examples of the essential entities (related to COVID-19) are shown here:

4.2.2 Relation extraction
Regarding the relationships between the concepts, the focus is on prioritizing reusing established connections in reusable ontologies. Relationships that were not mentioned in the above ontologies were defined using Protéé.

4.2.3 Class hierarchy
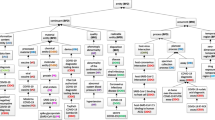
Due to the diversity of concepts in the field of COVID-19, it is a complex task to organize concepts and relationships in proper order while maintaining interoperability. To do this, we have followed the approach of the concept organization described in [17]. Then, we created a parse tree that represents the child–parent relationship. An example of the class hierarchy for CovidO is shown in Fig. 3.
4.2.4 Ontology formalization
Once ontology concepts and the relationship between the concepts have concluded, those things that remain are mainly design ontology. An operations-oriented approach [39] is the foundation of the formalization conceptual model, which sees the ontology development process as a series of tasks based on the outputs of the development process described as follows.
Conceptual Design: At the time of conceptual design the attention of domain is required because he has to be aware of key ontological distinction patterns [39] for the application. Keeping this in mind, we create a CovidO conceptual model based on the class hierarchy and extracted relationship of concept, which is shown in Fig. 4. The different color coding represents the dimension and concepts belonging to the respective dimension, and the dotted arrow shows the classes and sub-classes relationship. Solid arrows represent the relationship between the entities with min–max cardinality.
4.2.5 Integration/linking
Since there are numerous ontologies for the COVID-19 disease, it seems sensible to reuse their concepts when applicable (Table 4). Ultimately, what is required is to develop and reuse domain vocabularies. We have chosen the reuse entities based on usability and similarity. We have followed this best practice in the development of the CovidO ontology. We have integrated the concepts from the given prefixes.
Vocabulary | Prefix |
|---|---|
dbo | |
schema | |
obo | |
codo | |
foaf | |
snomed | |
symp | |
ogms | |
drug | |
ibo | http://www.semanticweb.org/archana/ontologies/2021/5/untitled-ontology-6 |
4.2.6 Implementation of ontology
Once the conceptual design is created, implementation can be done in two intermediate ways: formalization and ontology design. In formalization, transform the conceptual model into a formal or semi-compatible one [28]. In the ontology design, create ontology using some ontology editors like Protégé-2000 [40], OilEd, OntoStudio, Apollo, and Swoop with supported standard language. We have used Protégé version 5.5.0 to build the CovidO.
An example of the ontology implemented using the ODA approach has been given based on the data taken from WHO reports. The information is organized around a number of subdomains and related fields, including connectivity infrastructure and interfaces for coronavirus expression as shown in Fig. 5, which shows the relationship between the concepts of data. After implementation, the CovidO implementation hierarchy of the class is shown in Fig. 6, A class of Covid-19 is an example of how the ontology’s classes describe a concept in a domain. The impact is a term used to describe all agents and has implications in several areas. Figure 6 represents the hierarchy of CovidO classes, the hierarchy of CovidO object properties, and the data properties hierarchy of CovidO, respectively.


4.3 Evaluation of CovidO
Evaluating the quality of developed ontologies is one of the most crucial components of ontology learning. Such assessment can then guide and regulate the ontology learning process in pursuit of the “optimal” ontology. However, judging ontology learning tools is tricky because it is still not clear how to compare them with developed ontology. Since numerous researchers have worked on ontology evaluation, a variety of approaches, methods, and tools are available [25], but used for a specific purpose and depending on what kind of ontologies are being evaluated and for what purpose. This assessment divides approaches into four categories:
-
(1)
Technology-based approach is used to investigate structural characteristics (syntax, formal semantics, and consistency) and measure the correctness and usability.
-
(2)
The quality-based approach is used to check the formal and semantic redundancy and measure the quality of ontology, like inconsistency, and missing definitions.
-
(3)
Data-driven approach, focus on usability of ontology.
-
(4)
Application-based approach, used to explore the scope of ontology in the specific domain.
According to Mehla and Jain, [41], the use of combined approaches is good for domain ontology. Therefore, we evaluate the CovidO ontology based on verification (which refers to developing an ontology properly) and validation (that means building the correct ontology) measures that cover all the above category (Technology, quality, data, and application-based approach).
4.4 Post-development phase
This phase is again very important to make the ontology reused and release updates in the ontology to make it usable. This phase is required because the information changes over time and human has no control over it. Post-development phase executes when the ontology has received some new features.
5 Result evaluation of CovidO
We have used verification and validation to evaluate the proposed CovidO ontology.
5.1 Verification
The verification employs both quantitative and qualitative methods for verification [42]. While the qualitative technique evaluates the ontology’s quality using the criteria-based approach, which examines the ontology’s consistency, conciseness, completeness, accuracy, and clarity. In the criteria-based approach, we used an Ontology Pitfalls Scanner (OOPs) tool [43] to evaluate the quality of CovidO. OOPs show the 41 pitfalls or errors (starting from P01 to P41) classified into two groups: (1) Classification by dimension and (2) Classification by evaluation criteria is shown in Fig. 7. The pitfall fall in dimension indicates a minor error that impacts ontology usability and clarity and hence the quality of ontology design. On the other hand, the pitfall comes under the evaluation criteria, which describes how accurately the ontology is defined.
Table 5 shows a comparison of the existing COVID-19 ontologies pitfalls with CovidO. Ontology O14 represents CovidO. The number in the table represents cases corresponding to mentioned pitfall, for example, IDO has 24 cases of pitfall P11. This means IDO has 24 cases that have no inference corresponding to pitfall P11 describes “no inference” in the structural dimension. Large numbers in the pitfall table indicate more ontological errors that occur frequently, whereas the “x” symbol indicates no errors. Yes, represents the respective pitfall present instance. In the above table, we have mentioned only those pitfalls that have at least one case in any one of these ontologies. CovidO ontology has a few pitfalls (P22, P30) that cannot be eliminated because in our case, pitfall P22 represents a different naming convention in the ontology. These errors are because of reused entities, for example, Covid-19 has “-” which causes this pitfall to appear. In CovidO, three cases of Pitfall P30 are present that show equivalent classes not explicitly declared. Oops pitfall shows that cases (dbo:Village,dbo:Settlement), (dbo:State,dbo:Country) and (covido:Author,covido:Writer) might be equivalent but this is not true in our situation. Although they both belong to the same individual person, the terms “author” and “writer” have different meanings in the context of research publications and news stories, respectively. Also, State and Country have different contexts. Figure 8 gives a clearer picture of the pitfalls contained in the related ontology and demonstrates that the ontologies O5, O6, O8, O13, and O14 have fewer pitfalls. We can see that ontology O2 has the greatest number of potential pitfalls and CovidO has comparatively fewer pitfalls as compared to others. Ontology O11 creates a parsing error while validating to the oops scanner, hence the result is not presented.
In the quantitative approach, we measure the number of attributes in the ontology. A metric-based technique has been used to count how many attributes (node, depth, breadth, total number of levels) are present in the ontology. This method does not detect the anomalies inside the ontology. To measure the quantitative evaluation of CovidO, we have used the metric-based OntoMetric tool (https://ontometrics.informatik.uni-rostock.de/ontologymetrics/) which helps to show the richness of ontology. OntoMetric used five metrics-Schema, Instance, Base, Graphs, and Individual Axioms to categorize the features. Table 6 displays the comparison between CovidO and the current COVID-19 ontologies using the OntoMetric tool. In the table, Base metrics axioms (represent a true element in a domain), Logical axiom (define the logical meaning of axioms), Class Count (represents the cardinality of classes in ontology), Object Property Count (number of object properties that associate concept to concept or instance to instance), and Data Property Count (number of data properties hold data values) are given. In the Class axioms, Subclass of axioms (which show the number of subclasses in the ontology) and Equivalent class (which represent how many classes are equivalent in the ontology) are given. The number in the table for base metrics and class axioms indicates the cardinality of axioms, whereas schema metrics reflect the design of ontology by Attribute richness (which defines attributes corresponding to ontology classes), Inheritance Richness (demonstrating the distribution of information across different levels), and relation richness (illustrating the diversity of types of relations). The knowledge base features two average populations ( define the individuals who distribute the information within a domain) and Class richness (describe the distribution of the population for each class). Higher value in the schema metrics and knowledge base metrics represent good value. Ontology O11 and O13 have not been parsed by the OntoMetric tool, the value is blanked. Table 6 shows that O14 which is CovidO has a good level of information and has better class richness in the knowledge base. The results of the OntoMetric evaluation of CovidO (O14) against the COVID-19 ontologies are displayed in Fig. 9. The graph represents the different metadata points about the proposed and the available Covidd-19 ontologies. The metadata points are shown on two axes, with one axis representing values in an upward direction and the other in a downward direction. The values in the upward and downward directions are larger than one and less than one, respectively. The metadata categories that have values that are distant from the axis (values larger than one) indicate the goodness of the ontologies when they are oriented upward. But when the axis is moving in the other direction, the metadata categories with values far from the axis (less than 1) show that the ontology’s metadata categories are less striking. We can observe that the O14 has produced significant results in the graph.
5.2 Validation
Validation entails creating the correct ontology in the context of syntax, semantics, and consistency. Validation implies the correctness and usability of ontology. We have used existing accessible validators (W3C RDF validator, OWL validator, RDF Triple-Checker, and Vapor), and competence questions to validate the CovidO ontology.
5.2.1 Validators
We use the W3C RDF validator [32], OWL validator [33], RDF Triple-Checker [34], and Vapor [35] to validate the CovidO. While RDF TripleChecker and Vapor are used to check the correctness of namespaces in the document, the W3C RDF validator, and OWL validator ensure that the ontology is syntactically correct with regard to RDF and OWL syntax, respectively. The W3C RDF validator successfully extracts all of the CovidO tuples. The absence of any error messages during validation is evidence that the ontology is well-designed and syntactically sound. CovidO also validates using the OWL validator successfully and shows no error. CovidO is loaded on RDF Triple-Checker to validate via URI and shows all the classes and properties have a well-defined prefix.
5.2.2 Competency questions
The set of competency questions describes the requirement of the ontology as per the specified domain as well as checks the completeness of the developed ontology. A set of competency questions (some of them are mentioned in Sect. 3) that cover the scope of the dimension of CovidO are provided in natural language (English) and are translated into SPARQL queries. The SPARQL query engine is almost similar to OWL just like SQL is to relational DB. The underlying structure of OWL is graph-based rather than tables; therefore, SPARQL constructs graph patterns to infer the knowledge from the ontology. The competency question, corresponding SPARQL query, and retrieve results from CovidO are shown in Table 7.
Used Prefix: | |
|---|---|
PREFIX rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#> | |
PREFIX owl:<http://www.w3.org/2002/07/owl#> | |
PREFIX rdfs:<http://www.w3.org/2000/01/rdf-schema#> | |
PREFIX xsd:<http://www.w3.org/2001/XMLSchema#> | |
PREFIX covido:<http://w3id.org/CovidO#> | |
PREFIX schema:<http://schema.org/> | |
PREFIX foaf:<http://xmlns.com/foaf/0.1/> | |
PREFIX dbo:<https://dbpedia.org/page/> | |
PREFIX obo:<http://purl.obolibrary.org/obo/> | |
PREFIX dboo:<https://dbpedia.org/ontology/> | |
PREFIX codo:<http://w3id.org/codo#> |
6 Conclusion
The purpose of this work is to provide CovidO, an ontology for processing COVID-19 information, to facilitate the integration of data from diverse data sources, and to provide answers to interesting queries on all COVID-19-related aspects. CovidO has the advantage to integrate the biomedical field with real-world COVID-19 data in all possible dimensions. The dimensions we assign to COVID-19 help CovidO ontology to make it stronger and more transparent based on primary care. Also, play an important role in facilitating the bridge to integration and analysis of research in all branches related to COVID-19. This work presents Coronavirus Disease Ontology (CovidO): (1) Describes complete knowledge of COVID-19; (2) Maps existing ontologies and creates standard metadata to understand and share COVID-19 knowledge. (3) Acts as a vocabulary for researchers, engineers, and developers to find the commonly used COVID-19 gesticulation for particular accordance, and to use the scope and dynamics of a specific gesture. The result evaluation show quality and the quantity of CovidO are better than the other ontologies and have wild scope than other. As we evaluate our work and procedures, we believe that our effort will assist all healthcare professionals in taking precautions, following recommendations, and lowering the risk of COVID-19 transmission and infection.
Data availability
The data are available upon reasonable request to the corresponding authors.
References
Organization WH (2020) Modes of transmission of virus causing covid-19: implications for ipc precaution recommendations. Sci Brief
Westerski A (2013) Semantic technologies in idea management systems: a model for interoperability, linking and filtering. Adam Westerski
Francesco O, Motta E (2015) Klink-2: integrating multiple web sources to generate semantic topic networks. In: International Semantic Web Conference. Springer, pp 408–424
Dividino R, Soares A, Matwin S, Isenor AW, Webb S, Brousseau M (2018) Semantic integration of real-time heterogeneous data streams for ocean-related decision making. Defence Research and Development Canada= Recherche et développement pour la défense Canada
Kaye AD, Cornett EM, Brondeel KC, Lerner ZI, Knight HE, Erwin A, Charipova K, Gress KL, Urits I, Urman RD et al (2021) Biology of covid-19 and related viruses: epidemiology, signs, symptoms, diagnosis, and treatment. Best Pract Res Clin Anaesthesiol 35(3):269–292
Djurić D, Gašević D, Devedžić V (2005) Ontology modeling and mda. J Object Technol 4(1):109–128
Cui L, Tao S, Zhang G-Q (2016) Biomedical ontology quality assurance using a big data approach. ACM Trans Knowl Discov Data (TKDD) 10(4):1–28
Babcock S, Beverley J, Cowell LG, Smith B (2021) The infectious disease ontology in the age of covid-19. J Biomed Seman 12:1–20
Beverley J, Babcock S, Cowell L, Smith B (2021) The COVID 19 infectious disease ontology
He Y, Yu H, Ong E, Wang Y, Liu Y, Huffman A, Huang H-h, John B, Lin AY, Arabandi S, et al. (2021) Cido: the community-based coronavirus infectious disease ontology
Sharma S, Jain S (2023) The coronavirus disease ontology (covido). In: Semantic Intelligence: Select Proceedings of ISIC 2022. Springer, vol 964, pp 89–103
Beverley J, Babcock S, Cowell L, Smith B (2021) The COVID 19 infectious disease ontology
Xiao Y, Zheng X, Song W, Tong F, Mao Y, Liu S, Zhao D (2021) Cido-covid-19: an ontology for covid-19 based on cido. In: 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC). IEEE, pp 2119–2122
Sargsyan A, Kodamullil AT, Baksi S, Darms J, Madan S, Gebel S, Keminer O, Jose GM, Balabin H, DeLong LN et al (2020) The covid-19 ontology. Bioinformatics 36(24):5703–5705
Dutta B, DeBellis M (2020) Codo: an ontology for collection and analysis of covid-19 data. Preprint arXiv:2009.01210
de Lusignan S, Liyanage H, McGagh D, Jani BD, Bauwens J, Byford R, Evans D, Fahey T, Greenhalgh T, Jones N et al (2020) Covid-19 surveillance in a primary care sentinel network: In-pandemic development of an application ontology. JMIR Public Health Surveill 6(4):21434
Patel A, Debnath NC, Mishra AK, Jain S (2021) Covid19-ibo: a covid-19 impact on indian banking ontology along with an efficient schema matching approach. N Gener Comput 39(3):647–676
Bonino L (2020) Who covid-19 rapid version crf semantic data model. See https://bioportal.bioontology.org/ontologies/COVIDCRFRAPIDBioPortal
Reese JT, Unni D, Callahan TJ, Cappelletti L, Ravanmehr V, Carbon S, Shefchek KA, Good BM, Balhoff JP, Fontana T et al (2021) Kg-covid-19: a framework to produce customized knowledge graphs for covid-19 response. Patterns 2(1):100155
Badenes-Olmedo C, Chaves-Fraga D, Poveda-VillalÓn M, Iglesias-Molina A, Calleja P, Bernardos S, MartÍn-Chozas P, Fernández-Izquierdo A, Amador-Domínguez E, Espinoza-Arias P, et al. (2020) Drugs4covid: Drug-driven knowledge exploitation based on scientific publications. Preprint arXiv:2012.01953
Qundus JA, Schäfermeier R, Karam N, Peikert S, Paschke A (2021) Roc: an ontology for country responses towards covid-19. Preprint arXiv:2104.07345
Jacobsen A, de Miranda Azevedo R, Juty N, Batista D, Coles S, Cornet R, Courtot M, Crosas M, Dumontier M, Evelo CT, et al. (2020) FAIR principles: interpretations and implementation considerations. MIT Press One Rogers Street, Cambridge, MA 02142-1209, USA journals
Foundry O (2013) The open biological and biomedical ontologies. Data of access 10
Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ et al (2007) The obo foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol 25(11):1251–1255
Cristani M, Cuel R (2005) A survey on ontology creation methodologies. Int J Seman Web Inf Syst (IJSWIS) 1(2):49–69
Fernández-López M, Gómez-Pérez A, Juristo N (1997) Methontology: from ontological art towards ontological engineering
Gruninger M (1995) Methodology for the design and evaluation of ontologies. In: Proc. IJCAI’95, Workshop on Basic Ontological Issues in Knowledge Sharing
Uschold M, King M (1995) Towards a methodology for building ontologies. Citeseer, pp 1–13
Falbo RdA, Menezes CSd, Rocha ARCd (1998) A systematic approach for building ontologies. In: Ibero-American Conference on Artificial Intelligence. Springer, pp 349–360
Jones D, Bench-Capon T, Visser P (1998) Methodologies for ontology development
Sure Y, Staab S, Studer R (2004) On-to-knowledge methodology (otkm). In: Handbook on Ontologies. Springer, pp 117–132
Mann WC, Thompson SA (1987) Rhetorical Structure Theory: A Theory of Text Organization. University of Southern California, Information Sciences Institute Los Angeles
Pinto HS, Staab S, Tempich C (2004) Diligent: towards a fine-grained methodology for distributed, loosely-controlled and evolving engineering of ontologies. In: ECAI. Citeseer, vol 16, p 393
Noy NF, McGuinness DL, et al. (2004) Ontology development 101: a guide to creating your first ontology. 2001. See http://protege.stanford.edu/publications
Dalal S, Jain S, Dave M (2022) An ontology for social media data analysis. In: Proceedings of the 2nd International Semantic Intelligence Conference (ISIC)
Sharma S, Jain S (2021) Comprehensive study of semantic annotation: Variant and praxis. Advances in Computational Intelligence, its Concepts Applications (ACI 2021). 2823, 102–116
Alterovitz G, Xiang M, Hill DP, Lomax J, Liu J, Cherkassky M, Dreyfuss J, Mungall C, Harris MA, Dolan ME et al (2010) Ontology engineering. Nat Biotechnol 28(2):128–130
Subhashini R, Akilandeswari J (2011) A survey on ontology construction methodologies. Int J Enterprise Comput Bus Syst 1(1):60–72
Vegetti M, Roldán L, Gonnet S, Leone H, Henning G (2016) A framework to represent, capture, and trace ontology development processes. Eng Appl Artif Intell 56:230–249
Noy NF, Crubézy M, Fergerson RW, Knublauch H, Tu SW, Vendetti J, Musen MA (2003) Protégé-2000: an open-source ontology-development and knowledge-acquisition environment. In: AMIA... Annual Symposium Proceedings. AMIA Symposium, pp 953–953
Mehla S, Jain S (2021) Development and evaluation of knowledge treasure for emergency situation awareness. Int J Comput Appl 43(5):483–493
Tartir S, Arpinar IB, Sheth AP (2010) Ontological evaluation and validation. Theory Appl Ontol Comput Appl:115–130
Poveda-Villalón M, Gómez-Pérez A, Suárez-Figueroa MC (2014) OOPS! (OntOlogy Pitfall Scanner!): an on-line tool for ontology evaluation. Int J Seman Web Inf Syst (IJSWIS) 10(2):7–34
Acknowledgements
The authors would like to thank the National Institute of Technology Kurukshetra, Haryana, India, for providing grant and facilities to conduct research work.
Funding
National Institute of Technology, Kurukshetra, India, funded this research under the Institute fellowship.
Author information
Authors and Affiliations
Contributions
Both the authors have discussed and constructed the ideas, designed the ontology model and wrote the paper together.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest regarding the publication.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sharma, S., Jain, S. CovidO: an ontology for COVID-19 metadata. J Supercomput 80, 1238–1267 (2024). https://doi.org/10.1007/s11227-023-05509-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11227-023-05509-4