Abstract
This paper presents the Coronavirus Disease Ontology (CovidO), a superset of the available Coronavirus (COVID-19) ontologies, including all the possible dimensions. CovidO consists of an ontological network of thriving distinct dimensions for storing coronavirus information. CovidO has 175 classes, 169 properties, 4141 triples, 645 individuals with 264 nodes and 308 edges. CovidO is based on standard input of coronavirus disease data sources, activities, and related sources, which collects and validates records for decision-making used to set guidelines and recommend resources. We present CovidO to a growing community of artificial intelligence project developers as pure metadata and illustrate its importance, quality, and impact. The ontology developed in this work addresses grouping the existing ontologies to build a global data model.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction and Motivation
The World Health Organization declared a Public Health Emergency of International Concern on 30 January 2020 and a pandemic on 11 March 2020 [1]. At the same time, novel coronavirus (COVID-19) pandemic data has been collected by various data sources like the World Health Organization (WHO) and DXY.CN, BNO News, National Health Commission (NHC) of the People’s Republic of China, China Centers for Disease Control and Prevention (CCDC), Hong Kong Health Department, Macau Government, Taiwan CDC, US Centers for Disease Control and Prevention (CDC), Government of Canada, Australian Government Department of Health, European Centers for Disease Prevention and Control (ECDC), Ministry of Health Singapore (MOH), and others. The COVID-19 data sources and access links are shown below.
-
WHO. https://www.who.in.
-
DXY.cn. https://www.dxy.cn.
-
BNO News. https://bnonews.com.
-
National Health Commission (NHC). http://en.nhc.gov.cn.
-
China CDC (CCDC). https://www.chinacdc.cn/en/.
-
Hong Kong Health Department. https://www.dh.gov.hk.
-
Macau Government. https://www.gov.mo/en/.
-
Taiwan CDC. https://www.cdc.gov.tw/En.
-
US CDC. https://www.cdc.gov/.
-
Government of Canada. https://www.canada.ca/en.html.
-
Australian Government. Department of Health https://www.health.gov.au/.
-
Ministry of Health Singapore (MOH). https://www.moh.gov.sg/.
-
Ministry of Health and Family Welfare. https://www.mohfw.gov.in/.
A significant issue is that the data sources related to COVID-19 are heterogeneous, static, and broad in scope. So many heterogeneous and stationary data sources create situations where data is sometimes under-utilized or, in more extreme cases, not used for the decision-making process [2]. Another vital issue of COVID-19 is to provide semantic (machine understandable) representation of data from various exciting fields such as research, health, resources, drugs, and treatment. Ontology is emerging to solve these issues. Ontology solves the problem of changing user expectations and data integration demands driven by its volatility in a rapidly growing digital market and societal challenges related to resource efficiency [3]. Ontologies have proven practical tools for representing domain knowledge, integrating data from disparate sources, and supporting many semantic applications [4].
This paper presents an OWL-based Coronavirus disease ontology (CovidO) that defines all the possible aspects and relations to describe COVID-19. To develop CovidO, we have defined seven dimensions that cover all essential aspects of COVID-19: (1) About the COVID-19 infectious disease, symptoms, drugs, and treatment; (2) Information statistics of COVID-19 cases in a geographical region; (3) COVID-19 patients information with the cause of infection and exposure of pandemic; (4) COVID-19 related resources and their availability in a location; (5) Impact of COVID-19 in different verticals like education, finance, business, research and social; (6) Various guidelines and prevention and vaccine mandates by public authority; (7) Global and biomedical research on COVID-19. Several ontologies (e.g., Infectious Disease Ontology (IDO), Virus Infectious Disease Ontology (VIDO), Coronavirus Infectious Disease Ontology (CIDO), etc.) have been created for coronavirus one after another. They comprehensively and thoroughly describe coronavirus disease. These ontologies individually fail to cater to all the dimensions of coronavirus. The ontology developed in this paper aims to group existing ontologies to construct a common global data model with the unified purpose. The main contributions of this paper are:
-
to provide a list of dimensions to cover every aspect of coronavirus-related information.
-
to develop standard metadata (Providing a schema) called CovidO as a global data model to annotate the Covid-19 information.
This paper presents our work to be independent of external ontologies, we defined a new namespace https://w3id.org/CovidO with the prefix CovidO (registered entry at http://prefix.cc) for all classes used in the ontology. As a permanent URL service, we use w3id.org.
The content of this paper is organized as follows. The related work describes literature on the various existing COVID-19 associated ontologies and their scope boundary. The dimension section describes the incidence of the COVID-19 information dimensions in the primary analysis and provides a summary analysis. The conceptual design and scope of CovidO are described in the CovidO section to build the ontology. The ontology design section defines the competency questions to determine the scope of CovidO with abstract design. The method section outlines the model to develop CovidO. In the evaluation section, we present the evaluation for the COVID-19 schema, in addition to simple predictions for the future incidence of COVID-19. Some concluding remarks are given in the conclusion.
2 Related Work
Several ontologies represent the COVID-19 pandemic in different contexts. We have found some ontologies related to COVID-19, each representing a completely different scope of COVID-19. They are briefed here:
-
O1:
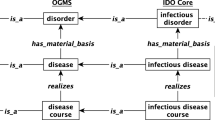
COVID-19 Infectious Disease Ontology (IDO-COVID-19): IDO-COVID-19 [5] is the most particular version of CIDO (Coronavirus Infectious Disease Ontology) [6], containing information about COVID-19 and its cause SARS-CoV-2. IDO-COVID-19 adhers to the OBO Foundry design philosophy by extending the CIDO in the same way as the CIDO extends VIDO (Virus Infectious Disease Ontology) [5] and the VIDO extends the IDO (Infectious Disease Ontology) [7]. IDO-COVID-19 also pulls concepts from other ontologies, such as SARS-CoV-2, imported from NCBITaxon. IDO-COVID-19, CIDO, and VIDO ontologies consist of concepts related to disease dimension, other dimensions are not covered.
-
O2:
COviD-19 Ontology for the case and patient information (called CODO): CODO [8] is an ontology that contains COVID-19 case data in a format that can be used by other ontologies and software systems and is based only on OWL and different W3C standards. CODO tracks specific pandemic cases, including information such as how the patient is considered to have been infected and potential further contacts who may be at risk owing to their association with the infected individual. CODO also tracks clinical tests, travel history, available resources, actual demand (e.g., ICU bed, invasive ventilators), trend analyses, and forecast increases. The CODO ontology covered the cases, patients, and resources dimensions.
-
O3:
COVID-19 surveillance Ontology (COVID-19): COVID-19 surveillance Ontology [9] is a COVID-19 application ontology that intends to provide COVID-19 cases and respiratory information by obtaining data from multiple medical records systems. This ontology is constructed as a taxonomy with only 32 classes. COVID-19 verified by a lab test, SARS-CoV-2 identified, Probable COVID-19, Clinical codes, Possible COVID-19, Suspected COVID-19, Under investigation, Exposure, COVID-19 excluded are the ten core ideas of COVID-19 ontology. The COVID-19 ontology was created using the protégé tool, and its format is based on the OWL language.
-
O4:
COVID19-IBO: The COVID-19 Impact on Banking Ontology (COVID-19-IBO) [10] is a knowledge graph which covers semantically the COVID-19’s impact on Indian banking industry under the “Impact on business vertical” dimension. In addition, the authors have provided a schema matching technique with satisfactory results for mapping the COVID-19 ontologies.
-
O5:
Kg-COVID-19: The KG-COVID-19 [11] framework is used to create customized COVID-19 knowledge graphs. The FAIR (Findable, Accessible, Interoperable, and Reusable) approach is followed by KG-COVID-19, which combines various COVID-19 heterogeneous biomedical data and covers the COVID-19 disease, symptoms, and treatment dimensions.
-
O6:
COVID-19 Ontology: The COVID-19 ontology includes the function of molecular and cellular entities in viral-host interactions throughout the virus life cycle and a wide range of medical and epidemiological concepts associated with COVID-19. A scalable new coronavirus (SARS-CoV-2) entity is represented as an ontology. As a prominent target of ongoing COVID-19 medicinal research, the ontology contains a broad scope on chemical entities ideal for drug repurposing. The ontology’s performance was evaluated using Medline and the Allen Institute’s COVID-19 corpus.
-
O7:
DRUGS4COVID19: DRUGS4COVID19 [12] identifies drugs and their associations with COVID-19. The ontology’s core concepts include drug, effect, disease, symptoms, disorder, chemical substance, and so on.
-
O8:
COVIDCRFRAPID: The World Health Organization’s (WHO) COVIDCRFRAPID [13] ontology is a semantic data model for the COVID-19 RAPID case record form. COVIDCRFRAPID ontology provides semantic references to the form filled by patients during the treatment as questions and responses. It shows a variety of application scenarios, including graph-based machine learning.
-
O9:
ROC: Ontology (Country Responses toward COVID-19) ROC [14] enables data integration from heterogeneous data sources and answers interesting questions. ROC was designed to assist statistical analysis in exploring and analyzing the efficacy and side effects of government responses to COVID-19 in various nations.
We investigate these existing ontologies, focusing on a group of individuals to discuss a specific topic like drug, protein interaction databases, protein function annotations, COVID-19 patients, and cases. So they have a limited scope that does not cover all aspects of COVID-19. We have found that these ontologies refer to the COVID-19 pandemic but represent different aspects and scopes. We fill this gap, and our work follows the same approach to ontology design and has a common motivation. We have conglomerated all these ontologies into a comprehensive design covering all the required distinct dimensions (COVID-19 cases information, patient information, disease-symptom-treatment, resources, COVID-19 impact, research, and event or news related to COVID-19) discussed in the next section. By adopting established models, we aim to facilitate integration, linking, and reuse across the data sources and make data accessible to a wide range of applications. In addition, new entities have been introduced required.
3 COVID-19 Information Dimensions
With a view to allowing stakeholders in the research community and application developers to reach out and benefit, CovidO has been created as a platform through specific dimensions. There are many domains and sub domains; we group these domains into seven significant divisions that cover all the aspects of COVID-19 related knowledge. These seven dimensions are shown in detail in Table 1. The CovidO ontology is developed based on these dimensions representing COVID-19 information in OWL format and other W3C standards utilized by other ontologies and software systems. The last column of Table 1 describes the core CovidO classes associated with a particular dimension. CovidO allows detailed tracking of specific pandemic dimensions. For example, the diseases and treatment dimension includes clinical test tracking, test report history, illness, symptoms, medication, and clinical measurement and diagnosis. Similarly, CovidO traces other dimensions as well. In brief and with overall dimensions, CovidO monitors the COVID-19 patient’s travel history, symptoms, medication, available healthcare facilities, resources, actual need (e.g., ambulance, invasive ICU bed with ventilators), trend study, impact on business verticals, research publications finding, guidelines for public health Safety, news, and growth projections. To the best of our knowledge, we have not found any ontology that describes all the seven dimensions D1 to D7. Nor was any such ontology found which could provide an interlink between them. All ontologies have different scopes and common goals to provide the schema for COVID-19 data.
4 The Coronavirus Disease Ontology (CovidO)
The CovidO knowledge representation model encodes knowledge in the form of classes, relationships, properties, instances, and axioms [12]. Our work is inspired by COviD-19 Ontology (codo), and we have taken their work forward with some new features and new dimensions that cover all aspects of COVID-19. We have significantly expanded the capabilities of the codo ontology model made for COVID-19 cases and patients information, i.e., changes to classes, properties, relations to be extensible. We are giving annotations for ontology concepts that have already been produced but neither annotated nor labeled.
4.1 Design Methodology
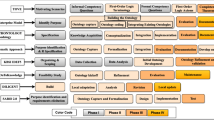
From the survey of the literature one can find that there are several ontology development methodology (ODM) that carried set of activities to create ontologies. In ODM, Knowledge acquisition, integration, and alignment drive the speed of building ontology and these come with the risk of redundancy, consistency, and conflicts. Invoking the existing ODM, we opted for a mixed approach of Diligent [15] and Methontology [16] to develop the CovidO. The general procedure, and functions to obtain CovidO describe in four phases as DataToMetadata (D2MD): 1. Ontology Requirement Specification (ORs): In the ORs process, set the goal for ontology development and study the feasibility of the scope of ontology. We examine heterogeneous data sources given in the introduction to building ORs for CovidO and state-of-the-art requirement analysis according to the dimension of CovidO given in Sect. 3. ORs document help to design competency questions (given in Table 4) to define CovidO’s scope. CovidO domain knowledge should be organized as a meaningful ORs model at the knowledge level. After gathering sufficient information and ORs, we create a CovidO conceptual model that describes the problem and its solution. 2. Ontology Development Phase (ODP): ODP phase is the 2nd phase in the methodology which is responsible for deciding the Ontology Architecture, Designing Conceptual Map, Encoding in the OWL format. We have assumed that initial ontology is already constructed in the form of codo. It saves initial ontology development time and will help expand ontology in a distributed manner with different stakeholders’ objectives as a dimensions. According to extendable objectives make the changes in the ontology locally and then revise updates to satisfy consistency and verify scope. The analysis attempts to find the similarities in changing requests and users’ ontology. The study looks for commonalities between the ontologies of changing requests and users. Instead, decide what changes should be made to construct conceptual design. Once have a conceptual design, then implementation can be done in two intermediate ways: formalization and ontology design. In formalization, to transform the conceptual model into a formal or semi-compatible model. In the ontology design, to make ontology use some ontology editor like protégé with supported formal language. 3. Validation & Evaluation: Validation step is performed to validate the ontology based on different criteria like content validity, application-based analysis, etc. 4. Deployment: In this phase makes the ontology to reuse and available for further use. Where the publication of the ontology is on the cloud or public portal with take care of maintenance and proper updates.
4.2 New Features
CovidO is formed by reusing existing ontologies adding new classes and properties to cover all the dimensions listed in Sect. 3. Table 2 represents the core classes of CovidO with referral namespace. We have divided the Statistics class of codo into two parts, codo: Statistics class and CovidO: Resource. Statistics class represents the actual cases information, and Resource class represents the resources utilized for covid-19. Similarly, other classes of codo like Status, Symptom, Disease, and CovidTestingFacility have been added according to dimensions. Table 3 describes some new relationships between the classes of CovidO that were not present in other Covid ontologies. Currently, CovidO contains 175 classes, 169 relationship types, and goes to evaluation. We applied the Pellet Reasoner to verify that CovidO is consistent.
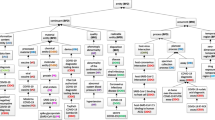
The top-level class structure diagram and relationships between core concepts are shown in Fig. 1. There are many types of relationships between concepts. The content ensures the semantic consistency of relationships between concepts and facilitates logical axioms and reasoning definitions.
4.3 CovidO Scope
Competency Questions (CQs) play an essential role in the ontology development lifecycle, representing the ontology requirements. Some CQs have been formalized through COVID-19 data sources described in the introduction section. We could structure and expand the conceptual modeling design of codo through CQs to obtain the CovidO. The CQs have broad coverage on COVID-19 data, So we are trying to map it into seven dimensions of COVID to mitigate it and cover all possible aspects of the pandemic so far. Table 4 represents some competency questions with the respective dimension that CovidO is expected to answer.
4.4 Reusing the Ontology Concept
According to [17], there are two perspectives to reuse ontology: (1) assembling, extending, specializing, and adapting other ontologies that are components of the final ontology, or (2) integrating other ontologies on the single concept that integrate all concept. We have used second approach, integration of other ontologies. The core concepts for CovidO are determined as prevention, vaccine, hospital facility, disease, infection, disorder, virus, SARS-Cov2, Coronavirus, agent, patient, disease, symptom, drug, treatment, organization, impact, host, diagnosis, statistics, place, etc. Which is already in use in other ontologies O1 to O12. CovidO integrates the concept with existing ontologies to pursue the basic principle of ontology implementation. Figure 2 represents the terms inherited by CovidO from available schema. Different colors define each schema. The solid and the dotted line show immediate and remote child-parent relations between classes. Most inherited entities belong to Schema, BFO, and CODO, while Kg4Grug, SYMP, and VO are the least inherited ontologies.
We use a permanent URL service, w3id.org, that makes it independent of external ontologies. We have defined a new namespace convention https://w3id.org/CovidO for all classes used in CovidO, with the prefix covido (the entry registered at http://prefix.cc). A common concept is integrated into a single concept that unifies them all. The concept selection is based on the best fit for the scope and dimension of CovidO. For determination of scope, competency questions were created and are publicly available via GitHub https://github.com/sumitsnit/CovidO.
5 Evaluation
We evaluated CovidO in two ways: using SPARQL query and OOPS! Pitfall Scanner. SPARQL query describes the accessibility of elements and OOPS! Pitfall represents the RDF quality of CovidO.
5.1 SPARQL Query Evaluation
A set of competency questions over CovidO is given in Table 4. For scope evaluation of the CovidO, answers to the SPARQL [18] queries built on the questions from Table 4 are considered. CovidO is schema-based ontology, so schema-level SPARQL query is allowed till now. Once the ordered representation is done as user requirement from the coronavirus distributed and heterogeneous data sources, CovidO instance-level questions are permitted to answer. We evaluate CovidO on schema-level competency questions based on the D3 dimension. The SPARQL queries and their results are shown in Table 5. The prefix and their namespaces used for SPARQL queries are shown in Table 6.
5.2 OOPS! Pitfall Evaluation
We have used OOPS! Pitfall scanner [19] to examine CovidO. OOPS! Pitfall ontology diagnosis online tool detects 40 different types of pitfalls in OWL ontologies, including semantic and structural checks and best practices verification. OOPS! Pitfall scanner has two components, Pitfall Scanner, and Suggestion Scanner. Pitfall Scanner checked the ontology syntax, and Pellet reasoner has analyzed the logical consistency of ontology. Whereas the Suggestion scanner has thrown some suggestions for possible errors of ontology elements. We resolved the problems reported by OOPS! in the CovidO and updated CovidO.
6 Conclusion
This work presents Coronavirus Disease Ontology (CovidO): (1) Describes complete knowledge of COVID-19; (2) Maps existing ontologies and create standard metadata to understand and share COVID-19 knowledge. (3) Acts as a vocabulary for researchers, engineers, and developers to find the commonly used COVID-19 gesticulation for particular accordance’s, and to use the scope and dynamics of a specific gesture. The intention and scope of the CovidO can be summarized in seven dimensions as discussed above. As future work, some possible use case will be added to make it more integrating linguistics. In addition, we intend to release and deploy the CovidO RESTful service in the Cloud and API clients to the leading coronavirus annotation [20]
References
World health organization. https://covid19.who.int/. Accessed 10 Feb 2022
Dividino R, Soares A, Matwin S, Isenor AW, Webb S, Brousseau M (2018) Semantic integration of real-time heterogeneous data streams for ocean-related decision making. Defense Research and Development Canada = Recherche et développement pour la ..
Osborne F, Motta E (2015) Klink-2: integrating multiple web sources to generate semantic topic networks. In: International semantic web conference. Springer, pp 408–424
Ding L, Kolari P, Ding Z, Avancha S (2007) Using ontologies in the semantic web: a survey. In: Ontologies. Springer, pp 79–113
Beverley J, Babcock S, Cowell L, Smith B (2021) The covid 19 infectious disease ontology
He Y, Yu H, Ong E, Wang Y, Liu Y, Huffman A, Huang HH, John B, Lin AY, Arabandi S et al (2021) Cido: the community-based coronavirus infectious disease ontology
Babcock S, Beverley J, Cowell LG, Smithd B (2020) The infectious disease ontology
Dutta B, DeBellis M (2020) Codo: an ontology for collection and analysis of covid-19 data. arXiv:2009.01210
de Lusignan S, Liyanage H, McGagh D, Jani BD, Bauwens J, Byford R, Evans D, Fahey T, Greenhalgh T, Jones N et al (2020) In-pandemic development of an application ontology for covid-19 surveillance in a primary care sentinel network. JMIR Publ Health Surveill
Patel A, Debnath NC, Mishra AK, Jain S (2021) Covid19-ibo: a covid-19 impact on Indian banking ontology along with an efficient schema matching approach. New Gener Comput 39(3):647–676
Reese JT, Unni D, Callahan TJ, Cappelletti L, Ravanmehr V, Carbon S, Shefchek KA, Good BM, Balhoff JP, Fontana T et al (2021) Kg-covid-19: a framework to produce customized knowledge graphs for covid-19 response. Patterns 2(1):100155
Badenes-Olmedo C, Chaves-Fraga D, Poveda-VillalÓn M, Iglesias-Molina A, Calleja P, Bernardos S, MartÍn-Chozas P, Fernández-Izquierdo A, Amador-Domínguez E, Espinoza-Arias P et al (2020) Drugs4covid: Drug-driven knowledge exploitation based on scientific publications. arXiv:2012.01953
Bonino L (2020) Who covid-19 rapid version crf semantic data model. BioPortal
Qundus JA, Schäfermeier R, Karam N, Peikert S, Paschke A (2021) Roc: an ontology for country responses towards covid-19. arXiv:2104.07345
Pinto HS, Staab S, Tempich C (2004) Diligent: towards a fine-grained methodology for distributed, loosely-controlled and evolving engineering of ontologies. In: ECAI, vol 16. Citeseer, p 393
Fernández-López M, Gómez-Pérez A, Juristo N (1997) Methontology: from ontological art towards ontological engineering
Pinto HS, Martins J (2000) Reusing ontologies. In: AAAI 2000 spring symposium on bringing knowledge to business processes, vol 2. AAAI, Karlsruhe, Germany, p 7
Seaborne A, Manjunath G, Bizer C, Breslin J, Das S, Davis I, Harris S, Idehen K, Corby O, Kjernsmo K et al (2008) Sparql/update: a language for updating rdf graphs. W3c Member Submission 15
Gómez-Péreza A, Oops!(ontology pitfall scanner!): supporting ontology evaluation on-line
Sharma S, Jain S (2021) Comprehensive study of semantic annotation: variant and praxis
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Sharma, S., Jain, S. (2023). The Coronavirus Disease Ontology (CovidO). In: Jain, S., Groppe, S., Bhargava, B.K. (eds) Semantic Intelligence. Lecture Notes in Electrical Engineering, vol 964. Springer, Singapore. https://doi.org/10.1007/978-981-19-7126-6_8
Download citation
DOI: https://doi.org/10.1007/978-981-19-7126-6_8
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-19-7125-9
Online ISBN: 978-981-19-7126-6
eBook Packages: Computer ScienceComputer Science (R0)