Abstract
The progressive damage of kidney function and structure exhibits a high risk of cardiovascular disease and is the leading cause of life-threatening chronic kidney disease (CKD). Early diagnosing is the only way to prevent CKD from worsening. Different data mining and machine learning approaches are implemented for predicting chronic kidney disease; however, the extraction of hidden details from the clinical data is a difficult challenge. To overcome this situation, a new model for accurate prediction of chronic kidney disease is proposed in this article. This work evaluated the proposed predictive model using a CKD dataset. The raw chronic kidney disease dataset is initially preprocessed using data cleaning, data reduction as well as data transformation steps. These steps make the classification system easier to predict diseased and normal classes by enhancing the data quality. The features are then selected using a hybrid flash butterfly optimization algorithm. Here, the weight functions of the kernel soft plus extreme learning machine (KSELM) method are optimally tuned using the hybrid flash butterfly optimization (HFBO) algorithm to increase prediction accuracy. The efficiency of the proposed HFBO-KSELM algorithm is examined using different evaluation measures namely precision, accuracy, and specificity, recall, F1-score, and computation time. The results of the experimental analysis display superior performance of the proposed HFBO-KSELM algorithm over other existing methods, particularly with an accuracy of 97.8%, precision of 97.4%, recall of 96.9%, specificity of 97.5%, F1-score of 97.1%, computational time of 2.3 s, training accuracy of 0.978, testing accuracy of 0.94 and ROC/AUC of 0.97. Finally, the KSELM algorithm accurately predicts and classifies them into benign and malignant classes. The experimental result showed that the proposed HFBO-KSELM algorithm provides the effectiveness and robustness to detect chronic kidney disease.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Chronic kidney disease (CKD) is mostly found among adults. It means kidney damage due to not filtering the blood. The main function of the kidney is filtering the excess waste and water out of the blood for the urine. Therefore, the kidney is the main organ that has been cared for more attentively. Moreover, chronic kidney disease (CKD) causes continuous loss of the Glomerular filtration rate (GRF) for 3 months is the early stage of kidney disease. For the past, some years, more than l million people are affected by renal failure and passed away due to frequent dialysis and kidney replacement surgery. So, kidney disease has to be predicted from the early stage by utilizing the Machine Learning technique. The prediction of machine Learning is done based on the patient’s disease’s hidden pattern [1, 2]. The development of kidney disease happens without manifesting the symptoms. So, doctors use the Machine Learning technique which is used for predicting CKD. Moreover, numerous machine learning techniques are trained based on the medical data of the patient. The research aspires to resolve several algorithms namely Linear Discriminant Analysis (LDA), Gradient Boosting (GB) Support Vector Machine (SVM), and AdaBoost (AB) to categorize kidney-diseased people. All the ignored value is solved via the imputation method of K-nearest Neighbors [3]. Categorization and Feature extraction is the crucial process of machine learning techniques. The feature extraction function helps in extracting the supreme and appropriate features from the dataset. By following the extraction of the feature, the classification function is performed [20]. Traditional machine learning spent additional time for calculation since two algorithms are employed for feature classification and extraction. Moreover, Convolution Neural Network (CNN) algorithm is utilized alternative to conventional machine learning in biomedical signal processing techniques. The integration process of extraction and classification has happened in the CNN model. This merge erases the necessity of individual feature extraction modules. Hence, Cross-correlation is utilized for enhancing the calculation speed and accuracy [4].
The diseases like high blood pressure, unhealthy lifestyles, and diabetes are also reasons for the cause of CKD. So, the damage has been found in the immune and nervous systems that lead to disability in people. So, another machine learning technique is utilized for diagnosing CKD and prevents from CKD. ANN is the numerical model that trains appropriately to produce a correct solution. SVM separates binary data and attained the increased distance between the labeled data. As compared with SVM, ANN has attained 97.75% [5]. A saliva-based diagnosis has attained several advantages by detecting in a non-invasive way. AU680 analyzer calculates the sample analyzed with appropriate reagents [6]. The optimization issues are solved by bio-inspired algorithms like The Fruit fly optimization algorithm for Feature selection (FS) [7]. Cloud Computing is also applicable to medical services by employing a hybrid intelligent model to predict CKD [8]. As a whole, the result reveals that DT, GBT, and SVM classifiers along with chosen features have achieved good performance at 100% accuracy [9]. The advantage of utilizing the neural networks is that this model uses the hidden layer for automatic feature combinations. It gains the physical indicators and semantic data in Electronic Health Records (EHR) to predict kidney disease [10]. This paper develops an automated intelligent chronic kidney disease prediction model to help medical practitioners in identifying normal and abnormal classes accurately. The important contributions of this article are described as follows:
-
A novel Hybrid Flash Butterfly Optimization-based Kernel Softplus Extreme Learning Machine (HFBO-KSELM) approach is proposed for the accurate prediction of chronic kidney disease.
-
The hybrid flash butterfly optimization (HFBO) algorithm selects optimal features and Kernel soft plus extreme learning machine (KSELM) is employed to improve the accuracy of the classifier.
The remaining part of this article is, in Sect. 2 the existing works are based on Chronic Kidneys. The proposed methodology based on Chronic Kidney Disease detection is presented in Sect. 3. The experimental results are explained in Sect. 4. Finally, Sect. 5, concludes the article.
2 Related works
As mentioned in Table 1, Ma et al. [11] developed the detection of CKD by using the deep learning model. In this paper, Heterogeneous Modified Artificial Neural Network (HMANN) was introduced to detect diseases early, segmental, and diagnose chronic kidney diseases by the Internet of Medical Things (IoMT) platform. The HMANN technique tested three classifiers namely, which were obtained to find a better solution for this issue. As a result, the HMANN method improved the accuracy rate and minimized the computational time in the process of kidney stone prediction. On the other hand, it requires high processing time for big neural networks. Khamparia et al. [12] developed a concept using a Deep Stacked AutoEncoder (DSAE) method to classify CKD with multimedia data learning techniques. The main aim of this paper was the prediction of CKD at an early stage and it helps to reduce the treatment cost for patients. This paper introduced the Deep Stacked autoencoder (SAE) method for diagnosing CKD. SAE consists of two autoencoders stacked with a softmax classifier. However, a single-stacked autoencoder was unable to reduce input feature dimensionality.
Qin et al. [13] illustrated a Diagnosing chronic kidney disease using a machine-learning methodology. The main objective of machine learning is to detect disease fast and accurately. The K-Nearest Neighbor (KNN) method was used for loading the values lost from the dataset. The established method achieved better performance by obtaining the KNN, RF, Feed-forward Neural Network (FNN), Logistic regression (LOG), Naïve Bayes classifier and SVM. Various parameters were obtained for evaluating the performance. But, this paper failed to collect more representative and complex data.
Bhaskar and Manikandan [14] established a deep-learning method for the automated diagnosis of chronic kidney disease. The CNN method was developed for detecting Chronic Kidney Disease (CKD) accurately. The introduced sensing approach is tested and evaluated by physicians. As the result, detecting CKD in deep learning techniques was more effective. Meanwhile, it does not perform very well when the data set has high noise.
Jerlin and Perumal [15] reviewed the diagnosis of Chronic Kidney Disease (CKD) classification by a Multi-Kernel Support Vector Machine and Fruit Fly Optimization (MKSVM-FLO) Algorithm. The CKD dataset contained 400 sample images for patients. Some evaluation parameters such as accuracy, specificity, and sensitivity were applied to predict the efficiency. On the other hand, this method missed classifying all data sets.
Rubini and Perumal [16] developed a classification of chronic kidney disease based on a Hybrid Kernel Support Vector Machine and Gray Wolf Optimization (HKSVM-GWO) method. Medical data required a lot of training and information for prediction and reconstruction. So, in this paper HKSVM-GWO algorithm was introduced to classify and predict Chronic Kidney Disease (CKD). The experimental analyses were carried out and demonstrated that HKSVM-GWO achieved a high accuracy rate of 97.26%. On the other hand, the system performance was not enhanced.
Siddhartha et al. [17] established a diagnosis of chronic kidney disease at an early stage using a Majority Vote-Grey Wolf Optimization (MV-GWO) algorithm. Controlling Chronic Kidney Disease (CKD) at an early stage was very important as it minimized the mortality rate and treatment cost. The MV-GWO algorithm was introduced to detect CKD using a CKD data set. The data set was collected from the UCI repository. They used different evaluation metrics to validate the introduced method, such as accuracy, sensitivity and F1-score to achieve better performance. Meanwhile, it is processed at in slow convergence speed and it falls easily into the local optimum.
Elhoseny et al. [18] reviewed a concept for diagnostic classification and prediction for chronic kidney disease. The Density-based feature selection with Ant Colony-based Optimization (D-ACO) algorithm was introduced for the detection and classification of Chronic Kidney Disease (CKD) in healthcare services. The obtained performance metrics evaluated the parameters and enhanced the performance. The main disadvantages of this optimization were premature consolidation and stagnant output.
Rady and Anwar [19] illustrated the data mining techniques for predicting the severity stages of chronic kidney disease. The Probabilistic Neural Network (PNN) algorithm was used to identify the diagnostic errors. As a result, the data mining algorithm classified the severity stage and achieved the highest classification accuracy of 96.7%. Meanwhile, the PNN algorithm was slow compared to other existing algorithms and it required more memory.
Pasadana et al. [20] discussed the kidney disease prediction by utilizing decision tree (DT). The objective of this paper was to compute the DT algorithms performance as well as comparing their performances. The UCIs CKD dataset was utilized for evaluating the performance and it contains attributes. The experimentation results showed that the design acquired higher accuracy for identifying the chronic kidney disease (CKD). But this paper failed to develop the other advanced DT algorithms.
Jongbo et al. [21] analyzed the ensemble approaches bagging and random subspace for the development of the chronic kidney disease diagnosis model. This paper aimed at enhancing the classification performance of the model by Naïve Bayes, Decision Tree and k Nearest Neighbors (KNN) classifiers. The result found that the KNN classifier based on a random subspace ensemble obtained a prediction accuracy. On the other hand, it was more expensive because it stored all the training data.
In recent studies, few papers based on chronic kidney disease prediction are referred and reviewed with certain drawbacks such as high expensive, high complexity, premature consolidation, stagnant output, slow convergence speed, misclassification of datasets, high dimensionality, high processing time, etc. Hence, the HFBO-KSELM algorithm is proposed for overcoming these limitations. The proposed HFBO-KSELM algorithm provides the effectiveness and robustness to detect chronic kidney disease.
3 Proposed methodology
Chronic Kidney Disease (CKD) is considered a serious health disorder and its prevalence is increasing constantly. Despite taking numerous efforts to control its progression, it remains a major health burden because it shows no symptoms until it reaches an advanced stage. However, chronic kidney disease can be detected on time using an efficient machine learning algorithm. To fulfill this requirement, this paper proposes a new chronic kidney disease prediction model using Hybrid Flash Butterfly Optimization-based Kernel Softplus Extreme Learning Machine (HFBO-KSELM) approach.
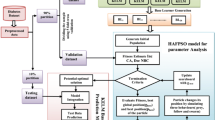
The raw chronic kidney disease dataset is initially preprocessed using data cleaning, data reduction as well as data transformation steps. These steps make the classification system easier to predict diseased and normal classes by enhancing the data quality. The features are then selected using a Hybrid Flash Butterfly Optimization algorithm. Finally, the KSELM algorithm accurately classifies benign and malignant classes separately. The architecture of the proposed model is presented in Fig. 1.
3.1 Data pre-processing
Before the process of classification, the data needs to be altered and cleaned [11]. Data pre-processing is defined as the transformation of raw data into an understandable design. The data preprocessing is considered the most significant phase and the data quality needs to be evaluated before providing it to the machine learning techniques. The data pre-processing includes some the factors such as data reduction, data transformation, and data cleaning.
Data Cleaning Data cleaning processes remove inaccurate, incomplete as well as incorrect data from the dataset. Also, the missing values are replaced during the cleansing process.
Data Reduction The data reduction process assists in minimizing the data volume which makes the evaluation easier. In addition to this, reducing data minimizes storage space. During the reduction process, the dimensionality of the data is minimized and the data is compressed.
Data Transformation Data transformation refers to the changes made in the data structure or data format. The data transformation may either be simple or complex based on input data. The data transformation process includes smoothing, normalization as well as aggregation.
3.2 Feature selection phase
The feature selection phase simplifies the overfitting issues, data realization as well as a storage size and minimizes the training cost to achieve high accuracy. The performances of the classifier are enhanced by selecting an appropriate feature and minimizing the computational time. In this paper, a Hybrid Flash Butterfly Optimization (HFBO) Algorithm is employed to select optimal features with greater accuracy.
3.2.1 Hybrid flash butterfly optimization (HFBO) algorithm
The firefly algorithm’s (FA) search strategy has taken and utilized butterfly vision for the local optimization in HFBO. The HFBOs every stage with the optimization phase is presented the switching parameter settings [22].
3.2.2 Initialization phase
The butterflies’ population initialization is performed as irregular data. The initial statistical function expression is given as follows:
The above equation, \(Y_{{{\text{mc}},k}}\) and \(Y_{{{\text{vc}},k}}\) represent the lower and upper boundary of the problem accordingly, \({\text{rand}}\) define a random number in [0, 1], and \(Y_{j,k}\) defines the \(j\) th solution for \(k\) th dimension. This strategy is utilized for the population initialization position of the swarm intelligence algorithms.
3.2.3 Solution encoding
The main objective of solution encoding is to minimize redundant features and error percentages between predicted and actual features for categorizing benign and malignant classes of chronic kidney disease datasets. If the dataset contains \(x\) number of features, \(1 + x\) decision variables are taken into account for determining bandwidth and selecting features in the HFBO algorithm. Among total variables, the initial one is assumed as kernel bandwidth while the remained ones are assumed as features. The variable’s value lies in the range [0, 1]. When the variable value becomes larger than 0.5, the related features are selected or else not.
3.2.4 Fitness determination
The HFBO algorithm then transforms the encoded solution into binary values as 0 and 1 to signify the data chosen from the data. If the feature is chosen from the data, the solution vector’s dimension \(z_{j}^{{{\text{Dim}}}}\) is represented by ‘1’ and if the feature is not chosen from the data, the solution vector’s dimension is depicted as’0’. The below expression signifies the transformation of the solution vector to binary values,
The fitness is numerically derived as,
Here the terms \(\omega_{1}\) \(\omega_{2}\) indicate the weight values of error rate and selected feature respectively where, \(\omega_{1} = [0,1]\) and \(\omega_{2} = 1 - \omega_{1}\). Also, the term \({\text{accuracy}}\;({\text{KSELM}})\) depicts the accurate verification rate utilizing KSELM; \(n^{w}\) and \(n^{ * }\) represents wrongly classified data samples and correctly classified data samples, respectively.
3.2.5 Optimization phase
The optimization phase \(G_{j}^{s + 1}\) is computed in the following equation.
During the HFBO search stage, sensory modality \(d\) sets to an irregular number in (0,1). The confused and disordered strategy is utilized for updating the one-dimensional chaotic mapping because of the interval (0, 1) of the parameter \(d\). This strategy is used for the swarm intelligence algorithm’s population initialization position.
3.2.6 Global search
\(\delta\) is replaced by the parameter \(q\) and the global search movements and are calculated in the below equation.
The above equation \(Y_{j}^{s}\) defines the solution vector \(Y_{j}\) of the \(j\) th iteration and \(\delta\) is represented as a random number in (0, 1), \(h_{b}\) representing the current best location identified in all the solutions in the present stage. The Parameter \(\delta\) is considered a scaling factor and is used for adjusting the distance between the best solution and \(j\) th butterfly.
3.2.7 Local search
The optimal values are searched by the individuals and the two phases of HFBO are changed. The butterfly’s vision is taken into the HFBO’s local search phase. The numerical expression of the butterfly’s search stage is given below.
From the above equation, \(Y_{j}^{l}\) and \(Y_{k}^{s}\) are the search agents in local search space. \(\varepsilon\) denotes random value like \(\varepsilon \in \left[ { - 0.5,\,0.5} \right]\) and \(\delta\) represents a random number in [0,1].
3.2.8 Switch parameter (SP)
Conversion of intensive local search and normal global search is done by setting the switch parameter. The random production of number in every iteration is [0, 1] is happened for conducting a local or global search with the comparison of SP. Then the local search stage is performed by setting the value to 0 and the global search is performed by SP to 1.
3.2.9 Chaotic map and parameter \(\delta\)
Chaos is the usual event in nonlinear systems. The one-dimensional map’s classical mapping is the logistical mapping and it is expressed in the below equation.
From the above equation, \(\alpha\) represents the chaotic factor, \(\alpha\) \(\alpha \, \in \,\left( {0,4} \right)\).
3.2.10 Complexity analysis
The time complexity manifests the algorithm’s performance and it supports computing the time complexity algorithm. These happen when there is a limited duration to identify the value of global optimum. The final complexity of HFBO is given in the equation as follows:
From the above equation,\(m\) represents maximum evaluation, \(S_{n}\) represents maximum evaluation, \(P\left( {{\text{me}}} \right)\) represents the time complexity of the initialization phase, \(P\left( {S_{n} {\text{me}}} \right)\) represents calculation fitness of all agents, \(P\left( {S_{n} m^{2} \log m} \right)\) represents position updating, \(P\left( {S_{n} m} \right)\) represents the quick sort, and \(P\left( {S_{n} {\text{me}}} \right)\) is updating parameter cost.
3.3 Data classification
This section provides a comprehensive illustration of the proposed KSELM-based chronic kidney disease prediction model for accurate classification of benign and malignant classes. An elaborate description of the KSELM algorithm is delineated in the following sub-sections.
3.3.1 Kernel-based soft plus extreme learning machine (KSELM)
A kernel-based soft plus extreme learning machine (KSELM) with selected features is utilized to classify the chronic kidney disease dataset as normal and abnormal [23, 24]. The most advanced learning approach is ELM. Sometimes this ELM’s input and hidden layers are unsuitable because it chooses biased parameters and weight arbitrarily.
To shortcoming, this problem of ELM, kernel-based ELM is introduced in this paper. KELM removes the input and invisible layer’s weight initialization conception. The framework of the KELM model is presented in Fig. 2.
\((Y_{P\,} ,Z_{P} )\) is a training set that consists of n samples.\(H\) represent the hidden later in KELM. \(F_{Y}\) represent the ELM’s implementation function. This scenario is mathematically formulated as follows:
The output of the ELM is formulated as follows:
From the above equation, \(W_{P} \,{\text{and}}\,\alpha_{P}\) is represented as the output of the hidden neuron. The invisible layer in terms of input \(Y_{{}}\) is denoted as \(H(Y)\).\(B_{P}\) is represented as the dependence of the secret neuron.
Sometimes, the total number of invisible neurons is very low compared to the total number of training samples due to the Non-square matrix of parameter S. Since the matrix β is not squared it is necessary to obtain β to estimate the weight of the output neuron
The \(s *\) is represented as the outcome of the KELM needs weight parameter. The matrix S is denoted as the Moore–Penrose generalized inverse S∗.
From the above equation, the evaluation \(\alpha\) (output weight) is added to ELM during the kernel process by computing a positive value \(\frac{1}{V}\). \(V\) is represented as a boundary user-defined. The feature mapping of the secret layer is unrecognized, and then they introduce the ELM’s kernel function. \(\varphi\) is represented as the kernel function of the EML. This scenario is simulated as follows:
From the above equation, the Kernel function is represented as \(k(Y_{P} ,Y_{Q} )\). The kernel function output is defined as follows:
The Different kinds of kernel functions namely polynomial and radial basis functions are used with ELM.
Soft plus ELM The soft plus ELM is represented as follows:
The KSELM algorithm is evaluated using a chronic kidney disease dataset containing multivariate characteristics with 400 instances and 25 attributes. The preprocessed data are split into two divisions testing data and training data. The data which are selected for training are extracted and classified using the KSELM algorithm while the remaining data are directly tested to evaluate the classification accuracy of the model. To obtain better classification performance with minimal error and enhanced generalization, the soft plus activation function is utilized at the output layer. Thus, the KSELM algorithm effectively predicts the kidney disease instances and accurately segregates them into two distinct classes (ie. benign and malignant) as mentioned in Fig. 3.
4 Results and discussion
The Hybrid Flash Butterfly Optimization-based Kernel Softplus Extreme Learning Machine (HFBO-KSELM) algorithm is proposed for diagnosing chronic kidney disease. The Chronic Kidney Disease dataset is chosen for predicting CKD. The metrics namely precision, specificity, recall, accuracy, F1-score and computational time were utilized for performance evaluation. The implementation is conducted under the Python platform which contains an Intel Core i5 processor, 8 GB RAM, windows10 and a 64-bit operating system.
4.1 Dataset description
The dataset of chronic kidney disease (CKD) is collected from the website https://archive.ics.uci.edu/ml/datasets/Chronic_Kidney_Disease. The dataset is acquired from the UCI repository which consists of sample data from 400 patients. Different classes of characteristics are considered Table 2 depicts various attributes of chronic kidney disease data.
4.2 Hyperparameter configuration
The parameter configuration of the proposed HFBO-KSELM algorithm is described in Table 3.
4.3 Evaluation metrics
To validate the performance of the proposed HFBO-KSELM technique, a few metrics namely precision, accuracy, specificity, recall, F1-score, and computational time were utilized. True positive (\(N_{{{\text{true}}\;{\text{positive}}}}\)) signifies the correctly predicted positive class, false negative (\(N_{{{\text{false}}\,{\text{negative}}}}\)) indicates the inaccurately diagnosed samples, false positive (\(N_{{{\text{false}}\,{\text{positive}}}}\)) represents the positive samples were mistakenly diagnosed as negative and true negative (\(N_{{{\text{true}}\,{\text{negative}}}}\)) denotes the correctly predicted negative class.
4.3.1 Accuracy (\(A_{c}\))
The accuracy refers to the probability of accurately classified samples to the total samples that are expressed as,
4.3.2 Precision (\(P_{r}\))
Precision is computed as the ratio between the number of true positives to the sum of false positives and true positives. The precision can be derived by using the below equation,
4.3.3 Recall (\(R\))
The ratio among the total number of accurately classified positive samples to the sum of true positive samples and false negative samples is referred to as recall. The recall is derived as,
4.3.4 F1-score ( \(F\) )
F1-score is the harmonic mean of precision and recall that is formulated as below,
4.3.5 Specificity (\(S\))
Specificity is calculated as the probability between the number of true positives to the sum of total false positives and true negatives. The sensitivity is denoted as,
4.3.6 Computational time
The computational time is the time required to complete their task or computational process and the unit of the computational time is seconds.
4.4 Performance evaluation
The ROC/AUC curve is plotted between recall and specificity which is described in Fig. 4. ROC represents the trade-off between recall and specificity as well as AUC is the probability of random positive samples being positioned to the right side of the random negative samples. The proposed HFBO-KSELM algorithm shows a ROC/AUC rate of 0.97 which is the best performance rate compared to other previous approaches. The HFBO-KSELM algorithm accurately predicts chronic kidney disease, due to its high performance of ROC/AUC.
Each class has two types of cases such as benign and malignant. The performance rate of the proposed HFBO-KSELM algorithm by using various metrics is described in Table 4.
The two different periods of training accuracy and testing accuracy are denoted in Fig. 5. Here, 80 percent of data is selected for training accuracy, and the residual 20 percent of data is given to testing accuracy. The training accuracy of the proposed HFBO-KSELM algorithm is 0.978 and the testing accuracy of the proposed HFBO-KSELM algorithm is 0.94.
The actual class and prediction class of the proposed HFBO-KSELM algorithm is explained in the confusion matrix formation that is represented in Fig. 6.
4.5 Comparative analysis
For performance evaluation, a few state-of-the-art methods namely KSELM [23], HKSVM-GWOalgorithm [16], MKSVM-FLOalgorithm [15], DL-HMANN [11], MV-GWO algorithm [17], are compared to proposed HFBO-KSELM algorithm.
Figure 7 portrays the precision rate of various methods namely KSELM, MKSVM-FLO, HKSVM-GWO, MV-GWO, DL-HMANN, and proposed KSELM-HFBO. From this comparative analysis, the proposed HFBO-KSELM algorithm attains a high-performance rate of 97.4% which effectively diagnoses chronic kidney disease and the MKSVM-FLO algorithm has a low precision rate of 80%. Remaining methods like KSELM, HKSVM-GWO, MV-GWO, and DL-HMANN attained the precision rate of 86%, 91%, and 93%, respectively. The proposed HFBO-KSELM algorithm gives better outcomes rather than other state-of-the-art methods.
Recall analysis is performed by using different methods namely KSELM, MKSVM-FLO, HKSVM-GWO, MV-GWO, DL-HMANN, and proposed HFBO-KSELM depicted in Fig. 8. The proposed HFBO-KSELM algorithm comprises a superior recall rate of 96.9% and the MKSVM-FLO algorithm has the lowest recall rate of 78%.
In Fig. 9, the analysis is carried out and the graph is plotted for the proposed HFBO-KSELM approach with other existing approaches like KSELM, MKSVM-FLO, HKSVM-GWO, MV-GWO, and DL-HMANN. The experimental evaluation is conducted for each respective method to effectively forecast chronic kidney disease and the results showed that the specificity rate is acquired at 97.5%.
Figure 10 illustrates the comparative analysis of the F1-score and the graph is plotted for the KSELM approach with other existing approaches like KSELM, MKSVM-FLO, HKSVM-GWO, MV-GWO, and DL-HMANN. The results and observations showed that the proposed HFBO-KSELM in classifying chronic disease achieves a high F1-score of 97.1%.
4.6 Discussion
Figure 11 denotes the accuracy of different methods like KSELM, MKSVM-FLO, HKSVM-GWO, MV-GWO, DL-HMANN, and proposed KSELM-HFBO. The proposed HFBO-KSELM algorithm has a high performance in the prediction of CKD because this proposed HFBO-KSELM technique achieves a high accuracy rate of 97.8%. Other state-of-the-art methods such as KSELM, MKSVM-FLO, HKSVM-GWO, MV-GWO, and DL-HMANN comprise the accuracy rate of 84%, 88%, 90%, and 95%, respectively. Figure 12 illustrates the comparative analysis of computational time the graph is plotted for the proposed KSELM approach with other existing approaches like KSELM, MKSVM-FLO, HKSVM-GWO, MV-GWO, and DL-HMANN. The performances are validated by the computational time for a selected classifier. The computational time for classifiers is considered. The Outcome showed that the proposed approach achieved a low computational time of 2.3 s when compared to other approaches.
5 Conclusion
The proposed HFBO-KSELM algorithm contains two kinds of classes for deleting chronic kidney disease. The CKD dataset is acquired from the UCI repository which consists of sample data from 400 patients. To validate the performance of the proposed HFBO-KSELM technique, a few metrics namely precision, F1-score recall, accuracy, specificity, and computational time were utilized. For performance evaluation, a few state-of-the-art methods namely the MKSVM-FLO algorithm, HKSVM-GWO algorithm, MV-GWO algorithm, and DL-HMANN are compared to the proposed HFBO-KSELM algorithm. In the comparative analysis, the proposed HFBO-KSELM algorithm attained an accuracy of 97.8%, a precision of 97.4%, a recall of 96.9%, a specificity of 97.5%, F1-score of 97.1%, a computational time of 2.3 s, training accuracy of 0.978, testing accuracy of 0.94 and ROC/AUC of 0.97, respectively. The experimental result showed that the proposed HFBO-KSELM algorithm provides the effectiveness and robustness to detect chronic kidney disease. Meanwhile, the cost required for implementation is high and hence various other chronic diseases cannot be determined. In future, we will predict various other chronic trouble factors such as diabetes, hypertension, and kidney failure family history by utilizing a new dataset.
Data availability
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
Code availability
Not applicable.
References
Wang W, Chakraborty G, Chakraborty B (2020) Predicting the risk of chronic kidney disease (ckd) using machine learning algorithm. Appl Sci 11(1):202. https://doi.org/10.3390/app11010202
Salkar C (2021) A detailed analysis on kidney and heart disease prediction using machine learning. J Comput Nat Sci 1:9–14
Ghosh P, Shamrat FJM, Shultana S, Afrin S, Anjum AA, Khan AA (2020) Optimization of prediction method of chronic kidney disease using machine learning algorithm. In: 2020 15th International Joint Symposium on Artificial Intelligence and Natural Language Processing (iSAI-NLP), pp 1–6. https://doi.org/10.1109/iSAI-NLP51646.2020.9376787
Bhaskar N, Suchetha M (2021) A computationally efficient correlational neural network for automated prediction of chronic kidney disease. IRBM 42(4):268–276. https://doi.org/10.1016/j.irbm.2020.07.002
Al-Wahsh H, Lam NN, Quinn RR, Ronksley PE, Sood MM, Hemmelgarn B, Tangri N, Ferguson T, Tonelli M, Ravani P, Liu P (2022) Calculated versus measured albumin-creatinine ratio to predict kidney failure and death in people with chronic kidney disease. Kidney Int. https://doi.org/10.1016/j.kint.2022.02.034
Navaneeth B, Suchetha M (2020) A dynamic pooling based convolutional neural network approach to detect chronic kidney disease. Biomed Signal Process Control 62:102068. https://doi.org/10.1016/j.bspc.2020.102068
Berchtold L, Crowe LA, Combescure C, Kassaï M, Aslam I, Legouis D, Moll S, Martin PY, de Seigneux S, Vallée JP (2022) Diffusion-magnetic resonance imaging predicts decline of kidney function in chronic kidney disease and in patients with a kidney allograft. Kidney Int 101(4):804–813
Abdelaziz A, Salama AS, Riad AM, Mahmoud AN (2019) A machine learning model for predicting of chronic kidney disease based internet of things and cloud computing in smart cities. Security in smart cities: models, applications, and challenges. Springer, Cham, pp 93–114
Abdel-Fattah MA, Othman NA, Goher N (2022) Predicting chronic kidney disease using hybrid machine learning based on apache spark. Comput Intell Neurosci
Ren Y, Fei H, Liang X, Ji D, Cheng M (2019) A hybrid neural network model for predicting kidney disease in hypertension patients based on electronic health records. BMC Med Inform Decis Mak 19(2):131–138
Ma F, Sun T, Liu L, Jing H (2020) Detection and diagnosis of chronic kidney disease using deep learning-based heterogeneous modified artificial neural network. Futur Gener Comput Syst 111:17–26. https://doi.org/10.1016/j.future.2020.04.036
Khamparia A, Saini G, Pandey B, Tiwari S, Gupta D, Khanna A (2020) KDSAE: chronic kidney disease classification with multimedia data learning using deep stacked autoencoder network. Multim Tools Appl 79(47):35425–35440
Qin J, Chen L, Liu Y, Liu C, Feng C, Chen B (2019) A machine learning methodology for diagnosing chronic kidney disease. IEEE Access 8:20991–21002
Bhaskar N, Manikandan S (2019) A deep-learning-based system for automated sensing of chronic kidney disease. IEEE Sens Lett 3(10):1–4. https://doi.org/10.1109/LSENS.2019.2942145
Jerlin Rubini L, Perumal E (2020) Efficient classification of chronic kidney disease by using multi-kernel support vector machine and fruit fly optimization algorithm. Int J Imaging Syst Technol 30(3):660–673
Rubini LJ, Perumal E (2020) Hybrid kernel support vector machine classifier and grey wolf optimization algorithm based intelligent classification algorithm for chronic kidney disease. J Med Imaging Health Inf 10(10):2297–2307
Siddhartha M, Kumar V, Nath R (2022) Early-stage diagnosis of chronic kidney disease using majority vote–Grey Wolf optimization (MV-GWO). Heal Technol 12(1):117–136
Elhoseny M, Shankar K, Uthayakumar J (2019) Intelligent diagnostic prediction and classification system for chronic kidney disease. Sci Rep 9(1):1–14
Rady EHA, Anwar AS (2019) Prediction of kidney disease stages using data mining algorithms. Inf Med Unlocked 15:100178. https://doi.org/10.1016/j.imu.2019.100178
Pasadana IA, Hartama D, Zarlis M, Sianipar AS, Munandar A, Baeha S, Alam ARM (2019) Chronic kidney disease prediction by using different decision tree techniques. J Phys Conf Ser 1255(1):012024
Jongbo OA, Adetunmbi AO, Ogunrinde RB, Badeji-Ajisafe B (2020) Development of an ensemble approach to chronic kidney disease diagnosis. Sci Afr 8:e00456. https://doi.org/10.1016/j.sciaf.2020.e00456
Chaki J, Dey N (2020) Texture feature extraction techniques for image recognition. Springer, Singapore
Sasank VVS, Venkateswarlu S (2021) Brain tumor classification using modified kernel based softplus extreme learning machine. Multim Tools Appl 80(9):13513–13534
Zhang M, Wang D, Yang J (2022) Hybrid-flash butterfly optimization algorithm with logistic mapping for solving the engineering constrained optimization problems. Entropy 24(4):525. https://doi.org/10.3390/e24040525
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
All authors agreed on the content of the study. PY and SCS collected all the data for analysis. PY agreed on the methodology. PY and SCS completed the analysis based on agreed steps. Results and conclusions are discussed and written together. The author read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Ethical approval
This article does not contain any studies with human participants.
Human and animal rights
This article does not contain any studies with human or animal subjects performed by any of the authors.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yadav, P., Sharma, S.C. HFBO-KSELM: Hybrid Flash Butterfly Optimization-based Kernel Softplus Extreme Learning Machine for Classification of Chronic Kidney Disease. J Supercomput 79, 17146–17169 (2023). https://doi.org/10.1007/s11227-023-05337-6
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11227-023-05337-6