Abstract
Hydrogen is an attractive energy carrier because of its high energy density, and used as a raw material in various chemical processes. Nowadays, hydrogen demand is supplied from non-renewable sources, and alternative sources are becoming mandatory. Hydrogen production by biological methods uses renewable resources as substrate and its production occurs at ambient temperature and atmospheric pressure. Thus, it is less energy intensive than the chemical and thermochemical methods used to produce hydrogen. This review is focused on fermentative hydrogen production by Escherichia coli. The hydrogen production pathway, the genetic manipulations, and expression of non-native pathways into this microorganism are reviewed. The hydrogen production using alternative substrates is a critical point to develop sustainable process by this reason the principal substrates for hydrogen production using E. coli are revised. Other strategies like two stages processes and immobilized cells are also discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Fossil fuels are the primary source of energy used to satisfy world’s energy demand, and their intensive use has caused an accelerated consumption of non-renewable resources (Davila-Vazquez et al. 2008). It has been suggested that depletion of fossil resources will lead to an energy crisis in the near future (Kapdan and Kargi 2006). In addition, there is now a general scientific consensus that observed trends in global warming are being caused by fossil fuel combustion and anthropogenic emissions of greenhouse gases (Luque et al. 2008). These issues have lead to explore new energy sources that could substitute fossil fuels, and be environmentally friendly and renewable.
Hydrogen is a promising fuel as it has a higher energy content than oil (142 MJ/kg for hydrogen vs. 42 MJ/kg for oil) (Nurul Islam et al. 2005) and its combustion results only in water and energy. Hydrogen is not only used as a fuel carrier; it is widely used by the chemical industry for the production of ammonia and methanol as well for the hydrogenation of fats and oils in the food industry, production of electronic devices, steel processing and re-formulation of gasoline in refineries (Ramachandran and Menon 1998; Kapdan and Kargi 2006; Fonseca et al. 2008).
At present, 40 % hydrogen is produced from natural gases, 30 % from heavy oil and naphtha, 18 % from coal, 4 % from electrolysis and only about 1 % from biomass (Sinha and Pandey 2011). These processes require high temperatures or pressures or both. If hydrogen production is based on fossil products or the processes to obtain this gas require high energy, then hydrogen is not the solution to solve the growing energy requirements. In the present scenario, biological hydrogen production processes are becoming important.
2 Biological hydrogen production
The main advantages of biological production are the use of renewable resources as substrate and its operation at ambient temperature and atmospheric pressure. Besides, it is less energy intensive than chemical and thermochemical methods used to produce hydrogen. Biological hydrogen production processes can be classified into three major categories: biophotolysis of water using algae and cyanobacteria, photofermentation of organic compounds by photosynthetic bacteria and dark fermentative production (Hallenbeck 2005).
In biophotolysis, photosynthetic organisms use solar energy to split water, producing O2 and reduced ferredoxin, the latter can reduce a hydrogenase or nitrogenase, producing hydrogen (Hallenbeck and Ghosh 2009). The main advantage of this process is the abundance of substrate and simple products, whereas the disadvantages are low conversion efficiencies, sensibility of hydrogenase to oxygen and light dependence (Hallenbeck and Ghosh 2009). The anaerobic photosynthesis carried out by non-sulfur purple bacteria is called photofermentation. In this process the solar energy is used to produce ATP and high-energy electrons that reduce ferredoxin. ATP and reduced ferredoxin drive proton reduction to hydrogen by nitrogenase enzyme. These organisms cannot obtain electrons from water and therefore use organic compounds. The main disadvantages are low conversion efficiencies and the expensive photo-bioreactors required. The advantage of this process is the use of organic acid wastes as substrate allowing the use of residues of dark fermentation as substrate (see below) and increasing the overall hydrogen yield.
3 The Dark fermentation pathway
A variety of different microorganisms can be used anaerobically to break down mainly carbohydrate-rich substrates into hydrogen and other products, principally organic acids (lactic, acetic, butyric, etc.) and alcohols (ethanol, butanol, etc.). Final products depend of type of microorganism, oxidation state of the substrate and environmental conditions (Hallenbeck and Ghosh 2009). For hydrogen production by dark fermentation both, axenic or non-axenic cultures could be used.
Hypothetically up to 12 mol of hydrogen can be obtained per mole of glucose, but there are no single metabolic pathways in nature that would allow this reaction. The theoretical yields of hydrogen from dark fermentations depend on the type of organisms that are used in fermentation (Mathews and Wang 2009). Facultative anaerobes such as Escherichia coli produce a maximum yield of 2 mol of hydrogen from each mole of glucose consumed, whereas other enterobacteria such as Enterobacter cloacae produce 4 mol (Redwood et al. 2009). Both of these microorganisms produce hydrogen from formate. Sequences analysis of hydrogenase 3 (Hyd 3) large subunit from E. coli and hydrogenases of E. cloacae (hydrogenase 3 large subunit and Fe-hydrogenase) is shown in Fig. 1. As noted, these large-subunit sequences of Hyd 3 of E. coli and E. cloacae show high identity. The presence of other hydrogenases in E. cloacae and the higher hydrogen yield of this microorganism implicate the simultaneous activity of NADH pathway in which the regeneration of NAD is coupled to the reduction of ferredoxin by NADH: ferredoxin oxidoreductase (NFOR), and the formate pathway to achieve this yield.
Dark fermentation seems to be the best promise for biohydrogen production due its low cost, rapid production rates, no direct solar input needed, and stable hydrogen-producing enzymes. Also, organic wastes from agriculture or sewage can be used into anaerobic bioreactors, achieving the dual goals of waste management and hydrogen production (Chittibabu et al. 2006). Dark fermentations also solve the problem of expensive photo-bioreactors, which are necessary for direct biophotolysis and photofermentations, whereas the weaknesses are the low hydrogen yields and the large quantities of side products formed (ethanol and organic acids such as acetate, lactate, succinate and butyrate).
4 Hydrogen production by Escherichia coli
E. coli can grow in the presence or absence of oxygen. In both conditions, glucose is transported and catabolized to pyruvate, but the further metabolism of pyruvate is different. In aerobic condition, the glycolysis and the Krebs cycle generate NADH, which it is reoxidized by the respiratory chain. Under anaerobic condition, NADH is still produced by glycolysis, but the respiratory chain is not working and NADH must be reoxidized to continue the glycolysis process. Thus, the key issue of fermentation is to recycle the NADH by the conversion of pyruvate to fermentation products (Clark 1989).
Figure 2 shows the fermentative pathway in E. coli. Carbohydrates are catabolized to phosphoenolpyruvate, which can be converted to oxaloacetate, by incorporation of CO2 by phosphoenolpyruvate carboxylase (PPC). Oxaloacetate is further converted to malate, fumarate and finally to succinate. A pathway from decarboxylate succinate to propionate was proposed (Haller et al. 2000; Froese et al. 2009), and some works reported propionate in E. coli fermentations (Jian et al. 2010; Zhang et al. 2010; Rosales-Colunga et al. 2010a; Redwood et al. 2012). Nevertheless, the metabolic function of this pathway remains uncertain. Most of the phosphoenolpyruvate is transformed to pyruvate, which is broken down into formate and acetyl-CoA by the pyruvate formate lyase (PFL). The formate is converted to hydrogen and CO2 by the formate-hydrogen-lyase complex (FHL), whereas acetyl-CoA yields acetate or ethanol. There is evidence for a pathway that uses acetyl-CoA to butyrate formation via crotonyl-CoA (Lugg et al. 2008), and butyrate production has been reported in E. coli fermentations under particular culture conditions (Blackwood et al. 1956; Redwood et al. 2012).
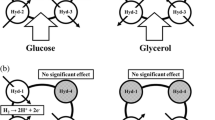
Under circumstances of high pyruvate accumulation, this may be converted to lactate by lactate dehydrogenase enzyme (LDH) (Clark 1989). As mentioned above the pathway to produce hydrogen involves formate production. Thus, formate metabolism is important for hydrogen production. There are three formate dehydrogenases (FDH) in E. coli, FDH-N, which is active when cells are growing anaerobically in the presence of nitrate, and it is encoded by the fdnGHI operon. FDH-O is induced under anaerobic growth and is encoded by the fdoGHI operon. The fdhF gene encodes FDH-H and it is only active under fermentative conditions. It forms part of the FHL complex and is responsible for the catabolism of formate in the hydrogen production pathway. Hydrogen is produced by the Hyd-3 enzyme, which also forms part of the FHL complex. Besides Hyd-3, E. coli has others 3 hydrogenases, Hyd-1, 2 and 4. Hyd-1 and Hyd-2 are considered as up-take hydrogenases; however, Kim et al. (2010) reported hydrogen production ability for Hyd-1 even under micro-aerobic conditions and Trchounian et al. (2012) found that Hyd-1, Hyd-2 and Hyd-3 can operate in reverse mode depending on pH and substrate type. Hyd-4 has not been biochemically characterized (Redwood et al. 2008).
FHL complex is shown in Fig. 3. This complex comprises seven proteins; six of them are encoded in the hyc operon. HycB, C, D, F and G are membrane electron transfer proteins, while HycE is the hydrogenase Hyd-3. FocA is not part of the complex, but is related to the formate metabolism because this is the formate channel and it exports the formate to prevent the acidification of the cytoplasm and then re-imports the formate when the pH is low in the culture medium. The formate regulon comprises genes that are involved in the formate metabolism. Besides the operon hyc and fdhf, the regulon also includes the hypA-E operon, fhlA, and hydN-hypF operon. The HypA-E, HypF, and HycI proteins are required for assembly of the Ni–Fe cofactor and the maturation of the three hydrogenases. FhlA is the transcriptional activator of the regulon, whereas HycA is the negative regulator. A detailed description of the formate regulon has been published by Leonhartsberger et al. (2002) and Sawers (2005).
5 Improvement of hydrogen production and yield by genetic manipulations of E. coli metabolic pathways
Among the genetically modified microorganisms reported for biohydrogen production, E. coli is one of the most widely used because its genome sequence is known, and the metabolic pathways and metabolism are the best understood of all bacteria. Also, there are molecular tools for its manipulation. Some examples of mutant strains of E. coli used for biohydrogen production are presented in Table 1.
Since HycA is the negative regulator of the formate regulon, the activity of FHL complex is increased when hycA gen is mutated (Leonhartsberger et al. 2002), thus HycA defective strains are hydrogen overproducer (Penfold et al. 2003; Yoshida et al. 2005). Yoshida et al. (2005) overexpressed fhlA and performed the hycA inactivation. With these genetic modifications, the transcription of fdhF and hycE increased 6.5- and 7-fold, respectively, and hydrogen production increased 2.8-fold compared with the wild-type strain. Hydrogenases 1 and 2 and formate dehydrogenase N and O are located in the periplasmic space (Fig. 3), whereas hydrogenase 3 and FDH-H are located in cytoplasm. The transport of these proteins to the periplasmic is performed by the Twin arginine translocation (Tat) protein system. Therefore, Tat mutant strains do not take formate up needed for hydrogen production. Penfold et al. (2006) reported that defective mutant strains of Tat transport (∆tatC and ∆tatA-E) showed a hydrogen production comparable to E. coli strain carrying a ∆hycA. However, double mutant strain ∆tatC ∆hycA did not increase hydrogen production. Thus, it is possible that discarding activities of the uptake hydrogenases that recycle a portion of hydrogen, and the formate hydrogenases N and O, which oxidize the formate without hydrogen production, could increase the hydrogen production by E. coli. Indeed, the effect of mutations in uptake hydrogenases, in lactate dehydrogenase gene (ldhA) and fhlA was studied by Bisaillon et al. (2006). They reported that each mutation contributed to a slight increase in hydrogen production, and the effect was synergistic. This same strain was used by Turcot et al. (2008) and gave the highest hydrogen production and yield in continuous cultures. The highest yields (at, or somewhat higher than 2 mol H2/mol glucose) were obtained with cultures limited for glucose (22 mM glucose); in a posterior work (Ghosh and Hallenbeck 2009b), a yield of 1.69 mol H2/mol glucose was achieved under 75 mM glucose.
Maeda et al. (2007a) performed multiple stable mutations to direct the metabolic flux toward hydrogen production. The best strain involves mutations on hyaB, hybC, hycA, fdoG, frdC, ldhA and aceE genes. The hyaB and hybC genes were deleted to abolish the uptake activity of hydrogenases 1 and 2. The fdoG and aceE genes code for the α subunit of formate dehydrogenase O and the pyruvate dehydrogenase respectively. The inactivation of frdC gene abolishes the succinate synthesis pathway. The same group reached the theoretical yield from formate with a strain with deletions of hyaB, hybC, hycA, fdoG, and overexpression of fhlA (Maeda et al. 2008). Yoshida et al. (2006) enhanced the hydrogen yield from 1.08 with the wild type strain to 1.82 mol H2/mol glucose using a ΔldhA, ΔfrdBC strain. The same yield was obtained by Mathews et al. (2010) using a strain with deletions on uptake hydrogenases (hyaAB, hybABC), hycA, lactate dehydrogenase (ldh) and fumarate reductase (frdBC), whereas Kim et al. (2009) reached 2.11 mol H2/mol glucose with a similar strain under low hydrogen partial pressure.
The synthesis of PFL, FHL, and FhlA is activated by the global transcriptional factor Fnr (Sawers 2005; Salmon et al. 2003; Perrenoud and Sauer 2005; Constantinidou et al. 2006), whereas the dual transcriptional regulator NarL repressed the synthesis of PFL and FHL (Overton et al. 2006). Fan et al. (2009) described increases in specific and molar yields of hydrogen achieved by the modification of focA, ppc, narL, and fnr genes. The strain ZF1 (ΔfocA) and ZF3 (ΔnarL) produced 14.9 and 14.4 µmol H2/mg dry cell weight, respectively, compared to 9.8 µmol H2/mg dry cell weight produced by the wild type strain. Strain ZF3 also displayed the best molar yield of 0.96 mol H2/mol of glucose compared to 0.54 for the wild type strain.
6 Improvement of hydrogen production by expression of heterologous pathways in E. coli
As discussed above, the low hydrogen yield of E. coli pathway is the main disadvantage. To overcome this drawback, some efforts have been made focused on the heterologous expression of hydrogenase genes to enhance hydrogen production (Table 2). The use of this strategy can be traced back more than 30 years ago to Karube et al. (1983). These authors cloned and expressed the hydrogenase from Clostridium butyricum into E. coli HK16. Since this first attempt, some other efforts have been made (Table 2). The overexpression of a Fe-hydrogenase from E. cloacae in a non-hydrogen-producing E. coli BL21 strain was made by Mishra et al. (2004) using degenerate primers designed from the conserved zone of hydA gene. The resultant recombinant strain showed the ability to produce hydrogen. King et al. (2006) reported the production in E. coli of active enzymes by the co-expression of proteins involved in maturation of hydrogenases from Clostridium acetobutylicum and Fe–Fe hydrogenases from C. acetobutylicum, C. pasteurianum, and Chlamydomonas reinhardtii. The purified enzymes showed similar specific activities to those purified from native sources. Akhtar and Jones (2008b) constructed a functional synthetic operon with the Fe–Fe hydrogenase (hydA) and its maturation factors (hydF, hydE and hydG) from C. acetobutylicum and demonstrated that the deletion of iscR gene, which codes for the transcriptional negative regulator of the iron-sulfur cluster, stimulated the recombinant Fe–Fe hydrogenase activity (Akhtar and Jones 2008a). Finally, they developed a synthetic hydrogen pathway by co-expression of a putative pyruvate flavodoxin/ferrodoxin oxidoreductase YdbK from E. coli, [4Fe–4S]-ferredoxin from C. pasteurianum and C. acetobutylicum HydF, HydE, HydG, and HydA reached a maximum yield of 1.88 mol H2/mol glucose consumed (Akhtar and Jones 2009). Kuchenreuther et al. (2010) described the production of active Fe–Fe hydrogenases from C. reinhardtii or C. pasteurianum using maturases from Shewanella oneidensis.
As discussed above, E. coli can perform the NADPH-dependent hydrogen production pathway if adequate hydrogenases from other microorganisms are expressed (King et al. 2006; Akhtar and Jones 2008b). Kim et al. (2011) introduced hydAEFG from C. acetobutiricum, fdxA and yumC from C. pasteurianum, and B. subtilis, respectively, in an E. coli BL21 (DE3) strain. Since NADPH is generated mainly by the pentose phosphate pathway, and the activation of this pathway is accompanied by activation of gluconeogenesis, FBPase II (a key enzyme in gluconeogenic pathway which is less sensitive to regulation, encoded by glpX), and glucose 6 phosphate 1 dehydrogenase (a key enzyme activating pentose phosphate pathway, encoded by zwf) were overexpressed in that E. coli strain. Overexpression of glpX increased the hydrogen yield 1.48-fold whereas the co-expression of the two genes increased the yield further 2.32-fold.
Agapakis et al. (2010) performed various hydrogen-producing electron circuits containing Fd-dependent hydrogenases from C. acetobutylicum, C. saccharobutylicum, C. reinhardtii, and S. oneidensis, ferredoxins from C. acetobutylicum, Spinacia olearcea, and Zea mays and PFORs from C. acetobutylicum, Desulfovibrio africanus, and E. coli. The E. coli BL21 (DE3) strain had multiple deletions in uptake hydrogenases and competing carbon pathways. The resulting hydrogen production yield was 0.4 mol H2/mol glucose.
Ni–Fe hydrogenases were also expressed in E. coli due to low oxygen sensitivity. Maeda et al. (2007b) cloned the bidirectional Ni–Fe hydrogenase (hoxEFUYH) from Synechocystis sp. in E. coli. This strain yielded 41-times more hydrogen than the strain with the empty vector after 18 h. This effect was due to the inhibition of the uptake activity. Sun et al. (2010) reported the co-expression of four structural genes for the NADP-dependent hydrogenase and nine genes for its maturation from Pyrococcus furiosus. The recombinant enzyme showed to be as functional as the native enzyme. They also observed that the maturation machinery of E. coli produces a functional hydrogenase when it only expressed the structural genes for the hydrogenase and a protease from P. furiosus. However, the hydrogenase activity was only reported in vitro. Weyman et al. (2011) cloned and expressed the structural gene for Ni–Fe hydrogenase, maturases and adjacent genes from Alteromonas macleodii “deep ecotype” in E. coli lacking native hydrogenases. The hydrogenase showed to be active in vitro in both aerobic and anaerobic conditions. They also demonstrated the activity of a Ni–Fe hydrogenase from Thiocapsa roseopersicina when co-expressed with the accessory proteins from A. macleodii. Wells et al. (2011) expressed the Synechocystis sp. hydrogen production pathway and its maturation factors in an E. coli strain in which the hydrogenases and formate production pathway were abolished. They reported in vivo production of 20 µmol of hydrogen per liter of culture.
7 Hydrogen production with E. coli using alternative carbon sources
Most of the research to improve hydrogen production has been conducted using glucose as substrate. However, to be competitive, biological hydrogen production must use carbohydrate rich wastes (Table 3). Penfold and Macaskie (2004) transformed to E. coli HD701, a hydrogenase-upregulated strain and FTD701 (a derivative of HD701 that has a deletion of the tatC gene), with the plasmid pUR400 carrying the scr regulon. This regulon contain the genes of Salmonella thompson to metabolize sucrose. The resulting E. coli strain produced hydrogen from sucrose. The parental strains did not produce hydrogen, whereas recombinant strains produced 1.27 and 1.38 ml H2/mg dry weight/l. Rosales-Colunga et al. (2010b) obtained a yield of 2.74 mol H2/mol lactose consumed, using cheese whey as substrate, and an E. coli ∆hycA ∆lacI strain (WDHL). In a subsequent work, hydrogen production from lactose, glucose and galactose was reported; the maximum yield was attained with galactose (Rosales-Colunga et al. 2012). Ghosh and Hallenbeck (2009a) reported the hydrogen yields from arabinose, fructose, gluconate, glucose, lactose, maltose, manitol, sorbitol, sucrose, trehalose, and xylose. The highest hydrogen yield obtained was 1.47 mol H2/mol substrate using sorbitol. Morsy (2011) used hydrolyzed molasses as substrate using the strain HD701. The highest hydrogen production of 570 ml of H2/l and a rate of 19 ml/l/h were obtained using a concentration of 10 g/l of reducing sugars. However, the maximal yield (132 ml of hydrogen/g of reducing sugars) was obtained from 2.5 g/l of reducing sugars.
Perego et al. (1998) used a corn starch hydrolysate (85 % glucose, dry basis) to produce hydrogen with E. coli and E. aerogenes; with E. coli a maximum yield of 0.36 mol/mol glucose was reached. In this study, E. aerogenes showed a better production from this substrate. Orozco et al. (2012) performed the hydrothermal hydrolysis of starch with carbon dioxide and detoxification of hydrolisate with activated coal. Hydrogen production using this hydrolysis strategy was equal to glucose controls. Akhtar and Jones (2009) reported an E. coli that expresses an amylase and it was used for hydrogen production from starch without previous hydrolysis.
Glycerol has become an abundant and inexpensive carbon source due to its generation as by-product from biodiesel fuel production. For this reason some efforts have been focused on obtaining hydrogen from glycerol. Yazdani and Gonzalez (2008) created the strain SY03 (pZSKLMgldA) in which the acetate and succinate pathways were minimized by inactivation of phosphate acetyltransferase (pta) and fumarate reductase (frdA), respectively. The enzymes responsible for the conversion of glycerol to dihydroxyacetonephosphate, a glycolytic intermediate, were overexpressed. The yield of ethanol and hydrogen reached was 95 % of the theoretical maximum. Trchounian et al. (2011) studied the glycerol fermentation and hydrogen production, they found that at pH of 5.5 the hydrogen production was 1.5-fold higher than at a pH 6.5. Starting with E. coli BW25113 frdC that lacks fumarate-reductase and by using both adaptive evolution and chemical mutagenesis combined with a selection method based on increased growth in glycerol. Hu and Wood (2010) obtained the strain HW2, that produced 20-fold more hydrogen in glycerol medium.
8 Fermentative approaches used to improve hydrogen production using E. coli
The main disadvantage of fermentative hydrogen production is the low yield due to the production of other metabolites, mainly organic acids. To improve the net yield of hydrogen a two-stage system can be used. In the first stage, a dark fermentation is used to produce hydrogen, and in the second stage the organic acids produced in the first step are used as substrate for photofermentation, increasing the total hydrogen yield. For example, Salih (1989) used cheese whey pretreated with E. coli to produce hydrogen by a photosynthetic Rhodospirillum rubrum. E. coli was only used to pretreat of cheese whey and not for hydrogen production; however, hydrogen production increased when pretreated whey was used (Table 4). Redwood and Macaskie (2006) tried to produce hydrogen in two stages, first, by fermentation of glucose by E. coli HD701 and then by photofermentation of the residual medium by R. sphaeroides. Nevertheless, hydrogen production did not occur during photofermentation of the residual liquor per se due to the presence of fixed nitrogen compounds. This issue was further solved by electroseparation of ammonium ion and the authors reported a continuous E. coli reactor and a continuous R. sphaeroides photobioreactor integrated by anion-selective electrodialysis, simultaneously transferring anionic fermentation products, while retaining repressive ammonium ion, E. coli cells and suspended solids (Redwood et al. 2009). This approach resulted in sustained hydrogen production by E. coli with a yield of 1.6 mol H2/mol hexose and sustained hydrogen photoproduction by R. sphaeroides. The overall yield was 2.4 mol H2/mol glucose. This electro-extractive strategy was also used to enhance continuous hydrogen and organic acid production by E. coli FTD67 (Redwood et al. 2012). The pH was controlled by separation of organic acids, which can be used in a further hydrogen production step by photofermentation. The maximal rate was 4.7 l/d/l of culture and yield of 0.7 mol/mol glucose.
Waks and Silver (2009) combined the industrial advantages of yeast with E. coli hydrogen production. They proposed biomass conversion to formate by S. cerevisiae and the subsequent conversion of formate to hydrogen by E. coli. The endogenous formate dehydrogenases of S. cerevisiae were deleted and the pyruvate formate lyase and alcohol dehydrogenase from E. coli were expressed; galactose was used in this first stage to produce formate. The formate-enriched medium was further used to produce hydrogen by E. coli. Abd-Alla et al. (2011) proposed the use of rotten dates to produce hydrogen in a 3-stage process. In the first stage, E. coli EGY was used to consume oxygen and maintain the anaerobic condition. In the second, stage hydrogen was produced using C. acetobutylicum ATCC 824 and finally photofermentation by R. capsulatus was used. The maximal total yield of the process was 7.8 mol hydrogen/mol sucrose. Seppälä et al. (2011) examined hydrogen production in a co-culture of E. coli and C. butyricum. They found that the total hydrogen production of the co-culture was higher compared to the monoculture of each strain. However, the co-culture yield (1.65 mol H2/mol glucose) was lower than that obtained by the pure culture of C. butyricum (2.09 mol H2/mol glucose).
Another strategy widely used to produce a variety of products is the use of immobilized cells. Its main advantages are increase in the biomass concentration, low risk of contamination, operational stability, and high productivity (Seol et al. 2011). Therefore, this strategy has been used in hydrogen production. For example, Ishikawa et al. (2006) probed the encapsulation of E. coli MC13-4 in alginate gel beads, and hydrogen increased 3-fold compared to a free cell system; nonetheless, the gas remained as bubbles in the interspace of the gel. This system was connected to a fuel cell and can produce electricity. In a later work (Ishikawa et al. 2008), a compact stacked flatbed reactor (CSFR) was developed to extract the produced gas easily. This reactor comprises pieces of agar plates containing E. coli MC13-4 at high density and reached the yield of 1.2 mol H2/mol glucose, and the production rate of 6.7 l of hydrogen/g dry cell/l of reactor/h. Seol et al. (2011) examined three different matrices (agar, agarose, and sodium alginate) to immobilize a hydrogen over-producer strain of E. coli (SH5). Using agar as matrix and optimal conditions a maximum production rate of 2.4 l of hydrogen/l/h and yield around 100 % of the theoretical from formate (1 mol/mol) were attained. They also probed a sustained production in a feed-batch operation mode.
Escherichia coli is a valuable microorganism in the study of hydrogen production as discussed above and is still a model that can provide useful information to know the hydrogen producer pathways and to improve them.
9 Conclusions
Genetic manipulations, the use of a diversity of carbohydrates, and redirection of the carbon flux to favor hydrogen production have been used to increase hydrogen yield. However, until now E. coli has been just an excellent microorganism to study processes to produce hydrogen in lab-scale. Additional efforts should be conducted to obtain suitable processes feasible to scaling-up to produce hydrogen for commercial purposes, for instance those where metabolites such as succinate or recombinant protein are the main products and hydrogen is a by-product. Novel and new approaches such as using synthetic biology to improve hydrogen production are still needed in a new generation of overproducing hydrogen strains.
References
Abd-Alla MH, Morsy FM, El-Enany A-WE (2011) Hydrogen production from rotten dates by sequential three stages fermentation. Int J Hydrog Energy 36:13518–13527
Agapakis C, Ducat D, Boyle P, Wintermute E, Way J, Silver P (2010) Insulation of a synthetic hydrogen metabolism circuit in bacteria. J Biol Eng 4:3
Akhtar M, Jones P (2008a) Deletion of iscR stimulates recombinant clostridial Fe–Fe hydrogenase activity and H2-accumulation in Escherichia coli BL21(DE3). Appl Microbiol Biotechnol 78:853–862
Akhtar MK, Jones PR (2008b) Engineering of a synthetic hydF-hydE-hydG-hydA operon for biohydrogen production. Anal Biochem 373:170–172
Akhtar MK, Jones PR (2009) Construction of a synthetic YdbK-dependent pyruvate: H2 pathway in Escherichia coli BL21(DE3). Metab Eng 11:139–147
Bisaillon A, Turcot J, Hallenbeck PC (2006) The effect of nutrient limitation on hydrogen production by batch cultures of Escherichia coli. Int J Hydrog Energy 31:1504–1508
Blackwood AC, Ledingham GA, Neish AC (1956) Dissimilation of glucose at controlled pH values by pigmented and non-pigmented strains of Escherichia coli. J Bacteriol 72:497–499
Chittibabu G, Nath K, Das D (2006) Feasibility studies on the fermentative hydrogen production by recombinant Escherichia coli BL-21. Process Biochem 41:682–688
Clark DP (1989) The fermentation pathways of Escherichia coli. FEMS Microbiol Lett 63:223–234
Constantinidou C, Hobman JL, Griffiths L, Patel MD, Penn CW, Cole JA, Overton TW (2006) A Reassessment of the fnr regulon and transcriptomic analysis of the effects of nitrate, nitrite, NarXL, and NarQP as Escherichia coli K12 adapts from aerobic to anaerobic growth. J Biol Chem 281:4802–4815
Davila-Vazquez G, Arriaga S, Alatriste-Mondragón F, de León-Rodríguez A, Rosales-Colunga LM, Razo-Flores E (2008) Fermentative biohydrogen production: trends and perspectives. Rev Environ Sci Biotechnol 7:27–45
Fan Z, Yuan L, Chatterjee R (2009) Increased hydrogen production by genetic engineering of Escherichia coli. PLoS One 4:e4432
Fonseca A, Sá V, Bento H, Tavares MLC, Pinto G, Gomes LACN (2008) Hydrogen distribution network optimization: a refinery case study. J Clean Prod 16:1755–1763
Froese DS, Dobson CM, White AP, Wu X, Padovani D, Banerjee R, Haller T, Gerlt JA, Surette MG, Gravel RA (2009) Sleeping beauty mutase (sbm) is expressed and interacts with ygfd in Escherichia coli. Microbiol Res 164:1–8
Ghosh D, Hallenbeck PC (2009a) Fermentative hydrogen yields from different sugars by batch cultures of metabolically engineered Escherichia coli DJT135. Int J Hydrog Energy 34:7979–7982
Ghosh D, Hallenbeck PC (2009b) Response surface methodology for process parameter optimization of hydrogen yield by the metabolically engineered strain Escherichia coli DJT135. Bioresour Technol 101:1820–1825
Hallenbeck PC (2005) Fundamentals of the fermentative production of hydrogen. Water Sci Technol 52:21–29
Hallenbeck PC, Ghosh D (2009) Advances in fermentative biohydrogen production: the way forward? Trends Biotechnol 27:287–297
Haller T, Buckel T, Retey J, Gerlt JA (2000) Discovering new enzymes and metabolic pathways: conversion of succinate to propionate by Escherichia coli. Biochemistry 39:4622–4629
Hu H, Wood TK (2010) An evolved Escherichia coli strain for producing hydrogen and ethanol from glycerol. Biochem Biophys Res Commun 391:1033–1038
Ishikawa M, Yamamura S, Takamura Y, Sode K, Tamiya E, Tomiyama M (2006) Development of a compact high-density microbial hydrogen reactor for portable bio-fuel cell system. Int J Hydrog Energy 31:1484–1489
Ishikawa M, Yamamura S, Ikeda R, Takamura Y, Sode K, Tamiya E, Tomiyama M (2008) Development of a compact stacked flatbed reactor with immobilized high-density bacteria for hydrogen production. Int J Hydrog Energy 33:1593–1597
Jian J, Zhang S-Q, Shi Z-Y, Wang W, Chen G-Q, Wu Q (2010) Production of polyhydroxyalkanoates by Escherichia coli mutants with defected mixed acid fermentation pathways. Appl Microbiol Biotechnol 87:2247–2256
Kapdan IK, Kargi F (2006) Bio-hydrogen production from waste materials. Enzyme Microb Technol 38:569–582
Karube I, Urano N, Yamada T, Hirochika H, Sakaguchi K (1983) Cloning and expression of the hydrogenase gene from Clostridium butyricum in Escherichia coli. FEBS Lett 158:119–122
Kim S, Seol E, Oh Y-K, Wang GY, Park S (2009) Hydrogen production and metabolic flux analysis of metabolically engineered Escherichia coli strains. Int J Hydrog Energy 34:7417–7427
Kim J, Jo B, Cha H (2010) Production of biohydrogen by recombinant expression of [NiFe]-hydrogenase 1 in Escherichia coli. Microb Cell Fact 9:54
Kim YM, Cho H-S, Jung GY, Park JM (2011) Engineering the pentose phosphate pathway to improve hydrogen yield in recombinant Escherichia coli. Biotechnol Bioeng 108:2941–2946
King PW, Posewitz MC, Ghirardi ML, Seibert M (2006) Functional studies of [FeFe] hydrogenase maturation in an Escherichia coli biosynthetic system. J Bacteriol 188:2163–2172
Kuchenreuther JM, Grady-Smith CS, Bingham AS, George SJ, Cramer SP, Swartz JR (2010) High-yield expression of heterologous [FeFe] hydrogenases in Escherichia coli. PLoS One 5:e15491
Leonhartsberger S, Korsa I, Bock A (2002) The molecular biology of formate metabolism in enterobacteria. J Mol Microbiol Biotechnol 4:269–276
Lugg H, Sammons R, Marquis P, Hewitt C, Yong P, Paterson-Beedle M, Redwood M, Stamboulis A, Kashani M, Jenkins M, Macaskie L (2008) Polyhydroxybutyrate accumulation by a Serratia sp. Biotechnol Lett 30:481–491
Luque R, Herrero-Davila L, Campelo JM, Clark JH, Hidalgo JM, Luna D, Marinas JM, Romero AA (2008) Biofuels: a technological perspective. Energy Environ Sci 1:542–564
Maeda T, Sanchez-Torres V, Wood T (2007a) Enhanced hydrogen production from glucose by metabolically engineered Escherichia coli. Appl Microbiol Biotechnol 77:879–890
Maeda T, Vardar G, Self W, Wood T (2007b) Inhibition of hydrogen uptake in Escherichia coli by expressing the hydrogenase from the cyanobacterium Synechocystis sp. PCC 6803. BMC Biotechnol 7:25
Maeda T, Sanchez-Torres V, Wood TK (2008) Metabolic engineering to enhance bacterial hydrogen production. Microb Biotechnol 1:30–39
Mathews J, Wang GY (2009) Metabolic pathway engineering for enhanced biohydrogen production. Int J Hydrog Energy 34:7404–7416
Mathews J, Li Q, Wang G (2010) Characterization of hydrogen production by engineered Escherichia coli strains using rich defined media. Biotechnol Bioprocess Eng 15:686–695
Mishra J, Khurana S, Kumar N, Ghosh AK, Das D (2004) Molecular cloning, characterization, and overexpression of a novel [Fe]-hydrogenase from a high rate of hydrogen producing Enterobacter cloacae IIT-BT 08. Biochem Biophys Res Commun 324:679–685
Morsy FM (2011) Hydrogen production from acid hydrolyzed molasses by the hydrogen overproducing Escherichia coli strain HD701 and subsequent use of the waste bacterial biomass for biosorption of Cd(II) and Zn(II). Int J Hydrog Energy 36:14381–14390
Nurul Islam M, Rafiqul Alam Beg M, Rofiqul Islam M (2005) Pyrolytic oil from fixed bed pyrolysis of municipal solid waste and its characterization. Renew Energy 30:413–420
Orozco RL, Redwood MD, Leeke GA, Bahari A, Santos RCD, Macaskie LE (2012) Hydrothermal hydrolysis of starch with CO2 and detoxification of the hydrolysates with activated carbon for bio-hydrogen fermentation. Int J Hydrog Energy 37:6545–6553
Overton TW, Griffiths L, Patel MD, Hobman JL, Penn CW, Cole JA, Constantinidou C (2006) Microarray analysis of gene regulation by oxygen, nitrate, nitrite, FNR, NarL and NarP during anaerobic growth of Escherichia coli: new insights into microbial physiology. Biochem Soc Trans 34:104–107
Penfold DW, Macaskie LE (2004) Production of H2 from sucrose by Escherichia coli strains carrying the pUR400 plasmid, which encodes invertase activity. Biotechnol Lett 26:1879–1883
Penfold DW, Forster CF, Macaskie LE (2003) Increased hydrogen production by Escherichia coli strain HD701 in comparison with the wild-type parent strain MC4100. Enzyme Microb Technol 33:185–189
Penfold DW, Sargent F, Macaskie LE (2006) Inactivation of the Escherichia coli K-12 twin-arginine translocation system promotes increased hydrogen production. FEMS Microbiol Lett 262:135–137
Perego P, Fabiano B, Ponzano GP, Palazzi E (1998) Experimental study of hydrogen kinetics from agroindustrial by-product: optimal conditions for production and fuel cell feeding. Bioprocess Eng 19:205–211
Perrenoud A, Sauer U (2005) Impact of global transcriptional regulation by ArcA, ArcB, Cra, Crp, Cya, Fnr, and Mlc on glucose catabolism in Escherichia coli. J Bacteriol 187:3171–3179
Ramachandran R, Menon RK (1998) An overview of industrial uses of hydrogen. Int J Hydrog Energy 23:593–598
Redwood MD, Macaskie LE (2006) A two-stage, two-organism process for biohydrogen from glucose. Int J Hydrog Energy 31:1514–1521
Redwood MD, Mikheenko IP, Sargent F, Macaskie LE (2008) Dissecting the roles of Escherichia coli hydrogenases in biohydrogen production. FEMS Microbiol Lett 278:48–55
Redwood M, Paterson-Beedle M, Macaskie L (2009) Integrating dark and light bio-hydrogen production strategies: towards the hydrogen economy. Rev Environ Sci Biotechnol 8:149–185
Redwood MD, Orozco RL, Majewski AJ, Macaskie LE (2012) Electro-extractive fermentation for efficient biohydrogen production. Bioresour Technol 107:166–174
Rosales-Colunga LM, González García R, De León Rodríguez A (2010a) Estimation of hydrogen production in genetically modified E. coli fermentations using an artificial neural network. Int J Hydrog Energy 35:13186–13192
Rosales-Colunga LM, Razo-Flores E, Ordoñez LG, Alatriste-Mondragón F, De León-Rodríguez A (2010b) Hydrogen production by Escherichia coli ∆hycA ∆lacI using cheese whey as substrate. Int J Hydrog Energy 35:491–499
Rosales-Colunga LM, Razo-Flores E, Rodriguez ADL (2012) Fermentation of lactose and its constituent sugars by Escherichia coli WDHL: impact on hydrogen production. Bioresour Technol 111:180–184
Salih FM (1989) Improvement of hydrogen photoproduction from E. coli pre-treated cheese whey. Int J Hydrog Energy 14:661–663
Salmon K, S-p Hung, Mekjian K, Baldi P, Hatfield GW, Gunsalus RP (2003) Global gene expression profiling in Escherichia coli K12. J Biol Chem 278:29837–29855
Sawers RG (2005) Formate and its role in hydrogen production in Escherichia coli. Biochem Soc Trans 33:42–46
Seol E, Manimaran A, Jang Y, Kim S, Oh Y-K, Park S (2011) Sustained hydrogen production from formate using immobilized recombinant Escherichia coli SH5. Int J Hydrog Energy 36:8681–8686
Seppälä JJ, Puhakka JA, Yli-Harja O, Karp MT, Santala V (2011) Fermentative hydrogen production by Clostridium butyricum and Escherichia coli in pure and cocultures. Int J Hydrogen Energy 36:10701–10708
Sinha P, Pandey A (2011) An evaluative report and challenges for fermentative biohydrogen production. Int J Hydrog Energy 36:7460–7478
Sun J, Hopkins RC, Jenney FE Jr, McTernan PM, Adams MWW (2010) Heterologous expression and maturation of an NADP-dependent [NiFe]-hydrogenase: a key enzyme in biofuel production. PLoS One 5:e10526
Trchounian K, Sanchez-Torres V, Wood TK, Trchounian A (2011) Escherichia coli hydrogenase activity and H2 production under glycerol fermentation at a low pH. Int J Hydrog Energy 36:4323–4331
Trchounian K, Poladyan A, Vassilian A, Trchounian A (2012) Multiple and reversible hydrogenases for hydrogen production by Escherichia coli: dependence on fermentation substrate, pH and the F0F1-ATPase. Crit Rev Biochem Mol Biol 47:236–249
Turcot J, Bisaillon A, Hallenbeck PC (2008) Hydrogen production by continuous cultures of Escherichia coli under different nutrient regimes. Int J Hydrog Energy 33:1465–1470
Waks Z, Silver PA (2009) Engineering a synthetic dual-organism system for hydrogen production. Appl Environ Microbiol 75:1867–1875
Wells MA, Mercer J, Mott RA, Pereira-Medrano AG, Burja AM, Radianingtyas H, Wright PC (2011) Engineering a non-native hydrogen production pathway into Escherichia coli via a cyanobacterial [NiFe] hydrogenase. Metab Eng 13:445–453
Weyman PD, Vargas WA, Chuang RY, Chang Y, Smith HO, Xu Q (2011) Heterologous expression of Alteromonas macleodii and Thiocapsa roseopersicina [NiFe] hydrogenases in Escherichia coli. Microbiology 157:1363–1374
Yazdani SS, Gonzalez R (2008) Engineering Escherichia coli for the efficient conversion of glycerol to ethanol and co-products. Metab Eng 10:340–351
Yoshida A, Nishimura T, Kawaguchi H, Inui M, Yukawa H (2005) Enhanced hydrogen production from formic acid by formate hydrogen lyase-overexpressing Escherichia coli strains. Appl Environ Microbiol 71:6762–6768
Yoshida A, Nishimura T, Kawaguchi H, Inui M, Yukawa H (2006) Enhanced hydrogen production from glucose using ldh- and frd-inactivated Escherichia coli strains. Appl Microbiol Biotechnol 73:67–72
Zhang H, Boghigian BA, Pfeifer BA (2010) Investigating the role of native propionyl-CoA and methylmalonyl-CoA metabolism on heterologous polyketide production in Escherichia coli. Biotechnol Bioeng 105:567–573
Acknowledgments
We thank CONACyT for partial funding of this work through SENER Grant 150001 and CONACyT-Básicas 178988. The authors thank Jennifer Eckerly Goss for English correction.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rosales-Colunga, L.M., De León Rodríguez, A. Escherichia coli and its application to biohydrogen production. Rev Environ Sci Biotechnol 14, 123–135 (2015). https://doi.org/10.1007/s11157-014-9354-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11157-014-9354-2