Abstract
Causal structure learning is one of the most exciting new topics in the fields of machine learning and statistics. In many empirical sciences including prevention science, the causal mechanisms underlying various phenomena need to be studied. Nevertheless, in many cases, classical methods for causal structure learning are not capable of estimating the causal structure of variables. This is because it explicitly or implicitly assumes Gaussianity of data and typically utilizes only the covariance structure. In many applications, however, non-Gaussian data are often obtained, which means that more information may be contained in the data distribution than the covariance matrix is capable of containing. Thus, many new methods have recently been proposed for using the non-Gaussian structure of data and inferring the causal structure of variables. This paper introduces prevention scientists to such causal structure learning methods, particularly those based on the linear, non-Gaussian, acyclic model known as LiNGAM. These non-Gaussian data analysis tools can fully estimate the underlying causal structures of variables under assumptions even in the presence of unobserved common causes. This feature is in contrast to other approaches. A simulated example is also provided.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The study of statistical causal reasoning can be roughly divided into two categories. First, if the causal structure of variables is known, the conditions under which the causal effects or intervention effects between variables can be inferred are investigated (Imbens and Rubin 2015; Pearl 2000). Second, if the causal structure is unknown, the conditions under which the causal structure or causal relationships of variables can be inferred are investigated (Spirtes et al. 1993; Shimizu 2014; Zhang and Hyvärinen 2016). The difference between the two tasks is whether the causal structure is known or unknown and reflects different purposes. The second category is called causal discovery or causal structure learning. The two categories are closely related. For example, suppose that the causal structure is unknown based on background causal knowledge. Then, the causal structure is inferred by using causal discovery methods from the latter category, and causal effects that can be inferred are identified based on the inferred causal structure. Causal effects are identified by combining the theories of the two categories as well as background knowledge.
Researchers in various fields, including prevention scientists, have hypothesized about the causal relationships for various phenomena. However, narrowing the candidate hypotheses to one based only on the background theory for a given field is usually difficult. In such cases, multiple candidates need to be compared based on data to determine which is better. Further, if the background theory is not sufficient, developing candidate hypotheses in the first place is difficult. In this case, candidate hypotheses should be generated based on experience or observed data. In either case, causal discovery or causal structure learning methods are useful.
Here is an example where causal discovery is required. People with depression have been reported to tend to have sleep problems. For example, according to an epidemiological survey (Raitakari et al. 2008), the correlation coefficient between depression and the degree of sleep disorder is 0.77 (Rosenström et al. 2012). Epidemiologic researchers may then want to find a causal model to explain this strong correlation. They may consider the following candidate causal models:
-
1.
Sleep problems causes depression.
-
2.
Depression causes sleep problems.
-
3.
There is no direct causal relationship between depression and sleep problems.
These three candidates are graphically represented in Fig. 1. Of course, a fourth candidate is that depression and sleep problems mutually cause each other, i.e., cyclic cases. In this paper, one-way causal relationships are assumed to simplify the illustrative examples. The concept can be further extended to cyclic cases (Lacerda et al. 2008).
If a sleep disorder causes depression, as shown by the causal structure on the left of Fig. 1, then reducing the degree of sleep problems of the subjects would decrease their depression. If the middle structure is the case, then reducing the degree of depression would decrease sleep problems. Lowering the severity of sleep problems would not change the degree of depression. Finally, if the right structure is the case, then depression and sleep problems are not causally related. Then, even if the severity of the sleep problems is lowered, the depression does not change.
By performing randomized experiments, the causal relationship between depression and sleep disturbance can be determined. However, actually performing randomized experiments is not easy. This paper discusses causal discovery methods based on observational data that do not need such randomized experiments to be performed. Note that several assumptions are needed in place of the randomization. Even if some assumptions are needed, they can generate specific causal hypotheses to be verified by further experiments. Therefore, these causal discovery methods do not aim to replace such experiments. Rather, they are intended to help prevention scientists hypothesize good causal model candidates before performing randomized experiments or to do their best when randomized experiments cannot be performed.
Causal structure learning methods aim to discover or infer causal graphs of variables based on data. Causal graphs illustrate the qualitative causal relations of variables. An example is given in Fig. S1 (available online). There are three variables to be analyzed: x1,x2, and z1. There are also two error variables: e1 and e2. x1 and x2, which are represented by boxes, are observed variables. z1, which is represented by a dotted circle, is an unobserved variable. The error variables e1 and e2 are unobserved, although they are not represented by dotted circles.
In the example graph, all edges between variables are directed. A directed edge starting from a variable and ending with another variable indicates that the former variable directly causes the latter. Based on the terminology of graph theory, the former variable is called a parent of the latter, and the latter variable is called a child of the former. In this causal graph, there is a directed edge from x1 to x2. This indicates that x1 directly causes x2. Thus, x1 is a parent of x2, and x2 is a child of x1. If there is no directed edge between two variables, then there is no direct causal relation between the two. The unobserved variable z1 directly causes both x1 and x2. Hence, it is called an unobserved common cause.
In causal structure learning, the objective is to infer the causal graph of variables based on their observed data. Note that this is done without actually intervening on any of the variables. A major topic in this field is understanding the conditions under which the causal graph can be uniquely estimated. This paper first reviews the framework for causal inference, which is also known as the identification of causal effects, and then introduces recent causal discovery methods based on the linear non-Gaussian acyclic model (LiNGAM). Examples of LiNGAM applications include epidemiology (Rosenström et al. 2012), economics (Moneta et al. 2013), finance (Zhang and Chan 2008), and neuroscience (Mills-Finnerty et al. 2014).
Framework of Causal Inference
This section provides a brief review of the causal inference framework based on the structural causal model (SCM) (Pearl 2000). First, structural equation models (SEMs) are introduced for describing data-generating processes (Bollen 1989), which are used to generate values of variables. This framework uses special types of equations known as structural equations to represent how the values of variables are determined.
The structural equations for the case described in Fig. S1 (available online) are given by
where the error variable e1 denotes all factors other than z1 that can contribute to determining the value of x1. Similarly, the error variable e2 denotes all factors other than x1 and z1.
Structural equations represent more than a simple mathematical equality. The left-hand sides of the equations are defined by their right-hand sides. For example, in Eq. 1, the value of x1 on the left-hand side is completely determined by that of z1 and e1 through the function f1.Footnote 1
In Eqs. 1 and 2, the value of e1 is first generated from the probability distribution p(e1). Then, the value of x1 is determined by those of z1 and e1 through the function f1. Subsequently, the value of e2 is generated from the probability distribution p(e2). Then, the value of x2 is determined by those of x1,z1, and e2 through the function f2. The variables z1,e1, and e2 are known as exogenous variables. The values of these exogenous variables are generated outside the model, and their data-generating processes are decided by the modeler to not be further modeled. In contrast, variables whose values are generated inside the model, such as x1 and x2, are known as endogenous variables.
Definition of Causality Based on Interventions
Next, causality is defined based on the interventions used in SCMs (Pearl 2000). First, interventions in SEMs are defined. Intervening on the variable x1 means forcing the value of x1 to be a constant c regardless of the other variables. This intervention is denoted by do(x = c). In SEMs, this means replacing the function determining x1 with the constant c, i.e., forcing all individuals in a population to take x = c. Suppose that x1 is intervened with and forced to take the value of c in the example given in Eqs. 1 and 2. This creates a new SEM denoted by Mx = c:
As a result, the causal graph shown on the left of Fig. Fig. S2 (available online) changes to that given on the right. The directed edge from the unobserved common cause z1 to the observed variable x1 in the causal graph of the original SEM given in Eqs. 1 and 2 disappears because x1 is forced to be c regardless of the other variables including z1. Note that the other functions are assumed to not change even if a function is replaced with a constant. Although this may be physically unrealistic in some cases, the revised SEM given in Eqs. 3 and 4 represents a hypothetical population where all individuals in the population are forced to take x = c but the other function f2 does not change.
Next, the post-intervention distribution is defined. When x1 is intervened with, the post-intervention distribution of x2 is defined by the distribution of x2 in the revised SEM, i.e., Mx = c:
The associated causal graph is shown on the right of Fig. Fig. S2 (available online).
Then, x1 is a cause of x2 in this population if there exist two different values c and d such that the post-intervention distributions are different, i.e., if the following holds:
A common method for quantifying the magnitude of causation from x1 to x2 is to assess the following average difference (Rubin 1974; Pearl 2000):
This is called the average causal effect. E denotes the expectation operator and is a shorthand for averaging according to a given distribution. This evaluates to what extent, on average, the value of x2 would change if the value of x1 has been changed from c to d. Other quantifying methods include assessing the ratio of the two averages or using the variance or other meaningful statistics that characterize the features of the post-intervention distribution.
As an example, assume that the function f2 in the SEM of Eqs. 1 and 2 is linear:
where b21,λ11, and λ21 are constants. Then, the post-intervened SEM \(M_{x_{1}=c}\) takes the form
Therefore, the average causal effect of x1 on x2 if the value of x1 has been changed from c to d is given by
The expected average change in x2 is thus the difference between d and c multiplied by the coefficient b21.
Similarly, the post-intervened model \(M_{x_{2}=c}\) shown on the right of Fig. S3 (available online) is written as
Then, the average causal effect of x2 on x1 when the value of x2 has been changed from c to d is given by
This is reasonable because x2 does not contribute to defining x1 in the original SEM shown in Eqs. 1 and 2 and on the left of Fig. S3 (available online).
Non-Gaussian Methods for Causal Discovery
In causal structure learning, the SCMs introduced above are used to represent model assumptions, including the background knowledge and hypotheses of the modeler. Model assumptions place constraints on the model and restrict the candidate causal structures. Among the structures that satisfy the model assumptions, the causal structure that is most consistent with the data distribution is searched for.
This section explains the basic setup (Pearl 2000; Spirtes et al. 1993) and then introduces the non-Gaussian causal discovery methods based on a model known as LiNGAM (Shimizu et al. 2006; Hoyer et al. 2008; Shimizu 2014). The focus remains on continuous variable cases.
A typical assumption is that the causal relations of variables are acyclic, i.e., there are no directed cycles in the causal graph. Further, the functional relations of the variables are assumed to be linear. The basic model for the continuous observed variables xi (i = 1,…,p) is therefore formulated as follows:
where pa(xi) is the set of parents of xi in the causal graph, ei (i = 1,…,p) are error variables, and bij (i,j = 1,…,p) are the coefficients that represent the magnitude of direct causation from xj to xi.
In the most basic setup, the error variables ei (i = 1,…,p) are assumed to be independent. The independence assumption between ei (i = 1,…,p) implies that there are no unobserved common causes. This means that unobserved common causes such as z1 in the causal graph of Fig. S1 (available online) must be observed. If there is an unobserved common cause, it is not part of the model (19) and generally makes some of the error variables in Eq. 19 dependent. This setup is discussed first. Then, an advanced model with unobserved common causes is presented.
In matrix form, a linear acyclic SCM with no unobserved common cause in Eq. 19 can be written as
where the coefficient matrix B collects the magnitudes of direct causation bij (i,j = 1,…,p) and the vectors x and e collect the observed variables xi (i = 1,…,p) and exogenous variables ei (i = 1,…,p), respectively. The zero/non-zero pattern of bij (i,j = 1,…,p) corresponds to the absence/existence pattern of the directed edges. In other words, if the coefficient bij≠ 0, there is a directed edge from xj to xi. If this is not the case, there is no directed edge from xj to xi (i,j = 1,…,p). Because of the acyclicity, the diagonal elements of B are all zeros.
Figure 2 provides an example of causal graphs for representing the linear acyclic SCMs with no unobserved common cause in Eq. 20. The SEM corresponding to the causal graph of the figure is written as
The goal of identifying causal structures with this basic setup is to estimate the unknown coefficient matrix B by using the data X. X is assumed to be randomly sampled from a linear acyclic SCM with no unobserved common cause, as represented by Eq. 20 above.
Classical Approach Based on Conditional Independence
Under the causal Markov condition and the faithfulness assumption (Spirtes et al. 1993), conditional independence relations provide a classical way to infer the causal structure of the linear acyclic SCM with no unobserved common causes in Eq. 20.Footnote 2 For any such linear acyclic SCM, the causal Markov condition holds (Pearl and Verma 1991) as follows. Each observed variable xi is independent of its non-descendants conditional on its parents, i.e., \(p(\boldsymbol {x}) = {\Pi }_{i = 1}^{p} p(x_{i}| pa(x_{i}))\). Thus, conditional independence between observed variables provides a clue as to what the underlying causal structure is.
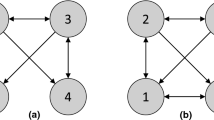
Unfortunately, in many cases, the causal Markov condition is insufficient for uniquely identifying the causal structure of the linear acyclic SCM with no unobserved common causes (Pearl 2000; Spirtes et al. 1993). An example of this is provided in Fig. 3. Suppose that data x are generated from the left causal graph shown in Fig. 3. According to the causal Markov condition, x2 and x3 are independent conditional on x1, and no other conditional independence holds. Therefore, the only information available for estimating the underlying causal structure is the conditional independence of x2 and x3. Within the class of linear acyclic SCMs with no unobserved common causes, the three causal graphs give the same conditional independence. In each of these three causal structures, only x2 and x3 are conditionally independent. However, only the left causal graph represents the right causal relations, and the other two causal graphs do not. The three causal structures are quite different, and there is no causal direction that is consistent across all three causal graphs. Thus, in this example, the causal Markov condition principle is not capable of uniquely estimating the underlying causal graph.
Basic LiNGAM
In this section, the basic LiNGAM is reviewed (Shimizu et al. 2006) before it is extended to cases with unobserved common causes (Hoyer et al. 2008). The assumptions of the basic LiNGAM may appear to be restrictive, and fortunately, they can be relaxed in many ways (Hoyer et al. 2008, 2009; Lacerda et al. 2008; Hyvärinen et al. 2010; Zhang and Hyvärinen 2009).
In Shimizu et al. (2006), a non-Gaussian version of the linear acyclic SCM was proposed with no unobserved common causes in Eq. 19. This is known as a LiNGAM:
where the error variables ei (i = 1,…,p) follow non-Gaussian continuous distributions and are independent. Without loss of generality, their means are assumed to be zeros.
LiNGAMs have been proven to be identifiable (Shimizu et al. 2006), i.e., the coefficients bij (i,j = 1,…,d) can be uniquely identified by using the non-Gaussianity of the data. Then, the causal graph can be drawn based on the zero/non-zero pattern of the coefficient matrix B that collects those coefficients bij (i,j = 1,…,p). In contrast, the classical approach in the previous subsection only uses the conditional independence of observed variables and does not use the non-Gaussian structure, even when they follow non-Gaussian distributions.
A principle for identifying the causal structure is presented below. First, the Darmois–Skitovitch theorem is referenced (Darmois 1953; Skitovitch 1953):
Theorem 1 (Darmois–Skitovitch theorem)
Define two random variablesy1andy2as linear combinations of the independent random variablessi(i = 1,…,Q):
Then, it can be shown that, ify1andy2are independent,all such variablessℓfor whichαℓβℓ≠ 0 areGaussian.
The contraposition of this theorem therefore shows that, if there exists a non-Gaussian sj for which αℓβℓ≠ 0,y1 and y2 are dependent.
To illustrate this, two variable LiNGAM cases are described. The number of observations is assumed to be large enough that estimation errors can be ignored. First, consider the case where x1 causes x2:
where b21≠ 0.
By regressing x2 on x1,
Thus, if x1(= e1) is the cause, because e1 and e2 are independent, x1 and \({r}_{2}^{(1)}(=e_{2})\) are also independent.
Next, consider the case where x2 causes x1:
where b12≠ 0. By regressing x2 on x1,
Thus, if x1 is not the cause, according to the Darmois–Skitovitch theorem, x1 and \({r}_{2}^{(1)}\) are dependent because e1 and e2 are non-Gaussian and independent. Furthermore, the coefficient of e1 on x1 and that of e1 on \({r}_{2}^{(1)}\) are non-zero because b12≠ 0 by definition. Therefore, the causal direction between x1 and x2 can be determined by examining the independence between explanatory variables and their residuals (Shimizu et al. 2011).
To evaluate independence, a measure that is not restricted to uncorrelatedness is needed because least-squares regression results in residuals that are always uncorrelated with but not necessarily independent of explanatory variables. For the same reason, non-Gaussianity is required for inferring the causal structure because uncorrelatedness is equivalent to independence for Gaussian variables. Common independence measures include HSIC (Gretton et al. 2005) and mutual information (Bach and Jordan 2002; Kraskov et al. 2004).
LiNGAM with Unobserved Common Causes
An extension of LiNGAM is now described for causal discovery in the presence of unobserved common causes (Hoyer et al. 2008). x1,…,xd denotes the observed variables, f1,…,fQ denotes the unobserved common causes, and e1,…,ed denotes the error variables. All of these variables are continuous. Then, the model is written as follows:
where bij and λiq are constants that represent the magnitudes of direct causation from xj and fq to xi, respectively (i,j = 1,…,p; q = 1,…,Q). The causal relations are assumed to be acyclic. The unobserved common causes fq (q = 1,…,Q) and error variables ei (i = 1,…,p) are further assumed to be non-Gaussian and independent. Although the assumption of independence for the unobserved common causes fq (q = 1,…,Q) looks strong, it can be made without loss of generality under the linearity assumption (Hoyer et al. 2008) because the observed variables are then linear combinations of error variables and hidden common causes.
By using the model in Eq. 34, the following two models with opposite directions of causation can be compared:
Figure 4 graphically represents these two models. Note that the number of unobserved common causes Q is assumed to be unknown.
In Shimizu and Bollen (2014), the model in Eq. 34 was related to a model with observation-specific intercepts instead of explicitly having unobserved common causes, as shown in Fig. 5. A major advantage of this approach is that neither the number of unobserved common causes Q nor number of coefficients λiq (i = 1,…,p; q = 1,…,Q) needs to be estimated. To explain the idea, the model in Eq. 34 for the observation m is rewritten as follows:
where \({x}_{i}^{(m)}, {f}_{q}^{(m)}\), and \({e}_{i}^{(m)}\) denote m-th observations of xi,fq, and ei, respectively (i = 1,…,p; q = 1,…,Q; m = 1,…,n).
Now, the sums of the unobserved common causes can be denoted by \({\mu }_{i}^{(m)}={\sum }_{q = 1}^{Q}\lambda _{iq}{f}_{q}^{(m)}\). Then, the following model is obtained with observation-specific intercepts:
where \({\mu }_{i}^{(m)}\) are observation-specific intercepts. The distributions of \({e}_{i}^{(m)}\) (m = 1,…,n) are assumed to be identical for every m. In this model, the observations are generated from the model with no unobserved common causes, possibly with different parameter values of the intercepts \({\mu }_{i}^{(m)}\). This model has the coefficients bij (i,j = 1,…,p) that are common to all observations as well as the observation-specific intercepts \({\mu }_{i}^{(m)}\). This is similar to mixed models (Demidenko 2004). Thus, it is called a mixed-LiNGAM.
Now, the problem of comparing Models 1 and 2 in Eqs. 35 and 36 becomes that of comparing Models 1′ and 2′:
where \({\mu }_{1}^{(m)} = {\sum }_{q = 1}^{Q}\lambda _{1q}{f}_{q}^{(m)}\) and \({\mu }_{2}^{(m)}={\sum }_{q = 1}^{Q}\lambda _{2q}{f}_{q}^{(m)}\) (m = 1,…,n).
A Bayesian approach is applied to compare Models 1′ and 2′ and estimate the possible causal direction between the two observed variables x1 and x2. The prior probabilities of the two candidate models are assumed to be uniform. Then, the log-marginal likelihoods of the two models may simply be compared to assess their plausibility. The model with the larger log-marginal likelihood is considered to be closest to the true model (Kass and Raftery 1995). Once the possible causal direction has been estimated, the coefficient b21 or b12 can be checked for its likeliness to be non-zero by examining its posterior distribution.
Error Distributions
The error distributions p(e1) and p(e2) can be modeled by using the generalized Gaussian distribution (Hyvärinen et al. 2001) as follows:
Here, the symbol Γ denotes the Gamma function:
where αi are the scaling parameters, and βi are the shape parameters (i = 1,2).
The error variances are
Thus, when the standard deviations of the errors are set to hi (i = 1,2), then the scaling parameters are automatically determined as follows:
Prior Distributions
Next, an informative prior distribution is used for the observation-specific intercepts \({\mu }_{i}^{(m)}\) (i = 1,2;m = 1,…,n). These observation-specific intercepts \({\mu }_{i}^{(m)}\) are the sums of many non-Gaussian independent unobserved common causes \({f}_{q}^{(m)}\) and are dependent. The central limit theorem states that the sum of independent variables becomes increasingly close to the Gaussian (Billingsley 1986). Based on this theorem, the non-Gaussian distributions of the observation-specific intercepts \({\mu }_{i}^{(m)}\) are approximated as the sums of many non-Gaussian independent unobserved common causes by using a bell-shaped curve distribution. The prior distribution of the observation-specific intercepts is modeled by the multivariate t-distribution as follows:
where τ1 and τ2 are constants, u ∼ tν(0,Σ), and Σ = [σab] is a symmetric scale matrix whose diagonal elements are 1s. C is a diagonal matrix whose diagonal elements give the variance of elements of u, i.e., \(\mathbf {C}=\frac {\nu }{\nu -2}\text {diag}(\boldsymbol {{\Sigma }})\) for ν > 2.
Numerical Examples
Experimental results using artificially generated data are presented here.Footnote 3 The parameters common to all of the observations were the coefficients b12 and b21 and the standard deviations of the error variables e1 and e2, which are denoted by h1 and h2. Then, the prior distributions of the parameters were modeled as follows:
The observation-specific intercepts \({\mu }_{i}^{(m)}\) (i = 1,2;m = 1,…,n) were generated as follows:
where the random variables u = [u1,u2]T followed the t-distribution with ν degrees of freedom ∼ tν(0,Σ). The parameters of the t-distribution Σ are given by the following positive definite matrix:
The standard deviations of the intercepts \({\mu }_{1}^{(m)}\) and \({\mu }_{2}^{(m)}\) are τ1 and τ2. σ12 determines the magnitude of covariance between the intercepts \({\mu }_{1}^{(m)}\) and \({\mu }_{2}^{(m)}\). The standard deviations of u1 and u2, which are denoted by std(u1) and std(u2), are \(\sqrt {\frac {\nu }{\nu -2}}\) because of the property of the t-distribution.
The hyper-parameters selected with the log-marginal likelihoods are the shape parameters β1 and β2 and the parameters of the prior distributions of the observation-specific intercepts \({\mu }_{1}^{(m)}\) and \({\mu }_{2}^{(m)}\), i.e., τ1,τ2, and σ21. An empirical Bayesian approach was used to select the hyper-parameters. The following were tested: β1,β2 = 0.5,1,2.0,6.0,τ1,τ2 = 0.4,0.6,0.8,σ12 = 0 ± 0.3,± 0.5,± 0.7,± 0.9. Then, the set of the hyper-parameters that achieved the largest log-marginal likelihood was selected. The naive Monte Carlo sampling approach was used to compute the log-marginal likelihoods with 10,000 samples for the parameters. The degree of freedom was fixed to eight.
Artificial datasets were generated with a sample size of 100 by using the following LiNGAM with unobserved common causes:
The Laplace or uniform distribution was randomly used for the distributions of the error variables e1 and e2. Their means were zero, and the standard deviations were \(\sqrt {3}\). The distributions of unobserved common causes fq were randomly selected from the 18 non-Gaussian distributions (Bach and Jordan 2002). The coefficient b21 was selected from the uniform distribution U(− 1.5,1.5). The constant c was 0.5 or 1.0. A larger c indicated a greater causal effect from the unobserved common cause fq. The number of unobserved common causes Q was 10. In this manner, 100 datasets were generated for every combination of the error distributions and constant c.
Subsequently, the log-marginal likelihoods of Models 1′ and 2′ were calculated, and the number of times the causal direction of the model with the largest log-likelihood was the same as that of the model used to generate the dataset was counted.
The Bayes factor was also computed. The Bayes factor of the two models being compared (Models 1′ and 2′) is denoted by K. To simplify the notation, K was assumed to be computed so that the larger likelihood was in the numerator and the smaller was in the denominator. Kass and Raftery (Kass and Raftery 1995) proposed that the Bayes factor is negligible if 2log K is 0–2, positive if 2log K is 2–6, strong if 2log K is 6–10, and very strong if 2log K is more than 10.
Overall, as the Bayes factor rose, so did the precision (i.e., the fraction of the number of findings that were successful) in both cases with the magnitudes of the effects of hidden common causes c = 0.5 and 1.0.
In the cases with the smaller magnitude of hidden common causes c = 0.5, for the model comparison indexes 2log K greater than 0 and no more than 2, the precision was 0.51, and the number of findings was 57. For the indexes 2log K greater than 2 and no more than 6, the precision was 0.67, and the number of findings was 96. For the indexes 2log K greater than 6 and no more than 10, the precision was 0.82, and the number of findings was 74. For the indexes 2log K greater than 10 and no more than 10, the precision was 0.82, and number of findings was 74. For the indexes 2log K greater than 10, the precision was 0.97, and number of findings was 173.
In the cases with the larger magnitude of hidden common causes c = 1.0, for the indexes 2log K greater than 0 and no more than 2, the precision was 0.58, and number of findings was 67. For the indexes 2log K greater than 2 and no more than 6, the precision was 0.57, and the number of findings was 131. For the indexes 2log K greater than 6 and no more than 10, the precision was 0.66, and the number of findings was 92. For the indexes 2log K greater than 10, the precision was 0.94, and the number of findings was 109.
This experimental result implies that considering the Bayes factor is useful when selecting a better model with the mixed-LiNGAM method. For the largest Bayes factor cases, the algorithm identified the correct model in more than 90% of the cases with a small sample size of 100.
Discussion
The main assumptions are the linearity and acyclicity of causal relations among observed variables and hidden common causes, non-Gaussian continuous errors, and such many hidden common causes whose sum can be approximated by a bell-shaped curve distribution. The effects of model violations have not yet been extensively studied and should be a good direction of future research. However, it should be possible to extend the proposed method to allow some types of nonlinearity and cyclicity based on the ideas of nonlinear and cyclic extensions (Hoyer et al. 2009; Zhang and Hyvärinen 2009; Lacerda et al. 2008) of basic LiNGAM.
Further, the effects of nonlinearly transforming observed variables should be investigated. Some transformations may make the observed variables more non-Gaussian, but they may also make the functional relations nonlinear. A promising way of modeling such transformations is to use the framework of post nonlinear causal models (Zhang and Hyvärinen 2009). The framework can handle variable-wise nonlinear transformations of observed variables generated from nonlinear and linear acyclic models with no hidden common causes, including the basic LiNGAM. The proposed method would benefit from such theoretical advances.
In the proposed approach, hidden common causes are assumed to be continuous. However, even if the hidden common causes are binary, their sum is approximated well by some bell-shaped curve distribution because of the central limit theorem if the number of hidden common causes is large enough. Therefore, the proposed Bayesian method should work better for more hidden common causes, as long as the noise levels including the magnitudes of effects of hidden common causes and those of error variables do not get too large. A natural way would be to use the Gaussian distributions to approximate the sums of hidden common causes motivated by the central limit theorem. However, in practice, the approximation may be not perfect, and there may be outliers. Thus, the t-distribution with heavier tails than the Gaussian distribution was used in the artificial data experiments in the hope that the inference would become more robust.
Further, in cases that all of the hidden common causes are known and measured, their effects can simply be removed by using regression. When only a smaller subset of the hidden common causes is known and measured, the current Bayesian approach for the two variable cases cannot fully benefit from the observed hidden common causes except when they are the only root variables, i.e., variables that have no parent variables. If they are the only root variables, the other variables only have to be conditioned on the root variables.
This study focused on two variable cases with hidden common causes. This is because analyzing only a smaller subset of observed variables does not lose validity if hidden common causes are allowed. For more than two variables, one approach is to apply the proposed method to every pair of the variables. Then, the estimation results can be combined to infer the entire causal graph.
Conclusion
The utilization of non-Gaussianity to estimate SEMs is useful for causal discovery because non-Gaussian methods are capable of uniquely estimating causal direction even in the presence of unobserved common causes under the model assumptions. Non-Gaussian data are widely encountered (Spirtes and Zhang 2016), and the non-Gaussian approach can be useful in such applications. Download links to papers and codes on this topic are available online: https://sites.google.com/site/sshimizu06/home/lingampapers.
Notes
These structural equations simply describe the data-generating processes and may be designed without the concept of causality.
Conditional independence-based approaches can also handle unobserved common causes, but their results usually contain many causal directed acyclic graphs, e.g., see the FCI algorithm (Spirtes et al. 1993).
Python codes written by Taku Yoshioka are freely available at https://github.com/taku-y/bmlingam
References
Bach, F.R., & Jordan, M.I. (2002). Kernel independent component analysis. Journal of Machine Learning Research, 3, 1–48.
Billingsley, P. (1986). Probability and measure. New York: Wiley-Interscience.
Bollen, K. (1989). Structural equations with latent variables. New York: Wiley.
Darmois, G. (1953). Analyse générale des liaisons stochastiques. Review of the International Statistical Institute, 21, 2–8.
Demidenko, E. (2004). Mixed models: Theory and applications. New York: Wiley-Interscience.
Gretton, A., Bousquet, O., Smola, A.J., Schölkopf, B. (2005). Measuring statistical dependence with Hilbert-Schmidt norms. In Proceedings of 16th international conference on algorithmic learning theory (ALT2005) (pp. 63–77).
Hoyer, P.O., Shimizu, S., Kerminen, A., Palviainen, M. (2008). Estimation of causal effects using linear non-Gaussian causal models with hidden variables. International Journal of Approximate Reasoning, 49, 362–378.
Hoyer, P.O., Janzing, D., Mooij, J., Peters, J., Schölkopf, B. (2009). Nonlinear causal discovery with additive noise models. Advances in Neural Information Processing Systems, 21, 689–696.
Hyvärinen, A., Karhunen, J., Oja, E. (2001). Independent component analysis. New York: Wiley.
Hyvärinen, A., Zhang, K., Shimizu, S., Hoyer, P. (2010). Estimation of a structural vector autoregression model using non-Gaussianity. Journal of Machine Learning Research, 11, 1709–1731.
Imbens, G.W., & Rubin, D.B. (2015). Causal inference in statistics, social, and biomedical sciences. Cambridge: Cambridge University Press.
Kass, R.E., & Raftery, A.E. (1995). Bayes factors. Journal of the American Statistical Association, 90, 773–795.
Kraskov, A., Stögbauer, H., Grassberger, P. (2004). Estimating mutual information. Physical Review E, 69, 066138.
Lacerda, G., Spirtes, P., Ramsey, J., Hoyer, P.O. (2008). Discovering cyclic causal models by independent components analysis. In Proceedings of the 24th conference on uncertainty in artificial intelligence (UAI2008) (pp. 366–374).
Mills-Finnerty, C., Hanson, C., Hanson, S.J. (2014). Brain network response underlying decisions about abstract reinforcers. NeuroImage, 103, 48–54.
Moneta, A., Entner, D., Hoyer, P., Coad, A. (2013). Causal inference by independent component analysis: theory and applications. Oxford Bulletin of Economics and Statistics, 75, 705–730.
Pearl, J. (2000). Causality: models, reasoning, and inference. Cambridge: Cambridge University Press.
Pearl, J., & Verma, T. (1991). A theory of inferred causation. In Allen, J., Fikes, R., Sandewall, E. (Eds.) Proceedings of the 2nd international conference on principles of knowledge representation and reasoning (pp. 441–452). San Mateo: Morgan Kaufmann.
Raitakari, O.T., Juonala, M., Rönnemaa, T., Keltikangas-Järvinen, L., Räsänen, L., Pietikäinen, M., Hutri-Kähönen, N., Taittonen, L., Jokinen, E., Marniemi, J., et al. (2008). Cohort profile: The cardiovascular risk in young finns study. International Journal of Epidemiology, 37, 1220–1226.
Rosenström, T., Jokela, M., Puttonen, S., Hintsanen, M., Pulkki-Råback, L., Viikari, J.S., Raitakari, O.T., Keltikangas-Järvinen, L. (2012). Pairwise measures of causal direction in the epidemiology of sleep problems and depression. PLoS ONE, 7, e50841.
Rubin, D.B. (1974). Estimating causal effects of treatments in randomized and nonrandomized studies. Journal of Educational Psychology, 66, 688–701.
Shimizu, S. (2014). LiNGAM: Non-gaussian methods for estimating causal structures. Behaviormetrika, 41, 65–98.
Shimizu, S., & Bollen, K. (2014). Bayesian estimation of causal direction in acyclic structural equation models with individual-specific confounder variables and non-Gaussian distributions. Journal of Machine Learning Research, 15, 2629–2652.
Shimizu, S., Hoyer, P.O., Hyvärinen, A., Kerminen, A. (2006). A linear non-Gaussian acyclic model for causal discovery. Journal of Machine Learning Research, 7, 2003–2030.
Shimizu, S., Inazumi, T., Sogawa, Y., Hyvärinen, A., Kawahara, Y., Washio, T., Hoyer, P.O., Bollen, K. (2011). DirectLiNGAM: A direct method for learning a linear non-Gaussian structural equation model. Journal of Machine Learning Research, 12, 1225–1248.
Skitovitch, W.P. (1953). On a property of the normal distribution. Doklady Akademii Nauk SSSR, 89, 217–219.
Spirtes, P., & Zhang, K. (2016). Causal discovery and inference: concepts and recent methodological advances. Applied Informatics, 3. https://doi.org/10.1186/s40535-016-0018-x.
Spirtes, P., Glymour, C., Scheines, R. (1993). Causation, prediction, and search. Berlin: Springer. (2nd edn. MIT Press 2000).
Zhang, K., & Chan, L. (2008). Minimal nonlinear distortion principle for nonlinear independent component analysis. Journal of Machine Learning Research, 9, 2455–2487.
Zhang, K., & Hyvärinen, A. (2009). On the identifiability of the post-nonlinear causal model. In Proceedings of the 25th conference on uncertainty in artificial intelligence (UAI2009) (pp. 647–655).
Zhang, K., & Hyvärinen, A. (2016). Nonlinear functional causal models for distinguishing causes form effect. In Wiedermann, W., & von Eye, A. (Eds.) Statistics and causality: methods for applied empirical research. Wiley.
Acknowledgments
The author thanks the guest editor Wolfgang Wiedermann and two reviewers for their helpful comments.
Funding
This work was supported by JSPS KAKENHI Grant Number 16K00045.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The author declares that there is no conflict of interest.
Ethical Approval
This article does not contain any studies with human participants or animals performed by the author.
Informed Consent
Informed consent was not required for this study.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Shimizu, S. Non-Gaussian Methods for Causal Structure Learning. Prev Sci 20, 431–441 (2019). https://doi.org/10.1007/s11121-018-0901-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11121-018-0901-x