Abstract
Chloroplast functional genomics, in particular understanding the chloroplast transcriptional response is of immense importance mainly due to its role in oxygenic photosynthesis. As a photosynthetic unit, its efficiency and transcriptional activity is directly regulated by reactive oxygen species during abiotic and biotic stress and subsequently affects carbon assimilation, and plant biomass. In crops, understanding photosynthesis is crucial for crop domestication by identifying the traits that could be exploited for crop improvement. Transcriptionally and translationally active chloroplast plays a key role by regulating the PSI and PSII photo-reaction centres, which ubiquitously affects the light harvesting. Using a comparative transcriptomics mapping approach, we identified differential regulation of key chloroplast genes during salt stress across Triticeae members with potential genes involved in photosynthesis and electron transport system such as CytB6f. Apart from differentially regulated genes involved in PSI and PSII, we found widespread evidence of intron splicing events, specifically uniquely spliced petB and petD in Triticum aestivum and high proportion of RNA editing in ndh genes across the Triticeae members during salt stress. We also highlight the role and differential regulation of ATP synthase as member of CF0CF1 and also revealed the effect of salt stress on the water-splitting complex under salt stress. It is worthwhile to mention that the observed conserved down-regulation of psbJ across the Triticeae is limiting the assembly of water-splitting complexes and thus making the BEP clade Triticeae members more vulnerable to high light during the salt stress. Comparative understanding of the chloroplast transcriptional dynamics and photosynthetic regulation will improve the approaches for improved crop domestication.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chloroplast as a photosynthetic powerhouse plays an important role in ATP harvesting and energy sequestration from light (Leister et al. 2017). With the rapidly advancing sequencing technologies, insights into the structural and functional organization of chloroplast and subsequent organization of photo-reaction centres in thylakoid membranes have been widely demonstrated and recently reviewed (Sablok et al. 2016). As compared to nuclear genomes, organelle genome with mono-, di- and polycistronic transcripts plays an important role in regulating the plant biomass as a measure of functional response during the abiotic stress (Suo et al. 2017). Chloroplast genome represents a typical quadripartite structure of 150–156 KB in angiosperms harbouring around 100–130 genes and repeats organized into large single copy (LSC) and short single copy (for a review see Sablok et al. 2016). Although the number of chloroplast-encoded proteins is relatively small as compared to nuclear proteins, enriched organization of functionally important genes in chloroplast play wide array of important roles such as photosynthetic carriers, maintaining the membrane fluidity, energy transport, and for maintaining the carbon assimilatory rate to boost biomass productivity (Leister et al. 2017). Among the cereal crops, in particular those which are contributing to the global economic food demand of 2050, understanding the physiological parameters that affect the water usage efficiency is of paramount importance (Araus et al. 2002). Previously, water usage efficiency, canopy structure which affects the chlorophyll concentrations and also the light scavenging mechanism have been widely demonstrated to a critical parameter for monitoring the mild or serve drought conditions (Araus et al. 2002). In case of drought and salinity stress conditions, photosynthetic apparatus, which not only scavenges the light and provides the required energy for state transitions but also optimizes the biomass accumulation has been widely studied (Leister et al. 2017). Apart from photosynthetic apparatus, there are other several subunits present in the thylakoid membrane of chloroplast, which play a key role in maintaining the osmotic conductance and the required pH for maintaining the sustainable membrane stability (Leister et al. 2017). Previously, much of the emphasis has been leveraged on understanding the genes involved in photosynthesis which is through the use of photosynthetic mutants (Spetea et al. 2014). However, the rapidly advancing filed of next generation sequencing, in particular RNA-seq has enabled to investigate the transcriptional analysis and highlighted the role of several processes such as RNA editing, intron splicing and promoters in maintaining the altered transcriptional landscape in chloroplast genomes under abiotic stress (Dinh et al. 2016; Sun and Guo 2016).
In addition to transcriptomics-based approaches, proteome-based investigations have been widely used to unravel the role of chloroplast translational differences under abiotic stress (Tamburino et al. 2017) highlighting reduction in the CO2 assimilation rates (Pintó-Marijuan and Munné-Bosch 2014). Apparent loss of the CO2 reduction has widely been linked to the biomass loss as a consequence of increase in the reactive oxygen species (Kwon et al. 2013; Pintó-Marijuan and Munné-Bosch 2014). In view of the ion transport and partitioning, it is apparent that selective accumulation of the Na+ ions has been shown to be a major factor regulating the transcription of the genes in the leaf and hence based on the ion accumulation of ions, turgor capacity can be seen to be fluctuated with Hordeum vulgare showing high propensity to maintain the turgor pressure during saline conditions as compared to Triticum aestivum (for a review see (Tang et al. 2015)). Previous reports highlight the increase rate of gluconeogensis as a measure to combat the salt stress, which is similar to the previously observed response in Arabidopsis thaliana (Guo et al. 2015), where increased concentration of mannose-6-phosphate reductase was found to be a measure to combat salt tolerance (Sickler et al. 2007).
Taking into account these previously illustrated functional changes, it is important to understand the transcriptional and translational variations in chloroplast under abiotic stress, particularly in those species, where the crop domestication has been widely affected due to the increasing salt concentration. Triticeae family represents an important family, with Aegilops and Triticum spp. representing the main food crops used globally around the world. Recent efforts have been mainly leveraged to understand the transcriptional profile of the nuclear genes in these species under abiotic stress to develop sustainable approaches to enhance the approaches for crop domestication (Ray et al. 2013). However, the lack in understanding the transcriptional landscape of chloroplast transcriptional plasticity across the Triticeae members at a comparative scale has not yet been established.
In the present research, we present a comparative transcriptional approach to highlight the functional differences in the chloroplast transcriptional machinery across the Triticeae members under salt stress. Using the comparative transcriptional approach, we identified differences not only in the transcriptional regulation of chloroplast-encoded genes but also revealed differences at the level of RNA editing and intron–exon splicing. Although the comparative assessment revealed conservation patterns across the transcriptional response in salt stress, distinct patterns of RNA editing and intron splicing differences were observed specifically in T. aestivum. The results observed in the present study using a comparative approach under salt stress, will allow to access the effects of salt stress on the transcriptional plasticity of chloroplast genes and will also elucidate the effects of photo-oxidation in salt stress, which will have global implication for crop domestication and for developing sustainable crop-breeding approaches.
Materials and methods
Plant growth and salt treatment
Seeds of a salt-tolerant cultivar of T. aestivum cv. Roshan were surface sterilized in a solution containing 3% (v/v) sodium hypochlorite for 5 min and then were rinsed three times with distilled water. Sterilized seeds were subsequently germinated on moistened filter paper (Whatman No. 1) in sterile petri dishes at a 24/20 °C cycle under a 16-h light/8-h dark photoperiod. Seedlings were transferred to pots (one seedling per pot) containing modified Hoagland’s nutrient solution (Munns 2013), which contains 6.5 mM KNO3, 4 mM Ca(NO3)2·4H2O, 2 mM MgSO4·7H2O, 0.1 mM NH4H2PO4, 0.045 mM FeCl3, 0.004 mM H3BO3, 0.0005 mM MnCl2·4H2O, 0.0002 mM ZnSO4·7H2O, 0.0002 mM CuSO4·5H2O and 0.0001 mM (NH4)6Mo7O24·4H2O, pH 6.5. The hydroponic system was based on the method of (Munns and James 2003). Solutions were changed every 7 days. Plants in trifoliate stage were divided into two groups representing the control and salt-treated group exposed to gradually increasing NaCl concentrations, respectively. In order to reduce the risk of plasmolysis due to osmotic shock, we used the “gradual step acclimation” method (Sanchez et al. 2008) during salt treatments. Salinity was increased from 50 mM at the start by 50 mM increments every day until the final level of 200 mM was reached. The experiment was performed with three replicates in a completely randomized design and root and leaf samples were subsequently harvested for ion measurements. For sequencing, only leaf samples were harvested as Na+-induced effect has been pre-dominantly shown previously in leaf and as reviewed in (Parida et al. 2002).
Ion measurement
Seven days after starting salt treatments (reaching a concentration of 200 mM), leaf and root samples were collected, washed in distilled water to remove any external salt and oven-dried at 60 °C for 48 h. The dried samples were ground into a fine powder using a mortar and pestle. Samples (1 g) were ashed at 600 °C in an electric furnace for 4 h, suspended in 5 mL of 2 N HCl, filtered through Whatman No. 1 filter paper and diluted with deionised water to a final volume of 50 mL. Concentrations of Na+ and K+ were measured according to standard flame photometer (Jenway PFP7, UK) procedure and reported as mg g-1 dry weight.
RNA extraction, cDNA library construction and sequencing
100 mg of leaf tissues were sampled from each of the three biological replicates and were used for extraction of the total RNA from the youngest leaves of both treated and control plants using the Trizol reagent (Invitrogen). Integrity of extracted RNAs was assessed by Bioanalyzer 2100 and RNA Nano 6000 Labchip kit (Agilent Technologies, Palo Alto, USA). RNA-Seq library preparation and sequencing were carried out at Beijing Genomics Institute BGI (Hong Kong, China). Briefly, for the RNA-seq libraries, 5 μg total RNA was treated with Ribo-Zero™ Magnetic Kit (Plant Leaf) (Epicentre) to deplete rRNA. The retrieved RNA is fragmented by adding First-Strand Master Mix (Invitrogen). First-strand cDNA is generated by First-Strand Master Mix and Super Script II reverse transcription (Invitrogen). Amplified products were purified using Agencourt RNAClean XP Beads, AGENCOURT, and then add Second-Strand Master Mix and dATP, dGTP, dCTP, dUTP mix were used to synthesize the second-strand cDNA (16 °C for 1 h). Following the purified cDNA was combined with end repair mix and incubated at 30 °C for 30 min. After purification with beads, A-Tailing mix was added and was further incubated at 37 °C for 30 min. Lastly, adenylated 3′ ends were combined and were incubated with index adapter and ligation mix at 30 °C for 10 min. Following, uracil-N-glycosylase enzyme was added into the purified ligation product and was further incubated at 37 °C for 10 min. The qualified libraries amplified on cBot were used to generate the cluster on the flow cell (TruSeq PE Cluster Kit V3-cBot-HS, Illumina) and were subsequently sequenced using HiSeq 2000 System (TruSeq SBS KIT-HS V3, Illumina). Sequencing was carried out in triplicates at BGI Shenzhen, China. The sequencing data have been deposited in NCBI Sequence Read Archive (SRA, http://www.ncbi.nlm.nih.gov/sra) under accession numbers SRR5648052 and SRR5647969.
Read mapping, coverage, splicing and editing in chloroplasts
FastQC software (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/) was used to assess the quality of raw sequence data. The raw reads were trimmed for quality using Trimmomatic software (Bolger et al. 2014). After evaluating the quality of the reads and removing low-quality reads and contaminated sequences, high-quality (Phred Score ≥ 30) reads were mapped to the chloroplast genome of T. aestivum (AB042240) and 6 closely related species of bread wheat, including Triticum urartu (KC912693), Triticum monocucum (KC912690), Aegilops tauschii (JQ754651), Aegilops speltoides (JQ740834), Aegilops cylindrica (KF534489), Aegilops geniculata (KF534490) using TOPHAT (Kim et al. 2013) with parameters (-g 2, --no-novel-juncs). For the coverage analysis, ChloroSeq (Castandet et al. 2016) with option –a 1 was used to identify the chloroplast transcriptional landscape from both DNA strands. ChloroSeq analysis reports a 100 nucleotide window coverage overlapping by 50 nt, normalized to the total number of reads. Splicing was quantified using ChloroSeq with option –a 2, which measures the ratio of spliced RNA to total RNA for a given gene (Castandet et al. 2016). RNA-editing sites were predicted using ChloroSeq option –a 3 and REDO tool (https://sourceforge.net/projects/redo/). To avoid false positives by ChloroSeq, we have kept only positions having efficiency > 5% and number of reads higher than 10.
Results and discussion
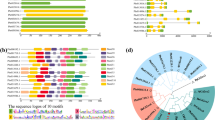
Among several of the ecological stresses, salt stress has been shown to be one of the most important factor affecting the distribution of the arable land globally and thus affecting the agricultural productivity per capita. Increasing evidences of salt stress globally is not only affecting the eco-physiological distribution and physiological adaptation of annual or perennial crops but has also resulted in the crop loss mainly due to the altered phenotypes with less biomass. Salt stress has been widely studied either from the transcriptional or post-transcriptional point of view in model and non-model plants including the economically important food and forage crops. However, limited reports address the fine scale resolution of salt stress at chloroplast level using comparative transcriptional approach across the Triticeae members (Fig. 1). In particular, in crop species much of the emphasis has been laid on the identification and elucidation of the nuclear genes involved in the salt stress, with reports identifying plethora of nuclear genes among which cation/anion transporters and anti-porters have been well defined (for a review see Tang et al. 2015). In the present research, we elucidate the chloroplast-based transcriptional regulation of salt stress using a comparative mapping approach.
Effect of salt on accumulation of ions and expression profiling
Na+ and K+ contents and K+/Na+ ratio in the upper leaves and roots of T. aestivum cv. Roshan treated with or without 200 mM NaCl are presented in Fig. 3. A significant accumulation of sodium ions as well as a reduction of potassium ions in the roots and leaves were observed in response to the salt treatment. As a result, the K+/Na+ ratio was significantly decreased by the salinity treatment in both roots and leaves. It is notable that roots accumulated Na+ at levels about three times higher than leaves. However, plasticity responses of Na+ accumulation have been widely demonstrated in leaves, which might reflect the ion transport (Parida et al. 2002) and its consequent effect on the electron balance across the thylakoid membrane (for a review see Tang et al. 2015). To analyse the organelle gene expression in long-term salt stress, we constructed and sequenced two cDNA libraries from the leaves of both salt-treated and control plants. After removing low-quality regions, adapters and all possible contaminations, a ~ 8.06 gigabase (GB) dataset was generated. Overall, 43,828,442 high-quality, clean reads with a GC of 44% for the control sample and 36,829,278 high-quality, clean reads with a GC of 44% for the salt-treated sample were obtained.
Photosynthetic efficiency during salt stress in Triticeae members
Significant changes in the abiotic-mediated stress expression have been widely demonstrated at the nuclear and organelle gene levels. Compared to nuclear gene expression level, organelle gene expression levels, in particular chloroplast gene expression levels mainly represent a subset of the genes mainly contributing towards the most essential process, photosynthesis, which plays a key role in maintaining the plant development (Leister et al. 2017) through light scavenging and ATP-mediated gene regulation (Fig. 2). To identify the comparative landscape of regulated genes across Triticeae members, we mapped the control and salt stress reads to 7 chloroplast genomes representing the Triticeae clade namely A. cylindrical (KF534489), A. geniculata (KF534490), A. speltoides (JQ740834), A. tauschii (JQ754651), T. aestivum (AB042240), T. monococcum (KC912690) and T. urartu (KC912693) using the ChloroSeq (Castandet et al. 2016). Chloroplast-embedded photosystem have been structurally, functionally and also biochemically explored for understanding and functionally elucidating the ionic exchange, membrane fluidity across the thylakoid membrane and also the synergistic and antagonistic effects on photosystem with respect to light harvesting (for a review see Allakhverdiev 2011; Najafpour and Allakhverdiev 2015). It has been widely shown that the photosynthetic organization in chloroplast has been divided into PSI and PSII at the levels of electron transport fluxes (Allakhverdiev 2011; Najafpour and Allakhverdiev 2015). PSII, a supramolecular pigment–protein complex plays an important role in light scavenging followed by water oxidation and plays an important role in maintaining the energy dissipation and also controls the reduction in the generation of singlet oxygen (Umate et al. 2007). Complex construction of the photosynthetic machinery and the regulation of the evolutionary-conserved genes in PSII has been a subject of immense importance to study the role of these low molecular weight (LMW) polypeptides in response to environmental responses. Although the PSII has been shown to be evolutionary conserved, the transcriptional and translational machinery of PSII has been shown to be distinct in Synechocystis (Allakhverdiev et al. 2002), where psbA genes have been shown to be suppressed, representing a damaged photo-oxidative state.
Among the 15 psb genes present in PSII, we observed up-regulation of psbA-D, psbM-N, psbT and psbZ, whereas the psbE, psbF, psbJ and psbL showed down-regulation (Table 1). psbA gene, which encodes the D1 protein has been defined as a major target of oxidative damage (Suo et al. 2017). However, to cope with salt stress or oxidative damage rapid denovo synthesis and degradation is required to maintain the PSII activation complex (Loll et al. 2007; Suo et al. 2017). Previously, salt stress-induced destabilization of PSI and complete loss of PSII have been shown and widely reviewed in Synechocystis (Allakhverdiev and Murata 2008; Allakhverdiev et al. 2000). Interestingly, both studies conclude that the loss was irreversible and was mainly affected by the increasing concentration of ions particularly Na+, which might be regulating the osmotic stress and also the membrane permeability, resulting in the loss of the cyclic electron flow. However, as compared to Synechocystis (Allakhverdiev et al. 2002), contrasting up-regulation of psbA genes has been observed, which allows for the maintenance of pH across membranes thus promoting the membrane stability (Saradhi et al. 1992). Up-regulation of psbA (D1) along with psbD, which encodes the D2 proteins was seen across all the BEP clade members, which indicates towards the rapid assimilation and degradation of D1 and D2 proteins as a counter measure to cope salt stress, and is in line with previous reports in tomato (Li et al. 2015). Up-regulation of the D1 and D2 proteins along with the lipid proteins indicates towards the conserved photoactivation of the damaged PSII across the BEP clade members.
Furthermore, we observed up-regulation of psbB and psbC, which encodes CP43 and CP47 enzymes, respectively, and plays a major role in the transfer of excitation energy as antenna proteins and have been shown to play major role in regulating the PSII photo-chemical efficiency by reducing the antenna size during the heat stress (Brestic et al. 2012) and salt stress (Singh et al. 2017). Interestingly, psbD was also found to be up-regulated, which has been shown to share translational coupling along with P680 complex and can be seen as a measure to combat the salt stress. Interestingly, we observed down-regulation of psbJ, which has been shown to play critical roles in the assembly of water-splitting complexes (Hager et al. 2002). Down-regulation of psbJ under salt stress hints towards the less stable assembly of the water-splitting complexes and thus high sensitivity to light during salt stress in BEP clade Triticeae members. Previously it has been shown that the assembly of psbJ also directly affects the PSI genes (Hager et al. 2002), which include psaA-J; however, we observed up-regulation of all psaA genes except psaJ (Table 1). We further examined the expression levels of rpoA, rpoB, rpoC1 and rpoC2, which encodes the subunit of plastid-encoded RNA polymerase (PEP) (Lgloi and Kössel 1992) and found all of them to be up-regulated (Table 1).
Plastid-encoded polymerase (PEP) in coordination with plastid-encoded promoters plays an important role in maintaining the active transcription of photosynthetic genes and has been shown to exist in diverse forms (Swiatecka-Hagenbruch et al. 2007). We observed up-regulation of psbB–psbT–psbH operon, which has a promoter for plastid-encoded RNA polymerase (PEP) (Stoppel and Meurer 2013) under salt stress suggesting a high interaction between the PEP polymerase and the promoter encoded by the psbB–psbT–psbH operon. It is worth to mention that psbH is a phosphoprotein encoded in cytochrome b 6 f complex and plays an important role in electron transport and has been shown to regulate the assembly of PSII (Bennett 1977; Stoppel and Meurer 2013).
Strikingly, up-regulation of D1, D2 and psbT across all BEP clade members clearly demonstrates that the post-translational repair required for the assembly of photodamaged PSII is supported by the up-regulated active transcription of psbT (Ohnishi et al. 2007). Previously, activation of repair system, which involves the synthesis of the precursor D1 protein has been shown as a defence system during low temperatures in Synechocystis sp. PCC 6803 (Mohanty et al. 2007). As compared to model plant Arabidopsis thaliana, where salt stress leads to the destruction of PSII and up-regulation of PSI, which in turn increases the cyclic electron flow, a dynamic regulation of PSII was observed across the BEP clade members. Similar down-regulation and damage to PSII have been previously been seen in Synechocystis sp. PCC 6803 under high light (Mohanty et al. 2007). Additionally, constant up-regulation of PSI genes psaA-I, expect psaJ, whose have functional roles are yet to be assigned suggesting a high cyclic electron flow in BEP clade members during the salt stress.
Previously dynamic up-regulation of PSI and PSII was observed in the Thellungiella halophila (Stepien and Johnson 2009), which is a salt-tolerant genotype and supports the observations from salt-tolerant phenotype of T. aestivum used in the present study.
Expression patterns of ATP subunits, cytochrome (Cyt) b6f and NADH genes
ATP synthase as a holo-subunit (CF0CF1) plays an important role in generating the required ATP for maintaining the proton motive force across the chloroplasts. We observed up-regulation of atpA genes across all the Triticeae members; however, down-regulation of rest of the atp subunits such as atpB and atpE, which are co-transcribed as di-cistronic was observed (Table 1). Interestingly, atpI was found to be up-regulated in salt stress, which is responsible for H+ translocations and acting as a H+ proton motive force across the thylakoid membrane to balance the osmotic pH tolerance (Table 1). Across all the Triticeae members, we further identified differential patterns of genes controlling the cytochrome b6f complex, which plays a key role in electron transfer and plastoquinone oxidation (Schöttler et al. 2007). We observed down-regulation of petA, which encodes cytochrome b6, whereas as up-regulation of petB, petD, which encodes 2 subunits of cytochrome (Cyt) b 6 f complex (Table 1). Cytochrome f complex loss has previously been shown in tobacco using petB and petD mutants illustrating that the expression of petB and petD is fundamental for the assembly of cytochrome b6f complex (Monde et al. 2000). However, the observed low expression of petA in Triticeae members indicated a relatively less stable assembly of dimeric cytochrome b 6 f. The observation is further supported with the down-regulation of petN in all the studied Triticeae members, which demonstrates the loss of the photosynthetic electrons and destabilization of Cytb6f complex during salt stress, which is in line with previously observed loss of functional mutants (Schwenkert et al. 2007). The observed destabilization is in line with a negligible increase in the expression of the petL in salt stress, which is a rate limiting gene for the confirmation of Rieske protein, playing a major role in Cytb6f complex stability (Schwenkert et al. 2007).
The observed down-regulation of genes involved in the Cytb6f complex and down-regulation of ATP synthase subunits reveals that overall salt stress resulted in the down-regulation of the proton motive force required for the ATP synthesis. However, the effect of the salt stress on polyploids is tolerant as compared to the diploids as revealed by few of the genes showing altered regulation pattern. This is further supported from the recent observation, which illustrates that polyploids are less susceptible to the chloroplast-mediated salt stress as compared to diploids (Meng et al. 2016). Triticeae members studied in this research represents a significant portion of the plants, which are C3 and play a major role in carbon assimilation. RUBISCO large subunit (Ribulose-1,5-bisphosphate carboxylase/oxygenase), rbcL plays a major role as carbon assimilatory enzyme. During salt stress, down-regulation of rbcL highlights the relatively less CO2-mediated carbon allocation as a result of oxidative damage, which indirectly down-regulates the crop yield.
Chloroplast NADH-like dehydrogenases play a critical role by recycling the electrons from ferredoxin (Fd) to PSI through plastoquinone and Cytb6f complex as intermediators (Peng et al. 2012). During salt stress across the Triticeae members, we observed most of the ndh genes up-regulated expect ndhC indicating a higher cyclic electron flow during the salt stress and thus improving the plant fitness (Table 1). Taking into account the studied genotype especially in case of T. aestivum, the present observation corroborates with the previous observation in soybean, which showed improved higher cyclic electron flow and plant fitness in salt-tolerant genotype (He et al. 2015). Up-regulation of the ndh genes along with the PSI genes during the salt stress strongly indicates the active formation of NDH-PSI supercomplex, which is a critical component of chlororespiration and alleviates the stromal over-reduction in salt stress (Peng et al. 2011) and thus sequestering more Na+ as a result of the available ATP through higher cyclic electron flow (He et al. 2015). High transcriptional activity of the ndh genes is also supported by the high abundance of the Na+/K+ ions during the salt stress (Fig. 3), which indicates the interplay of ATP demand and ndh-dependent cyclic electron flow. Previously, stromal over-reduction in stress due to NDH activity has been established in photosynthesis (Munekage et al. 2004). Taking into account these considerations, it can be concluded that NDH complex genes play an important role in the studied Triticeae members to circumvent the oxidative stress, which is in line with the observed NDH response in other perennial grasses (Van Den Bekerom et al. 2013).
Salt stress-mediated RNA splicing
Chloroplast-mediated gene regulation has widely been correlated with altered photosynthetic rates and membrane permeability during abiotic stress (Nouri et al. 2015). However, recently the role of alternative splicing in chloroplasts, which encodes both mono- and di-cistronic transcripts has been demonstrated. At nuclear level, the role of introns and alternative splicing has been widely demonstrated in land plants, with major representation of intron retention as the dominant splice events (Reddy et al. 2013). In chloroplast, presence of introns, specifically group II introns has been widely demonstrated to be a result of self-splicing ribozymes (Ostersetzer et al. 2005). We observed high splicing efficiency in atpF, ndhA and ndhB during salt stress across all Triticeae members (Fig. 4). Although the petB and petD also showed splicing efficiency, the observed change in the splicing efficiency was only observed in T. aestivum. To confirm, whether the observed splicing efficiency in the atpF, ndhA, ndhB was present across all members and petB and petD uniquely in T. aestivum, we further checked the intron expression levels (FPKM values), which revealed high expression and subsequent up-regulation of introns under salt stress (Table 2). The observed splicing-based regulation in T. aestivum is further supported by the corresponding low level of exon-based expression, where the exons were found to be down-regulated as compared to other members of the studied Triticeae members (Table 1). Higher intron expression and lower exonic expression during salt stress in T. aestivum highlight the role of group II intron-mediated splicing, which might regulate the assembly of Cytb6f complex (Monde et al. 2000). The observed splicing and the consecutive expression in atpF both at the exonic and intronic levels, which is a member of the H+ATPase (CF0 subunit of the CF0CF1) suggest the role of aberrant transcript production in response to salt stress. Taking together these observations, it can be presumed that introns play an important role in allopolyploids and may require species-specific chloroplast RNA splicing 1 (CRS1) interactions for effective splicing. Previously, in Zea mays, role and specificity of CRS1 as a splicing factor has been shown for group II-mediated intron splicing in atpF (Ostersetzer et al. 2005).
Salt stress-mediated RNA editing
RNA editing is a post-transcriptional phenomenon, which involves the conversion of cytosine to uridine and has been shown be regulated under the control of pentatricopeptide repeat (PPR) proteins (Kotera et al. 2005). Recent investigations across the organelle genomes of land plants revealed this post-transcriptional mechanism to be universally conserved across the land plants, and mainly relies on the editing factors CRR28 and RARE1 for post-transcriptional editing (Hein et al. 2016). Although the editing patterns vary across the land plants and even between the early branching plants and flowering plants, chloroplast genes such as ndh and accD display conserved pattern of editing across plant lineage (Hein et al. 2016). Differential patterns of RNA editing have been previously observed either at the gene level or at the ecotypic level, however, correlation between the RNA editing at the species level and at the level of the ecotypes has yet to be established. A recent example in perennial grass Lolium perenne demonstrated no differential patterns of RNA editing in ndh genes during drought stress (Van Den Bekerom et al. 2013), however ecotypes specific RNA editing was observed. Similarly, Δvac1 mutants (mutation in pentatricopeptide gene) showed partial editing of accD and ndH genes (Tseng et al. 2010). In contrast, recently Rodrigues et al. (2017) showed increased RNA editing of ndh, rps14 and rps16 genes in response to salt stress.
To investigate the potential effect of salt stress on RNA editing in studied Triticeae members, we identified editing sites across all the studied Triticeae members. RNA-editing events with over 90% efficiency was observed across ndh, psbZ, psbD, psbC, petL, petD and atpF (Table S1). Concurrent increase in the editing sites in ndh genes along with the up-regulation of ndh genes in salt stress strongly indicate that editing plays a crucial role in editing the transcriptionally active bases for up-regulation during the salt stress. It is worth to mention that ndh genes play a critical role as NADH-like dehydrogenases and also regulates the PSI cyclic electron flow. Previously, salt tolerance and cyclic electron flow around PSI have been found to be positively correlated in Glycine max (Lu et al. 2008; Rodrigues et al. 2017). Recently, in soybean, an enhanced editing rate has been reported in ndhA and ndhB during the salt stress revealing the role of editing in maintaining the homeostasis during the salt stress (Rodrigues et al. 2017). Moreover, conservation and editing of ndh genes have been widely observed across all land plant lineage including the basal Amborella suggesting that these groups of genes show variable patterns of RNA editing.
Interestingly, we didn’t observe high frequency editing events in ribosomal genes, which is in previously observed rps14 and rps16 editing by Rodrigues et al. (2017). Ribosomal genes are required for the translation of chloroplast-encoded peptides and loss of editing in these genes might suggest that the ribosomal editing is not affected during salt stress in studied Triticeae members. However, we indeed observe intronic editing events in rps16 and rpl2, during the salt stress and these events might be necessary for the splicing of the rps16 and rpl2 as evident from the splicing efficiency (Table S1). Previously, RNA editing of group II introns has been established as a pre-requisite for increasing the self-splicing in Oenothera (Börner et al. 1995). Additionally, editing in rps16 and rpl2 might be increasing the splicing efficiency by increasing the base pair frequency required for splicing (Ichinose and Sugita 2016). High rate of RNA editing in psb genes, which represents the PSII and Cytb6f complex indicates induced RNA editing as a consequence of salt stress and may be required for the effective translation of PSII genes to maintain the electron excitation levels and to help the assembly of Cytb6f complex to mediate the electron flow between the PSII and PSI. Nonetheless, we also observed RNA-editing events in rpoC1 and rpoC2, which play an important role in the assembly of the plastid-encoded polymerase (PEP). Taking into account the high expression of PEP polymerase during the salt stress, it can be concluded that the editing in the rpoC1 and rpoC2 contributes towards the effective translation of PEP polymerase and its interaction with the psbB–T–H-encoded promoter so as to circumvent the salt-induced oxidative stress.
Conclusion
In conclusion, we demonstrate the transcriptional landscape of chloroplasts across several species of Triticeae and highlights the key role of the photosynthetic genes including the water-splitting complex. We found up-regulation of genes associated with the plastid-encoded polymerase and psbB–psbT–psbH operon and down-regulation of petA. We observed conserved patterns of RNA editing in ndh genes and also unique patterns of petB and petD genes in T. aestivum. On a comparative assessment with previous reports of salt stress in diploids, it can be presumed that polyploids have less substantial effects of salt stress on the photodamage of photosynthetic reaction centres, which might be due to the altered Na+ translocations in vacuoles during salt stress in polyploids as compared to diploid progenitors. We also concluded that the up-regulation of the genes involved in the PSII as a defence measure to combat the negative oxidative damage is caused by salt stress.
References
Allakhverdiev SI (2011) Recent progress in the studies of structure and function of photosystem II. Elsevier, Amsterdam
Allakhverdiev SI, Murata N (2008) Salt stress inhibits photosystems II and I in cyanobacteria. Photosynth Res 98:529–539
Allakhverdiev SI, Sakamoto A, Nishiyama Y, Inaba M, Murata N (2000) Ionic and osmotic effects of NaCl-induced inactivation of photosystems I and II in Synechococcus sp. Plant Physiol 123:1047–1056
Allakhverdiev SI, Nishiyama Y, Miyairi S, Yamamoto H, Inagaki N, Kanesaki Y, Murata N (2002) Salt stress inhibits the repair of photodamaged photosystem II by suppressing the transcription and translation of psbAGenes in Synechocystis. Plant Physiol 130:1443–1453
Araus J, Slafer G, Reynolds M, Royo C (2002) Plant breeding and drought in C3 cereals: what should we breed for? Ann Bot 89:925–940
Bennett J (1977) Phosphorylation of chloroplast membrane polypeptides. Nature 269:344–346
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Börner GV, Mörl M, Wissinger B, Brennicke A, Schmelzer C (1995) RNA editing of a group II intron in Oenothera as a prerequisite for splicing. Mol Gen Genet 246:739–744
Brestic M, Zivcak M, Kalaji HM, Carpentier R, Allakhverdiev SI (2012) Photosystem II thermostability in situ: environmentally induced acclimation and genotype-specific reactions in Triticum aestivum L. Plant Physiol Biochem 57:93–105
Castandet B, Hotto AM, Strickler SR, Stern DB (2016) ChloroSeq, an optimized chloroplast RNA-Seq bioinformatic pipeline, reveals remodeling of the organellar transcriptome under heat stress. G3 Genes Genom Genet 6:2817–2827
Dinh SN, Sai TZT, Nawaz G, Lee K, Kang H (2016) Abiotic stresses affect differently the intron splicing and expression of chloroplast genes in coffee plants (Coffea arabica) and rice (Oryza sativa). J Plant Physiol 201:85–94
Guo R et al (2015) Comparative metabolic responses and adaptive strategies of wheat (Triticum aestivum) to salt and alkali stress. BMC Plant Biol 15:170
Hager M, Hermann M, Biehler K, Krieger-Liszkay A, Bock R (2002) Lack of the small plastid-encoded PsbJ polypeptide results in a defective water-splitting apparatus of photosystem II, reduced photosystem I levels, and hypersensitivity to light. J Biol Chem 277:14031–14039
He Y et al. (2015) Increasing cyclic electron flow is related to Na + sequestration into vacuoles for salt tolerance in soybean. J Exp Bot 66:6877–6889
Hein A, Polsakiewicz M, Knoop V (2016) Frequent chloroplast RNA editing in early-branching flowering plants: pilot studies on angiosperm-wide coexistence of editing sites and their nuclear specificity factors. BMC Evol Biol 16:23
Ichinose M, Sugita M (2016) RNA editing and its molecular mechanism in plant organelles. Genes 8:5
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36
Kotera E, Tasaka M, Shikanai T (2005) A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433:326
Kwon K-C, Verma D, Jin S, Singh ND, Daniell H (2013) Release of proteins from intact chloroplasts induced by reactive oxygen species during biotic and abiotic stress. PLoS ONE 8:e67106
Leister D, Wang L, Kleine T (2017) Organellar gene expression and acclimation of plants to environmental stress. Front Plant Sci 8:387
Lgloi G, Kössel H (1992) The transcriptional apparatus of chloroplasts. Crit Rev Plant Sci 10:525–558
Li J, Hu L, Zhang L, Pan X, Hu X (2015) Exogenous spermidine is enhancing tomato tolerance to salinity–alkalinity stress by regulating chloroplast antioxidant system and chlorophyll metabolism. BMC Plant Biol 15:303
Loll B, Kern J, Saenger W, Zouni A, Biesiadka J (2007) Lipids in photosystem II: interactions with protein and cofactors. Biochim Biophys Acta 1767:509–519
Lu K, Yang Y, He Y, Jiang D (2008) Induction of cyclic electron flow around photosystem 1 and state transition are correlated with salt tolerance in soybean. Photosynthetica 46:10–16
Meng F et al. (2016) Physiological and proteomic responses to salt stress in chloroplasts of diploid and tetraploid black locust (Robinia pseudoacacia L.). Sci Rep. https://doi.org/10.1038/srep23098
Mohanty P, Allakhverdiev SI, Murata N (2007) Application of low temperatures during photoinhibition allows characterization of individual steps in photodamage and the repair of photosystem II. Photosynth Res 94:217–224
Monde RA, Zito F, Olive J, Wollman FA, Stern DB (2000) Post-transcriptional defects in tobacco chloroplast mutants lacking the cytochrome b6/f complex. Plant J 21:61–72
Munekage Y, Hashimoto M, Miyake C, Tomizawa K-I, Endo T, Tasaka M, Shikanai T (2004) Cyclic electron flow around photosystem I is essential for photosynthesis. Nature 429:579–582
Munns R (2013) Hoagland’s nutrient solution. http://prometheuswiki.publish.csiro.au/tiki-index.php?page=Hoagland's+nutrient+solution
Munns R, James RA (2003) Screening methods for salinity tolerance: a case study with tetraploid wheat. Plant Soil 253:201–218
Najafpour MM, Allakhverdiev SI (2015) Recent progress in the studies of structure and function of photosystems I and II. J Photochem Photobiol B 152:173
Nouri M-Z, Moumeni A, Komatsu S (2015) Abiotic stresses: insight into gene regulation and protein expression in photosynthetic pathways of plants. Int J Mol Sci 16:20392–20416
Ohnishi N, Kashino Y, Satoh K, Ozawa S-i, Takahashi Y (2007) Chloroplast-encoded polypeptide PsbT is involved in the repair of primary electron acceptor QA of photosystem II during photoinhibition in Chlamydomonas reinhardtii. J Biol Chem 282:7107–7115
Ostersetzer O, Cooke AM, Watkins KP, Barkan A (2005) CRS1, a chloroplast group II intron splicing factor, promotes intron folding through specific interactions with two intron domains. Plant Cell 17:241–255
Parida A, Das AB, Das P (2002) NaCl stress causes changes in photosynthetic pigments, proteins, and other metabolic components in the leaves of a true mangrove, Bruguiera parviflora, in hydroponic cultures. J Plant Biol 45:28–36
Peng L, Yamamoto H, Shikanai T (2011) Structure and biogenesis of the chloroplast NAD (P) H dehydrogenase complex. Biochim Biophys Acta 1807:945–953
Peng L, Fukao Y, Fujiwara M, Shikanai T (2012) Multistep assembly of chloroplast NADH dehydrogenase-like subcomplex A requires several nucleus-encoded proteins, including CRR41 and CRR42, in Arabidopsis. Plant Cell 24:202–214
Pintó-Marijuan M, Munné-Bosch S (2014) Photo-oxidative stress markers as a measure of abiotic stress-induced leaf senescence: advantages and limitations. J Exp Bot 65:3845–3857
Ray DK, Mueller ND, West PC, Foley JA (2013) Yield trends are insufficient to double global crop production by 2050. PLoS ONE 8:e66428
Reddy AS, Marquez Y, Kalyna M, Barta A (2013) Complexity of the alternative splicing landscape in plants. Plant Cell 25:3657–3683
Rodrigues NF, Fonseca GC, Kulcheski FR, Margis R (2017) Salt stress affects mRNA editing in soybean chloroplasts. Genet Mol Biol 40:200–208
Sablok G, Mudunuri SB, Edwards D, Ralph PJ (2016) Chloroplast genomics: expanding resources for an evolutionary conserved miniature molecule with enigmatic applications. Curr Plant Biol 7:34–38
Sanchez DH et al (2008) Integrative functional genomics of salt acclimatization in the model legume Lotus japonicus. Plant J 53:973–987
Saradhi A, Pardha P, Mohanty P (1992) Enhancement of photosystem II photoreactions and high pH stability in thylakoids from cotyledonary leaves of Brassica juncea raised under sodium chloride stress. Physiol Plant 86:189–196
Schöttler MA, Flügel C, Thiele W, Bock R (2007) Knock-out of the plastid-encoded PetL subunit results in reduced stability and accelerated leaf age-dependent loss of the cytochrome b6f complex. J Biol Chem 282:976–985
Schwenkert S, Legen J, Takami T, Shikanai T, Herrmann RG, Meurer J (2007) Role of the low-molecular-weight subunits PetL, PetG, and PetN in assembly, stability, and dimerization of the cytochrome b6f complex in tobacco. Plant Physiol 144:1924–1935
Sickler CM, Edwards GE, Kiirats O, Gao Z, Loescher W (2007) Response of mannitol-producing Arabidopsis thaliana to abiotic stress. Funct Plant Biol 34:382–391
Singh RP, Runthala A, Khan S, Jha PN (2017) Quantitative proteomics analysis reveals the tolerance of wheat to salt stress in response to Enterobacter cloacae SBP-8. PLoS ONE 12:e0183513
Spetea C, Rintamäki E, Schoefs B (2014) Changing the light environment: chloroplast signalling and response mechanisms. Philos Trans R Soc Lond B 369:20130220
Stepien P, Johnson GN (2009) Contrasting responses of photosynthesis to salt stress in the glycophyte Arabidopsis and the halophyte Thellungiella: role of the plastid terminal oxidase as an alternative electron sink. Plant Physiol 149:1154–1165
Stoppel R, Meurer J (2013) Complex RNA metabolism in the chloroplast: an update on the psbB operon. Planta 237:441–449
Sun A-Z, Guo F-Q (2016) Chloroplast retrograde regulation of heat stress responses in plants. Front Plant Sci 7:398
Suo J, Zhao Q, David L, Chen S, Dai S (2017) Salinity response in chloroplasts: insights from gene characterization. Int J Mol Sci 18:1011
Swiatecka-Hagenbruch M, Liere K, Börner T (2007) High diversity of plastidial promoters in Arabidopsis thaliana. Mol Genet Genomics 277:725–734
Tamburino R et al (2017) Chloroplast proteome response to drought stress and recovery in tomato (Solanum lycopersicum L.). BMC Plant Biol 17:40
Tang X, Mu X, Shao H, Wang H, Brestic M (2015) Global plant-responding mechanisms to salt stress: physiological and molecular levels and implications in biotechnology. Crit Rev Biotechnol 35:425–437
Tseng C-C, Sung T-Y, Li Y-C, Hsu S-J, Lin C-L, Hsieh M-H (2010) Editing of accD and ndhF chloroplast transcripts is partially affected in the Arabidopsisvanilla cream1 mutant. Plant Mol Biol 73:309–323
Umate P et al (2007) Deletion of PsbM in tobacco alters the QB site properties and the electron flow within photosystem II. J Biol Chem 282:9758–9767
Van Den Bekerom RJ, Dix PJ, Diekmann K, Barth S (2013) Variations in efficiency of plastidial RNA editing within ndh transcripts of perennial ryegrass (Lolium perenne) are not linked to differences in drought tolerance. AoB Plants 5:plt035
Acknowledgements
We thank Institute of Science, High technology and Environmental Sciences, Graduate University of Advanced Technology for financial support (No. 94/5149). We also thank Iran National Foundation Science for financial support (No. 90003984).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mirzaei, S., Mansouri, M., Mohammadi-Nejad, G. et al. Comparative assessment of chloroplast transcriptional responses highlights conserved and unique patterns across Triticeae members under salt stress. Photosynth Res 136, 357–369 (2018). https://doi.org/10.1007/s11120-017-0469-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11120-017-0469-5