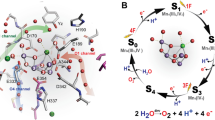
Abstract
In the cyanobacterial photosystem II (PSII), the O4-water chain in the D1 and CP43 proteins, a chain of water molecules that are directly H-bonded to O4 of the Mn4Ca cluster, is linked with a channel that connects the protein bulk surface along with a membrane-extrinsic protein subunit, PsbU (O4-PsbU channel). The cyanobacterial PSII structure also shows that the O1 site of the Mn4Ca cluster has a chain of H-bonded water molecules, which is linked with the channel that proceeds toward the bulk surface via PsbU and PsbV (O1-PsbU/V channel). Membrane-extrinsic protein subunits PsbU and PsbV in cyanobacterial PSII are replaced with PsbP and PsbQ in plant PSII. However, these four proteins have no structural similarity. It remains unknown whether the corresponding channels also exist in plant PSII, because water molecules are not identified in the plant PSII cryo-electron microscopy (cryo-EM) structure. Using the cyanobacterial and plant PSII structures, we analyzed the channels that proceed from the Mn4Ca cluster. The cyanobacterial O4-PsbU and O1-PsbU/V channels were structurally conserved as the channel that proceeds along PsbP toward the protein bulk surface in the plant PSII (O4-PsbP and O1-PsbP channels, respectively). Calculated protonation states indicated that in contrast to the original geometry of the plant cryo-EM structure, protonated PsbP-Lys166 may form a salt-bridge with ionized D1-Glu329 and protonated PsbP-Lys173 may form a salt-bridge with ionized PsbQ-Asp28 near the O1-PsbP channel. The existence of these channels might explain the molecular mechanism of how PsbP can interact with the Mn4Ca cluster.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In photosystem II (PSII), the Mn4CaO5 cluster catalyzes the water-splitting reaction: 2H2O→O2 + 4H++4e− (reviewed in refs. (Dau et al. 2012; Cox and Messinger 2013; Shen 2015)). Protons are released in response to changes in the oxidation state (the S n state, where the subscript represents the number of oxidation steps accumulated) of the oxygen-evolving complex, and it occurs with a typical stoichiometry of 1:0:1:2 for the S0→S1→S2→S3(→S4)→S0 transitions, respectively. Candidates for the relevant proton transfer pathways (e.g., (Renger 2001; Murray and Barber 2007; Ho and Styring 2008; Vassiliev et al. 2012; Ogata et al. 2013; Linke and Ho 2014) and proton-releasing sites (e.g., (Saito et al. 2015)) have been reviewed recently. The energetically lowest process for proton release from the Mn4CaO5 cluster is the S0-to-S1 transition, where the electron transfer occurs prior to H+ release and is thus rate limiting (Dau and Haumann 2008). It has been argued that the exchangeable μ-oxo bridge is possibly either O4 (linking Mn4 and Mn3 in Mn3CaO4-cubane) or O5 (in one of the corners of the cubane linking Mn4 and the cubane) (Yamanaka et al. 2011; Rapatskiy et al. 2012; McConnell et al. 2012; Galstyan et al. 2012; Cox et al. 2013) (see also relevant articles published prior to the detailed crystal structure (Robblee et al. 2002; Messinger 2004; Kulik et al. 2007)). The PSII crystal structures (Umena et al. 2011; Suga et al. 2015) possess a chain of strongly H-bonded water molecules (O4-water chain) directly linked to O4. The cyanobacterial PSII crystal structure also identified a chain of water molecules near the O1 site of the Mn4Ca cluster (O1-water chain). The potential energy profile of the H-bonds along the O4-water chain indicated that the proton released from OH− at O4 could be transferred along the O4-water chain (Saito et al. 2015). The activation energy of proton transfer is the lowest when all the water molecules are strongly H-bonded in the H-bond network (Stuchebrukhov 2009; Takaoka et al. 2016). In the cyanobacterial PSII crystal structure (Umena et al. 2011), the O4-water chain is connected with a channel that extends to the protein bulk surface via the PsbU protein subunit (O4-PsbU channel) (Takaoka et al. 2016). This channel may correspond to a previously proposed possible proton channel of PSII (Gabdulkhakov et al. 2009; Linke and Ho 2014). However, membrane-extrinsic protein subunits PsbU and PsbV in cyanobacteria are replaced with PsbP and PsbQ in higher plants and green algae (Bricker et al. 2012; Ifuku and Noguchi 2016). These extrinsic proteins are associated with the efficiency of O2-evolving activity in PSII (Ifuku et al. 2005). They are also involved in the retention of Ca2+ and Cl− (Ghanotakis et al. 1984a; Shen and Inoue 1993) or play a role in protecting the Mn4Ca cluster (Ghanotakis et al. 1984b). Notably, the crystal structures of the isolated PsbP (Ifuku et al. 2004) and PsbQ (Calderone et al. 2003) proteins show that the plant PsbP and PsbQ proteins and the cyanobacterial PsbU and PsbV proteins (Ferreira et al. 2004; Umena et al. 2011) have no structural similarities. This raises questions regarding the function of the O4-water chain and the continuous PsbU-like channel in plant PSII.
Recently, the cryo-electron microscopy (cryo-EM) structure of the plant PSII from spinach was reported at a resolution of 3.2 Å (Wei et al. 2016). The plant cryo-EM structure also shows membrane-extrinsic protein subunits, including PsbP and PsbQ. On the other hand, the existence of the channels in PsbP and PsbQ is unclear, because the positions of water molecules are not reported in the plant cryo-EM structure. Here, we present the structural properties of the channels that proceed from the Mn4Ca cluster toward the protein bulk surface in plant PSII, using the plant cryo-EM structure.
Methods
Coordinates and atomic partial charges
As a basis for the computations, the X-ray crystal structure of cyanobacterial PSII (PDB ID: 3ARC) (Umena et al. 2011) and cryo-EM structure of plant PSII (PDB ID: 3JCU) (Wei et al. 2016) were used. Hydrogen atoms were generated and energetically optimized using CHARMM version 40b (Brooks et al. 1983), where the positions of all non-hydrogen atoms were fixed and all titratable groups were kept in their standard protonation states (i.e., acidic and basic groups were ionized). Atomic partial charges of the amino acids were adopted from the all-atom CHARMM22 (MacKerell et al. 1998) parameter set. The atomic charges of cofactors were taken from our previous studies on PSII (Saito et al. 2011).
Analysis of the channel space and distribution of water molecules
The channel space was analyzed using the program CAVER (Petrek et al. 2006). To analyze the possible distribution of water molecules and Cl− in the channels, including the region where these molecules and ions are not identified in the crystal structure, we used a three-dimensional reference interaction site model (3D-RISM) with Placevent analysis (Beglov and Roux 1997; Kovalenko and Hirata 1999; Luchko et al. 2010; Case et al. 2012; Sindhikara et al. 2012), as previously used for the inner channel of channelrhodopsin (Watanabe et al. 2013; Wietek et al. 2014) and PSII (Takaoka et al. 2016). It should be noted that the distribution pattern of water molecules obtained from the 3D-RISM with Placevent analysis was consistent with the positions of the water molecules identified in the PSII crystal structure (Takaoka et al. 2016).
MD simulations
The PS II assembly described above was embedded in a lipid bilayer consisting of 546 1-palmitoyl-2-oleyl-sn-glycero-3-phosphocholine (POPC) and soaked in 80,353 flexible water models (SPC-Fw) (Wu et al. 2006). After structural optimization with position restraints on heavy atoms of the PSII assembly, the system was heated from 0.001 to 300 K during 5.0 ps, and the restraints were gradually released over 1.0 ns. After an equilibrating MD run for 45 ns, a production run was conducted over 5.0 ns for B-factor evaluation. All the equilibrating simulations were conducted by MD engine AMBER 14 (Case et al. 2014) with SHAKE algorithm for hydrogen constraint (Ryckaert et al. 1977), while NAMD version 2.10 (Phillips et al. 2005) was used for the production run with an MD time step of 0.5 fs without hydrogen constraint. For temperature and pressure control, Berendsen thermostat and barostat were employed in the equilibrating process (Berendsen et al. 1984), while Langevin thermostat and piston were used in the production run (Kubo et al. 1991; Feller et al. 1995).
Protonation pattern and pK a values
The present computation was based on the electrostatic continuum model created by solving the linear Poisson–Boltzmann equation with the MEAD program (Bashford and Karplus 1990). To obtain the protonation pattern of all titratable residues in PSII, we calculated the differences in electrostatic energies between protonated and deprotonated states in a reference model system using an experimentally measured pK a value. The experimentally measured pK a values employed as references were 12.0 for Arg, 4.0 for Asp, 9.5 for Cys, 4.4 for Glu, 10.4 for Lys, 9.6 for Tyr (Nozaki and Tanford 1967), and 7.0 and 6.6 for the Nε and Nδ atoms of His, respectively (Tanokura 1983a, b, c). The ensemble of the protonation patterns was sampled using a Monte Carlo method with Karlsberg (Rabenstein and Knapp 2001). The dielectric constants were set to ε p = 4 for inside the protein and ε w = 80 for water. All computations were performed at 300 K, pH 7.0 at an ionic strength of 100 mM. The linear Poisson–Boltzmann equation was solved using a three-step grid-focusing procedure at the resolutions of 2.5, 1.0, and 0.3 Å. The Monte Carlo sampling yielded the probabilities of the two protonation states, [protonated] and [deprotonated], for each titratable site.
Results and discussion
The O4-water chain and the connecting PsbU and PsbP channels
The channel space of the O4-water chain, originally reported in the cyanobacterial PSII structure (Saito et al. 2015; Takaoka et al. 2016), was also present in the D1 and CP43 proteins of the plant cryo-EM structure (Fig. 1). Channel space analysis suggested that the plant O4-water chain also proceeds from the Mn4Ca cluster toward CP43-Pro334, CP43-Thr335, and D1-Asn338, similar to the reported cyanobacterial O4-water chain (Saito et al. 2015; Takaoka et al. 2016). Analysis of the plant cryo-EM structure using 3D-RISM with Placevent suggested that water molecules could be present along the O4-water chain, as previously reported in the cyanobacterial structure (Takaoka et al. 2016). The distribution patterns of water molecules in the plant cryo-EM structure were consistent with the distribution patterns and the positions of water molecules in the cyanobacterial PSII structure (Takaoka et al. 2016) (Fig. 1). The shapes of O4-water chains of the cyanobacterial and plant PSII are not remarkably different, as the D1 and CP43 proteins, where the Mn4Ca cluster and the H-bonded O4-water chain are located, are highly conserved in the two PSII proteins.
In the cyanobacterial PSII, the O4-water chain in the D1 and CP43 proteins is further linked with the O4-PsbU channel (Takaoka et al. 2016). We found that the corresponding channel was also present along the interface of PsbP (O4-PsbP channel) in the plant cryo-EM structure, irrespective of the low structural similarity of PsbP with PsbU (Fig. 2a). The cyanobacterial O4-PsbU channel is mainly composed of D1, D2, CP43, CP47, PsbO, and PsbU (Takaoka et al. 2016), whereas the plant O4-PsbP channel is composed of only four protein subunits, D1, D2, CP43, and PsbP (Table 1). Both the O4-PsbP and O4-PsbU channels commonly start at the terminus of the O4-water chain (e.g., the D1-Asn338 moiety). On the other hand, the orientations of the channels toward the protein bulk surface are different. Superposition of the two channel spaces suggests that the plant O4-PsbP channel would not exist in cyanobacterial PSII due to the presence of PsbU-Tyr98 (Fig. 2c), whereas the cyanobacterial O4-PsbU channel would not exist in plant PSII due to the presence of PsbP-Val108 and PsbP-Asp137 (Fig. 2b). Notably, near the channel branch, there exists a salt-bridge between CP43-Lys339 and PsbP-Asp137 in plant PSII and between CP43-Lys339 and PsbU-Asp96 in cyanobacterial PSII. The two salt-bridges are most likely conserved and associated with the channel orientation (Fig. 2c).
Location of the cyanobacterial O4-PsbU (blue mesh) and plant O4-PsbP (green mesh) channels. a Overview. b Superposition of the cyanobacterial PsbU channel space into the plant cryo-EM structure (Wei et al. 2016). c Superposition of the plant PsbP channel space into the cyanobacterial structure (Umena et al. 2011). Residues that interrupt the channels are circled in red
The entire lengths and radii of the O4-PsbU and O4-PsbP channels were similar (Fig. 3). The radii of both the channels were smaller than ~1.4 Å in most of the regions, implying that water molecules are arranged as a single chain. These analyses suggest that plant PsbP may correspond to cyanobacterial PsbU in terms of connecting the O4-water chain in the D1 protein with the PSII protein bulk surface.
The channel space of the O4-water chain and the connecting O4-PsbU channel comprise several loop regions located in the protein–subunit interface with D1, CP43, and PsbU, e.g., residues D1−167 to 170, D1−333 to 337, CP43−333 to 339, CP43−353 to 357, PsbU−13 to 17, PsbU−21 to 22, PsbU−30 to 34, and PsbU−91 to 102. To evaluate the structural stability of the loop regions, MD simulations were performed for the cyanobacterial PSII complex (Umena et al. 2011). The calculated B-factors in the D1, CP43, and PsbU in the cyanobacterial PSII complex were highly associated with the original B-factors of the crystal structure (Fig. 4). Remarkably, the MD simulations suggested significantly high structural stability in the loop regions of D1, CP43, and PsbU proteins (Fig. 4). The calculated B-factors of the backbone Cα atoms in the loop regions were as low as those in the helices (e.g., trans-membrane helices D1−34 to 53, D1−110 to 136, D1−143 to 165, D1−196 to 221, and D1−268 to 292). Although MD simulations were not performed for the plant cryo-EM structure (Wei et al. 2016), high structural stability would be expected for the plant O4-water chain because of the structural similarity between the D1 and CP43 protein subunits in the cyanobacterial and plant PSII structures.
Calculated B-factors of the backbone Cα atoms in the cyanobacterial D1, CP43, and PsbU proteins, using the cyanobacterial 1.9 Å structure (closed circles) (Umena et al. 2011). The protein-loop regions of D1, CP43, and PsbU (D1−167 to 170, D1−333 to 337, CP43−333 to 339, CP43−353 to 357, PsbU−13 to 17, PsbU−21 to 22, PsbU−30 to 34) are colored red, while the other regions are colored black. For comparison, the B-factors stated in the PDB file (PDB ID: 3ARC) are shown (black open circles). Calculated B-factors are highly correlated with the original B-factors of the crystal structure
The O1-water chain and the connecting PsbU/V and PsbP channels
The cyanobacterial PSII crystal structure shows that the O1 site of the Mn4Ca cluster has a chain of H-bonded water molecules, the O1-water chain (Fig. 5) (Umena et al. 2011). Channel space analysis suggested that the O1-water chain in D1, which terminated near D1-Glu329, was linked with the channel that initiated near the CP43-Glu413…PsbV-Lys47 salt-bridge (NPsbV−Lys47–OCP43−Glu413 = 2.8 Å) and proceeded toward the bulk surface via PsbU and PsbV (O1-PsbU/V channel, Fig. 5). Remarkably, the corresponding water chain and the linked channel were conserved in the plant PSII; the O1-water chain was linked with the channel that initiated near the CP43-Glu413…PsbP-Lys170 salt-bridge (NPsbP−Lys170–OCP43−Glu413 = 2.7 Å) and proceeded toward the bulk surface via PsbP (O1-PsbP channel, Fig. 5). From the comparison, it seems likely that the salt-bridge between CP43-Glu413 and PsbP-Lys170 in plant PSII is structurally conserved as the salt-bridge between CP43-Glu413 and PsbV-Lys47 in cyanobacterial PSII.
PsbV-Lys47 seems to be involved in the region that affects the redox potential (E m) of heme in PsbV (i.e., cytochrome c550), in response to binding of the isolated PsbV protein to the PSII complex (Ishikita and Knapp 2005). The exact role of cytochrome c550 in the PSII complex is unclear, but the E m value of the heme differs significantly, by ~160 mV between the isolated cytochrome c550 form (Kerfeld et al. 2003) (–240 mV) and the PSII-bound cytochrome c550 form (–80 mV) (Roncel et al. 2003). It has been pointed out that the difference in the protein backbone C=O orientations in the two PsbV forms could explain 113 mV of the total E m difference, the majority of which (~80 mV) originated from the region of PsbV− 43 to 53 (Ishikita and Knapp 2005). In particular, the backbone C=O orientation differs significantly at PsbV-Lys47 in the two cytochrome c550 forms (Ishikita and Knapp 2005), which is obviously due to the formation of the CP43-Glu413…PsbV-Lys47 salt-bridge upon binding of the PsbV subunit to the PSII complex. The cyanobacterial CP43-Glu413…PsbV-Lys47 salt-bridge seems to correspond to the plant CP43-Glu413…PsbP-Lys170 salt-bridge (Fig. 5).
The cyanobacterial O1-PsbU/V and plant O1-PsbP channels have larger channel radii and shorter channel lengths than the cyanobacterial O4-PsbU and O4-PsbP channels (Fig. 3). In the narrow pore of the proton-conducting O4-water chain, 8 or 9 water molecules, in particular the initial four O moieties on the proton donor side (i.e., O4…OW539…OW538…OW393), are tightly H-bonded (Takaoka et al. 2016). The larger channel radii in the O1-PsbU/V and O1-PsbP channels imply that water molecules are probably more mobile than those in the O4-PsbU and O4-PsbP channels. Therefore, the O1-PsbU/V and O1-PsbP channels are likely to serve as a water-intake channel rather than a proton-conducting channel.
Protonation states near the channels in extrinsic proteins
The extrinsic proteins, cyanobacterial PsbU and PsbV proteins and plant PsbP and PsbQ proteins, are rich in charged residues. The calculated protonation states of titratable residues obtained using the cyanobacterial PSII structure indicated that in cyanobacterial PsbU and PsbV, all charged residues were in the standard protonation states, i.e., protonated basic and deprotonated acidic residues. However, the calculated protonation states of titratable residues obtained using the plant cryo-EM structure (Wei et al. 2016) indicated that 9 charged residues in PsbP and 7 charged residues in PsbQ were not in the standard protonation states, i.e., protonated acidic residues and deprotonated basic residues (Table 2). None of these residues were located along the O4-PsbP channel, while three of them, PsbP-Lys166, PsbP-Lys173, and PsbQ-Asp28, were located along the O1-PsbP channel.
PsbP-Lys166 is exposed to the protein bulk surface (i.e., protonated) in the isolated form of the PsbP crystal structure (isolated PsbP structure) (Ifuku et al. 2004), whereas it was calculated to be deprotonated in the plant cryo-EM structure. In the plant cryo-EM structure, PsbP-Asp165, next to PsbP-Lys166, is involved in the salt-bridge with CP43-Arg320 (OAsp165–NArg320 = 3.1 Å) at the interface of PsbP with CP43 (Wei et al. 2016). This could be the underlying reason why in the plant cryo-EM structure PsbP-Lys170 needs to be oriented toward PsbP-Lys166 (NLys166–NLys170 = 3.9 Å) in the interface with D1 and CP43, leading to deprotonation of PsbP-Lys166 (Table 2). The side-chain orientation of PsbP-Lys170 differs significantly between the isolated PsbP (Ifuku et al. 2004) and plant cryo-EM (Wei et al. 2016) structures, whereas the side-chain orientations of PsbP-Lys166 and PsbP-Asp165 resemble in the two structures. PsbP-Asp165 forms an H-bond with PsbP-His144 in the two structures, the presence of which was already suggested by Ido et al. (Ido et al. 2012; 2014) before the plant cryo-EM structure was reported.
The following three cases may be considered as the plausible reasons for the apparently energetically unstable face-to-face side-chain orientation of the PsbP-Lys166/PsbP-Lys170 pair:
(i) Case 1: the side-chain orientations are unreasonable in the plant cryo-EM structure
The apparently energetically unstable face-to-face side-chain orientation of the PsbP-Lys166/PsbP-Lys170 pair might possibly be due to structural uncertainties in the plant cryo-EM structure analyzed at 3.2 Å resolution (Wei et al. 2016). To test this hypothesis, we optimized the side chains of the D1, CP43, PsbP, and PsbQ proteins in the plant cryo-EM structure at the molecular mechanical level (MM-optimized structure, see SI for the atomic coordinates). In the MM-optimized structure, PsbP-Lys166 formed a salt-bridge with D1-Glu329 and PsbP-Lys173 formed a salt-bridge with PsbQ-Asp28 (Fig. 6). These results imply that some side-chain orientations are energetically unstable in the plant cryo-EM structure (Wei et al. 2016). Notably, the shape of the plant O1-PsbP channel essentially remained unchanged upon geometry optimization of the side chains (Fig. 6). The plant O1-PsbP channel seems to exist in plant PSII, irrespective of structural uncertainties in the plant cryo-EM structure.
Side-chain orientations in the original plant cryo-EM structure (Wei et al. 2016) (cyan) and in the MM-optimized geometry (magenta) near the O1-PsbP channel. In contrast to the original plant cryo-EM structure, the PsbP-Lys166/D1-Glu329 pair and the PsbP-Lys173/PsbQ-Asp28 pair formed salt-bridges (dotted lines) in the MM-optimized geometry. The shape of the plant O1-PsbP channel was essentially the same in the original plant cryo-EM structure (green mesh) and the MM-optimized geometry (magenta mesh). See SI for the atomic coordinates of the MM-optimized structure
(ii) Case 2: the side-chain orientations are reasonable in the plant cryo-EM structure, and the basic residues listed in Table 2 are actually deprotonated in PsbP of the PSII complex
Face-to-face side-chain orientation of the PsbP-Lys166/PsbP-Lys170 pair might be the case for the plant cryo-EM structure. However, it should also be noted that chemical modification of PsbP-Lys166 and PsbP-Lys170 affected the PsbP binding to the PSII complex (Tohri et al. 2004), implying that the two Lys residues form salt-bridges with acidic residues in the other protein subunits, in contrast to the geometry of the plant cryo-EM structure (Wei et al. 2016).
(iii) Case 3: the side-chain orientations are reasonable in the plant cryo-EM structure, but the basic residues are not deprotonated
This would be possible if anions (e.g., Cl−), which have not been identified in the plant cryo-EM structure, are present near the basic residues. The presence of Cl− would also stabilize the positively charged PsbP-Lys166/PsbP-Lys170 pair in the plant cryo-EM structure. This case seems to be energetically more favorable than case 2 and might possibly be associated with a role of PsbP in Cl− retention (Ghanotakis et al. 1984a). Based on observations of the isolated PsbP crystal structure, Ifuku et al. pointed out that structural rearrangement, possibly including these Lys residues that may occur upon the PsbP binding to the PSII complex, could be required for Cl− retention (Ifuku et al. 2004).
The calculated distribution pattern of Cl− covers the two Cl− binding sites, Cl-1 (Fig. 7c) and Cl-2 (Fig. 7d), identified in the cyanobacterial PSII crystal (Umena et al. 2011) and plant cryo-EM (Wei et al. 2016) structures. However, the Cl− distribution pattern of the plant cryo-EM structure obtained using 3D-RISM with Placevent suggested that Cl− is unlikely to exist in the PsbP-Lys166/PsbP-Lys170 moiety in the plant cryo-EM structure (Fig. 7b). The absence of the Cl− distribution in the PsbP-Lys166/PsbP-Lys170 moiety seems to be due to the insufficient binding space for Cl−. Possible Cl− binding sites in PsbP could be elsewhere, including the inner space of the O1-PsbP channel, as indicated in Fig. 7b. These results suggest that Cl− binding is determined not only by the positive charges in the protein environment, but also by the binding space size.
3D distributions of Cl− are shown as orange mesh obtained via 3D-RISM a in the cyanobacterial O1-PsbU/V and b the plant O1-PsbP channels. The focusing space between the PsbP-Lys166 and PsbP-Lys170 side chains in the plant cryo-EM structure (Wei et al. 2016) is indicated using red dotted circle c near the Cl-1 and d Cl-2 ions identified in the cyanobacterial PSII crystal structure (Umena et al. 2011). The backbone N atoms that provide the Cl− binding sites are labeled in brackets. The threshold of 3D distribution functions is 1.0 in all cases
Considering the three cases, it seems most likely that the unusual protonation states found for the residues listed in Table 2 are highly associated with structural uncertainties in the cryo-EM structure at a resolution of 3.2 Å (i.e., case 1), because the orientations of side chains cannot be determined unambiguously at this level of resolution. Indeed, the energetically optimized side-chain orientations in the MM-optimized structure show that protonated PsbP-Lys166 forms a salt-bridge with ionized D1-Glu329 and protonated PsbP-Lys173 forms a salt-bridge with ionized PsbQ-Asp28 (Fig. 6). It can be concluded that the unusual protonation states of PsbP-Lys166, PsbP-173, and PsbQ-Asp28 (Table 2) originate from the energetically unfavorable side-chain orientations that prevent the formation of salt-bridges in the original geometry of the plant cryo-EM structure.
Conclusions
The results of the present analysis suggested that the O4-PsbU channel in cyanobacterial PSII that proceeds from the O4 site of the Mn4Ca cluster toward the protein bulk surface was structurally conserved as the O4-PsbP channel in plant PSII (Figs. 1, 2). The salt-bridge between CP43-Lys339 and PsbP-Asp137 in the plant O4-PsbP channel corresponds to the salt-bridge between CP43-Lys339 and PsbU-Asp96 in the cyanobacterial O4-PsbU channel (Fig. 2). The radii of both channels were smaller than ~ 1.4 Å in most of the regions, implying that water molecules are arranged as a single chain (Fig. 3). The O1-PsbU/V channel in cyanobacterial PSII was also structurally conserved as the O1-PsbP channel in plant PSII (Fig. 5). The salt-bridge between CP43-Glu413 and PsbP-Lys170 near the plant O1-PsbP channel corresponds to that between CP43-Glu413 and PsbV-Lys47 near the cyanobacterial O1-PsbU/V channel. These salt-bridges seem to contribute to the observed high structural stability of the channel regions (Fig. 4). None of these salt-bridges exist in the channel inner spaces, as demonstrated in Fig. 3.
In the calculated protonation states, PsbP and PsbQ showed several deprotonated basic residues and protonated acidic residues in the original side-chain orientations of the plant cryo-EM structure (Table 2). The protonation states of the deprotonated basic residues and protonated acidic residues in the two extrinsic proteins (e.g., deprotonated PsbP-Lys166, Table 2) would be altered in the presence of Ca2+ and Cl−. This effect might possibly be associated with the retention of Cl− (e.g., deprotonated PsbP-Lys166, Table 2) and Ca2+ in PsbP and PsbQ, which are the known roles of the two proteins (Ghanotakis et al. 1984a). However, Cl− is unlikely to exist in the PsbP-Lys166/PsbP-Lys170 moiety in the plant cryo-EM structure due to the insufficient binding space for Cl− (Fig. 7b). The unusual protonation states of the residues listed in Table 2 are most likely to originate from structural uncertainties in the cryo-EM structure at a resolution of 3.2 Å (Wei et al. 2016). The energetically optimized side-chain orientations suggest that protonated PsbP-Lys166 forms a salt-bridge with ionized D1-Glu329 and protonated PSbP-Lys173 forms a salt-bridge with ionized PsbQ-Asp28 (Fig. 6), in contrast to the original geometry of the plant cryo-EM structure.
The existence of the O4-PsbP channel in plant PSII and the O4-PsbU channel in cyanobacterial PSII, irrespective of the low structural similarities between PsbP and PsbU, is remarkable. If PsbP were not present in plant PSII, the O4-water chain might be able to release protons directly toward the protein bulk surface. However, the removal of the extrinsic, highly charged PsbP subunit also affects the PSII protein assembly. Indeed, thermoluminescence studies showed that the Mn4Ca cluster was significantly unstable in the absence of PsbP (Ifuku et al. 2005). Remarkably, in cyanobacterial PSII, the Mn4Ca cluster was also significantly unstable in the absence of PsbU (Inoue-Kashino et al. 2005). The properties of the proton-conducting O4-water chain (Saito et al. 2015; Takaoka et al. 2016), which directly forms a short H-bond with the Mn4Ca cluster (Umena et al. 2011; Suga et al. 2015), would also be affected by removal of these extrinsic proteins. To maintain both the stability of the Mn4Ca cluster and the activity in the proton transfer along the O4-water chain, employing PsbP (or PsbU) that possesses the O4-PsbP channel (or the O4-PsbU channel) would be important. Tomita et al. reported that PsbP, neither PsbQ nor PsbO, affects the properties of the Mn4Ca cluster, possibly protein conformation near the Mn4Ca cluster, without altering the ligand structure (Tomita et al. 2009), e.g., assumingly, with altering the H-bond network or the ion localizations. The PsbP channels identified in the present study might be associated with the observed direct functional link of PsbP (Tomita et al. 2009) with the Mn4Ca cluster.
References
Bashford D, Karplus M (1990) pK a’s of ionizable groups in proteins: atomic detail from a continuum electrostatic model. Biochemistry 29:10219–10225
Beglov D, Roux B (1997) An integral equation to describe the solvation of polar molecules in liquid water. J Phys Chem B 101:7821–7826
Berendsen HJC, Postma JPM, Vangunsteren WF, Dinola A, Haak JR (1984) Molecular-dynamics with coupling to an external bath. J Chem Phys 81(8):3684–3690. doi:10.1063/1.448118
Bricker TM, Roose JL, Fagerlund RD, Frankel LK, Eaton-Rye JJ (2012) The extrinsic proteins of photosystem II. Biochim Biophys Acta 1817(1):121–142. doi:10.1016/j.bbabio.2011.07.006
Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M (1983) CHARMM: a program for macromolecular energy minimization and dynamics calculations. J Comput Chem 4(2):187–217
Calderone V, Trabucco M, Vujicic A, Battistutta R, Giacometti GM, Andreucci F, Barbato R, Zanotti G (2003) Crystal structure of the PsbQ protein of photosystem II from higher plants. EMBO Rep 4(9):900–905. doi:10.1038/sj.embor.embor923
Case DA, Darden TA, Cheatham I, T E, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Roberts B, Hayik S, Roitberg A, Seabra G, Swails J, Götz AW, Kolossváry I, Wong KF, Paesani F, Vanicek J, Wolf RM, Liu J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye X, Wang J, Hsieh M-J, Cui G, Roe DR, Mathews DH, Seetin MG, Salomon-Ferrer R, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA (2012) AMBER 12. University of California, San Francisco
Case DA, Babin V, Berryman JT, Betz RM, Cai Q, D.S. Cerutti, Cheatham I, T. E., Darden TA, Duke RE, Gohlke H, Goetz AW, Gusarov S, Homeyer N, Janowski P, Kaus J, Kolossváry I, Kovalenko A, Lee TS, LeGrand S, Luchko T, Luo R, Madej B, Merz KM, Paesani F, Roe DR, Roitberg A, Sagui C, Salomon-Ferrer R, Seabra G, Simmerling CL, Smith W, Swails J, Walker RC, Wang J, Wolf RM, Wu X, Kollman PA (2014) AMBER 14, University of California, San Francisco
Cox N, Messinger J (2013) Reflections on substrate water and dioxygen formation. Biochim Biophys Acta 1827(8–9):1020–1030. doi:10.1016/j.bbabio.2013.01.013
Cox N, Pantazis DA, Neese F, Lubitz W (2013) Biological water oxidation. Acc Chem Res 46(7):1588–1596
Dau H, Haumann M (2008) The manganese complex of photosystem II in its reaction cycle? Basic framework and possible realization at the atomic level. Coord Chem Rev 252:273–295
Dau H, Zaharieva I, Haumann M (2012) Recent developments in research on water oxidation by photosystem II. Curr Opin Chem Biol 16(1–2):3–10
Feller SE, Zhang YH, Pastor RW, Brooks BR (1995) Constant-pressure molecular-dynamics simulation: the Langevin piston method. J Chem Phys 103(11):4613–4621. doi:10.1063/1.470648
Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S (2004) Architecture of the photosynthetic oxygen-evolving center. Science 303(5665):1831–1838
Gabdulkhakov A, Guskov A, Broser M, Kern J, Muh F, Saenger W, Zouni A (2009) Probing the accessibility of the Mn4Ca cluster in photosystem II: channels calculation, noble gas derivatization, and cocrystallization with DMSO. Structure 17(9):1223–1234. doi:10.1016/j.str.2009.07.010
Galstyan A, Robertazzi A, Knapp EW (2012) Oxygen-evolving Mn cluster in photosystem II: the protonation pattern and oxidation state in the high-resolution crystal structure. J Am Chem Soc 134(17):7442–7449
Ghanotakis DF, Topper JN, Babcock GT, Yocum CF (1984a) Water-soluble 17-Kda and 23-Kda polypeptides restore oxygen evolution activity by creating a high-affinity binding-site for Ca2+ on the oxidizing side of photosystem-II. FEBS Lett 170(1):169–173. doi:10.1016/0014-5793(84)81393-9
Ghanotakis DF, Topper JN, Yocum CF (1984b) Structural organization of the oxidizing side of photosystem-II. Exogenous reductants reduce and destroy the Mn-complex in photosystems II membranes depleted of the 17 and 23 Kda. Biochim Biophys Acta 767(3):524–531. doi:10.1016/0005-2728(84)90051-3
Ho FM, Styring S (2008) Access channels and methanol binding site to the CaMn4 cluster in photosystem II based on solvent accessibility simulations, with implications for substrate water access. Biochim Biophys Acta 1777(2):140–153
Ido K, Kakiuchi S, Uno C, Nishimura T, Fukao Y, Noguchi T, Sato F, Ifuku K (2012) The conserved His-144 in the PsbP protein is important for the interaction between the PsbP N-terminus and the Cyt b559 subunit of photosystem II. J Biol Chem 287(31):26377–26387. doi:10.1074/jbc.M112.385286
Ido K, Nield J, Fukao Y, Nishimura T, Sato F, Ifuku K (2014) Cross-linking evidence for multiple interactions of the PsbP and PsbQ proteins in a higher plant photosystem II supercomplex. J Biol Chem 289(29):20150–20157. doi:10.1074/jbc.M114.574822
Ifuku K, Noguchi T (2016) Structural coupling of extrinsic proteins with the oxygen-evolving center in photosystem II. Front Plant Sci 7:84. doi:10.3389/fpls.2016.00084
Ifuku K, Nakatsu T, Kato H, Sato F (2004) Crystal structure of the PsbP protein of photosystem II from Nicotiana tabacum. EMBO Rep:362–367
Ifuku K, Yamamoto Y, Ono TA, Ishihara S, Sato F (2005) PsbP protein, but not PsbQ protein, is essential for the regulation and stabilization of photosystem II in higher plants. Plant Physiol 139(3):1175–1184. doi:10.1104/pp.105.068643
Inoue-Kashino N, Kashino Y, Satoh K, Terashima I, Pakrasi HB (2005) PsbU provides a stable architecture for the oxygen-evolving system in cyanobacterial photosystem II. Biochemistry 44(36):12214–12228. doi:10.1021/bi047539k
Ishikita H, Knapp E-W (2005) Redox potential of cytochrome c550 in the cyanobacterium Thermosynechococcus elongatus. FEBS Lett 579:3190–3194
Kerfeld CA, Sawaya MR, Bottin H, Tran KT, Sugiura M, Cascio D, Desbois A, Yeates TO, Kirilovsky D, Boussac A (2003) Structural and EPR characterization of the soluble form of cytochrome c-550 and of the psbV2 gene product from cyanobacterium Thermosynechococcus elongatus. Plant Cell Physiol 44:697–706
Kovalenko A, Hirata F (1999) Potential of mean force between two molecular ions in a polar molecular solvent: a study by the three-dimensional reference interaction site model. J Phys Chem B 103:7942–7957
Kubo R, Toda M, Hashitsume N (1991). Statistical Physics II, Springer, Berlin
Kulik LV, Epel B, Lubitz W, Messinger J (2007) Electronic structure of the Mn4OxCa cluster in the S0 and S2 states of the oxygen-evolving complex of photosystem II based on pulse 55Mn-ENDOR and EPR spectroscopy. J Am Chem Soc 129(44):13421–13435
Linke K, Ho FM (2014) Water in Photosystem II: Structural, functional and mechanistic considerations. Biochim Biophys Acta 1837(1):14–32
Luchko T, Gusarov S, Roe DR, Simmerling C, Case DA, Tuszynski J, Kovalenko A (2010) Three-dimensional molecular theory of solvation coupled with molecular dynamics in amber. J Chem Theory Comput 6:607–624
MacKerell AD Jr, Bashford D, Bellott RL, Dunbrack RL Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE III, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B 102(18):3586–3616
McConnell IL, Grigoryants VM, Scholes CP, Myers WK, Chen PY, Whittaker JW, Brudvig GW (2012) EPR-ENDOR characterization of (17O, 1H, 2H) water in manganese catalase and its relevance to the oxygen-evolving complex of photosystem II. J Am Chem Soc 134(3):1504–1512
Messinger J (2004) Evaluation of different mechanistic proposals for water oxidation in photosynthesis on the basis of Mn4OxCa structures for the catalytic site and spectroscopic data. Phys Chem Chem Phys 6:4764–4771
Murray JW, Barber J (2007) Structural characteristics of channels and pathways in photosystem II including the identification of an oxygen channel. J Struct Biol 159(2):228–237
Nozaki Y, Tanford C (1967) Acid-base titrations in concentrated guanidine hydrochloride. Dissociation constants of the guamidinium ion and of some amino acids. J Am Chem Soc 89(4):736–742
Ogata K, Yuki T, Hatakeyama M, Uchida W, Nakamura S (2013) All-atom molecular dynamics simulation of photosystem II embedded in thylakoid membrane. J Am Chem Soc 135(42):15670–15673
Petrek M, Otyepka M, Banas P, Kosinova P, Koca J, Damborsky J (2006) CAVER: a new tool to explore routes from protein clefts, pockets and cavities. BMC Bioinform 7:316. doi:10.1186/1471-2105-7-316
Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K (2005) Scalable molecular dynamics with NAMD. J Comput Chem 26:1781–1802
Rabenstein B, Knapp EW (2001) Calculated pH-dependent population and protonation of carbon-monoxy-myoglobin conformers. Biophys J 80(3):1141–1150
Rapatskiy L, Cox N, Savitsky A, Ames WM, Sander J, Nowaczyk MM, Rögner M, Boussac A, Neese F, Messinger J, Lubitz W (2012) Detection of the water-binding sites of the oxygen-evolving complex of photosystem II using W-band 17O electron-electron double resonance-detected NMR spectroscopy. J Am Chem Soc 134(40):16619–16634
Renger G (2001) Photosynthetic water oxidation to molecular oxygen: apparatus and mechanism. Biochim Biophys Acta 1503(1–2):210–228
Robblee JH, Messinger J, Cinco RM, McFarlane KL, Fernandez C, Pizarro SA, Sauer K, Yachandra VK (2002) The Mn cluster in the S0 state of the oxygen-evolving complex of photosystem II studied by EXAFS spectroscopy: are there three di-µ-oxo-bridged Mn2 moieties in the tetranuclear Mn complex? J Am Chem Soc 124:7459–7471
Roncel M, Boussac A, Zurita JL, Bottin H, Sugiura M, Kirilovsky D, Ortega JM (2003) Redox properties of the photosystem II cytochromes b559 and c550 in the cyanobacterium Thermosynechococcus elongatus. J Biol Inorg Chem 8:206–216
Ryckaert J-P, Ciccotti G, Berendsen HJC (1977) Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys 23(3):327–341. doi:10.1016/0021-9991(77)90098-5
Saito K, Shen J-R, Ishida T, Ishikita H (2011) Short hydrogen-bond between redox-active tyrosine YZ and D1-His190 in the photosystem II crystal structure. Biochemistry 50:9836–9844
Saito K, Rutherford AW, Ishikita H (2015) Energetics of proton release on the first oxidation step in the water-oxidizing enzyme. Nat Commun 6:8488. doi:10.1038/ncomms9488
Shen JR (2015) The structure of photosystem II and the mechanism of water oxidation in photosynthesis. Annu Rev Plant Biol 66:23–48. doi:10.1146/annurev-arplant-050312-120129
Shen J-R, Inoue Y (1993) Binding and functional properties of two new extrinsic components, cytochrome c-550 and a 12-kDa protein, in cyanobacterial photosystem II. Biochemistry 32:1825–1832
Sindhikara DJ, Yoshida N, Hirata F (2012) Placevent: an algorithm for prediction of explicit solvent atom distribution. J Comput Chem 33:1536–1543
Stuchebrukhov AA (2009) Mechanisms of proton transfer in proteins: localized charge transfer versus delocalized soliton transfer. Phys Rev E Stat Nonlin Soft Matter Phys 79(3 Pt 1):031927
Suga M, Akita F, Hirata K, Ueno G, Murakami H, Nakajima Y, Shimizu T, Yamashita K, Yamamoto M, Ago H, Shen JR (2015) Native structure of photosystem II at 1.95 A resolution viewed by femtosecond X-ray pulses. Nature 517:99–103
Takaoka T, Sakashita N, Saito K, Ishikita H (2016) pK a of a proton-conducting water chain in photosystem II. J Phys Chem Lett 7(10):1925–1932. doi:10.1021/acs.jpclett.6b00656
Tanokura M (1983a) 1H-NMR study on the tautomerism of the imidazole ring of histidine residues. I. Microscopic pK values and molar ratios of tautomers in histidine-containing peptides. Biochim Biophys Acta 742(3):576–585
Tanokura M (1983b) 1H-NMR study on the tautomerism of the imidazole ring of histidine residues. II. Microenvironments of histidine-12 and histidine-119 of bovine pancreatic ribonuclease A. Biochim Biophys Acta 742(3):586–596
Tanokura M (1983c) 1H nuclear magnetic resonance titration curves and microenvironments of aromatic residues in bovine pancreatic ribonuclease A. J Biochem 94(1):51–62
Tohri A, Dohmae N, Suzuki T, Ohta H, Inoue Y, Enami I (2004) Identification of domains on the extrinsic 23 kDa protein possibly involved in electrostatic interaction with the extrinsic 33 kDa protein in spinach photosystem II. Eur J Biochem 271(5):962–971
Tomita M, Ifuku K, Sato F, Noguchi T (2009) FTIR evidence that the PsbP extrinsic protein induces protein conformational changes around the oxygen-evolving Mn cluster in photosystem II. Biochemistry 48(27):6318–6325. doi:10.1021/bi9006308
Umena Y, Kawakami K, Shen J-R, Kamiya N (2011) Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473:55–60
Vassiliev S, Zaraiskaya T, Bruce D (2012) Exploring the energetics of water permeation in photosystem II by multiple steered molecular dynamics simulations. Biochim Biophys Acta 1817(9):1671–1678
Watanabe HC, Welke K, Sindhikara DJ, Hegemann P, Elstner M (2013) Towards an understanding of channelrhodopsin function: simulations lead to novel insights of the channel mechanism. J Mol Biol 425(10):1795–1814. doi:10.1016/j.jmb.2013.01.033
Wei X, Su X, Cao P, Liu X, Chang W, Li M, Zhang X, Liu Z (2016) Structure of spinach photosystem II-LHCII supercomplex at 3.2 Å resolution. Nature 534(7605):69–74. doi:10.1038/nature18020
Wietek J, Wiegert JS, Adeishvili N, Schneider F, Watanabe H, Tsunoda SP, Vogt A, Elstner M, Oertner TG, Hegemann P (2014) Conversion of channelrhodopsin into a light-gated chloride channel. Science 344(6182):409–412. doi:10.1126/science.1249375
Wu YJ, Tepper HL, Voth GA (2006) Flexible simple point-charge water model with improved liquid-state properties. J Chem Phys 124 (2). doi:10.1063/1.2136877
Yamanaka S, Isobe H, Kanda K, Saito T, Umena Y, Kawakami K, Shen J-R, Kamiya N, Okumura M, Nakamura H, Yamaguchi K (2011) Possible mechanisms for the O–O bond formation in oxygen evolution reaction at the CaMn4O5(H2O)4 cluster of PSII refined to 1.9 Å X-ray resolution. Chem Phys Lett 511:138–145
Acknowledgements
This research was supported by JST CREST, JSPS KAKENHI (JP15H00864, JP16H06560, JP26105012, and JP26711008), Materials Integration for engineering polymers of Cross-ministerial Strategic Innovation Promotion Program (SIP), and Interdisciplinary Computational Science Program in CCS, University of Tsukuba. Theoretical calculations were partly performed using Research Center for Computational Science, Okazaki, Japan.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sakashita, N., Watanabe, H.C., Ikeda, T. et al. Structurally conserved channels in cyanobacterial and plant photosystem II. Photosynth Res 133, 75–85 (2017). https://doi.org/10.1007/s11120-017-0347-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11120-017-0347-1