Abstract
The precise use of specific types of herbicides according to the weed species and density in fields can effectively reduce chemical contamination. A weed species and density evaluation method based on an image semantic segmentation neural network was proposed in this paper. A combination of pre-training and fine-tuning training methods was used to train the network. The pre-training data were images that only contain one species of weeds in one image. The weeds were automatically labeled by an image segmentation method based on the Excess Green (ExG) and the minimum error threshold. The fine-tuning dataset was real images containing multiple weeds and crops and manually labeled. Due to the limitation of computational resources, larger images were difficult to segment at one time. Therefore, this paper proposed a method of cutting images into sub-images. The relationship between sub-image size and segment accuracy was studied. The results showed that the training method reduced the workload of labeling training data while effectively avoiding overfitting. The accuracy of image segmentation decreases as the sub-image size decreases. Considering the limitation of computational resources, a subgraph of \(256 \times 256\) size was selected. The semantic segmentation network achieved 97% overall accuracy. The coefficient of determination (\(R^2\)) of weed density calculated by the algorithm and manually assessed was 0.90, and the root means square error (\(\sigma \)) was 0.05. The method could effectively assess the density of each species of weeds in complex environments. It could provide a reference for accurate spraying herbicides based on weed density and species.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Weeds are one of the most common agricultural damages (Hamuda et al., 2016). They compete with crops for water, sunlight, and nutrients in the field, which leads to crop yield reduction (Berge et al., 2008). To overcome the problem, researchers and farmers have made a lot of efforts. Since 1940, chemical weed control has become the most popular method (Hamuda et al., 2016). This method usually sprays herbicides uniformly regardless of the presence or absence of weeds. Spraying herbicides in weed-free areas not only wastes herbicides but also pollutes the agroecology (Rodrigo et al., 2014). Precise spraying herbicides according to the density of the weeds is an effective way to reduce agroecological pollution and herbicide waste. Also, choosing specialized herbicides according to the species of weeds is a way to enhance effectiveness. Accurate assessment of density and species of weeds is an important step in precision spraying. Therefore it is crucial to develop a method to assess weed species and density.
In recent years, along with the development of machine vision techniques, many scholars have researched precision weed control servo. Devices such as three-dimensional cameras, spectral cameras, and thermal cameras were used to collect images of the field. Several scholars have studied algorithms to segment weeds on these images to achieve weed identification and segmentation. Li and Tang (2018), Khan et al. (2018), Lammie et al. (2019), Stroppiana et al. (2018), Kazmi et al. (2015), Ge et al. (2019), Alenya et al. (2013), Kusumam et al. (2017), and Kazmi et al. (2014). However, the high price of such devices, and the requirement for a stable working environment, make it difficult to use them in complex agricultural industries nowadays (Zhang et al., 2020). The color camera is a simple, reliable, and cheap imaging device. It can acquire visible light images in complex farmland environments. Therefore, many scholars have studied using color cameras to do vision-serviced precision weeding (Sabzi et al., 2018; Bakhshipour & Jafari, 2018; Wang et al., 2019; Abdalla et al., 2019). Crop and weed segmentation by color difference and threshold segmentation is a simple method for weed segmentation. Researchers have also proposed a series of color indexes for weed segmentation (Lee et al., 2021; Rico-Fernández et al., 2019). However, most weeds, such as dogwood, oxalis, and quinoa, are green, the same as crops. Therefore, it is usually difficult to achieve better image segment results by color index and threshold segmentation methods. Commonly, colors can also form feature groups with other features, such as texture and shape, to segment the image by classifying pixels or regions with a classifier. Machine-learning algorithms are the most common classifiers, such as Artificial Neural Network(ANN), Support Vector Machine (SVM), and Naive Bayes (Zhang et al., 2021; Zou et al., 2019; Bakhshipour & Jafari, 2018; Chen et al., 2021). In a stable environment, the combination of image features and machine learning classifiers can achieve wonderful results with an 80–98% classification accuracy (Lottes et al., 2018). However, the field working environment is complex, the lighting conditions, the density of weeds, and the growth stages of plants are all changeable (Wang et al., 2019). And, occlusion, clustering and changing lighting conditions in a natural environment remain major challenges in weed detection and localization (Gongal et al., 2015). Using the traditional machine learning method to classify different species of crops is still difficult. Therefore, it is necessary to research an efficient, automated, and robust algorithm to deal with such situations (Abdalla et al., 2019).
In recent years, deep learning has been more widely used due to the decreasing cost of computer hardware and the improvement of GPU performance. It also has a wide range of applications in agriculture (Fu et al., 2020; Aversano et al., 2020; Too et al., 2019; Tiwari et al., 2019; Kamilaris & Prenafetaboldu, 2018; Kalin et al., 2019). Some scholars have studied crop weed segmentation using deep learning networks. Huang et al. (2018) proposed a weed cover map-generating workflows based on unmanned aerial vehicle (UAV) imagery and Fully convolutional networks (FCN). The segmentation accuracy was 94% . Le et al. (2021) proposed a weed detection method based on Faster-RCNN. However, the detection time of this method was 0.38s per image. The detection speed was slow. An instance segmentation-based weed detection algorithm was proposed by Champ et al. (2020). However, the accuracy of this algorithm is only 10–60%. From the above studies, it can be seen that deep learning models, especially semantic segmentation neural networks, can achieve better results in weed estimation than other methods. However, due to the limitation of computing resources, the performance of deep learning can be improved.
Deep learning requires a large amount of training data. These data usually need to be labeled manually. It is often difficult to manually label large amounts of data, especially for semantic segmentation tasks. Because this type of task requires labeling each pixel with a category (Deng et al., 2018; Kemker et al., 2018; Pan et al., 2017). Transfer learning is an effective way to reduce training samples (Abdalla et al., 2019). Transfer learning usually consists of two steps: pre-training and fine-tuning. The weights obtained in the pre-training stage are applied in the fine-tuning stage. This approach allows the fine-tuning stage to find the optimal solution faster than the random initialization of parameters. In order to make the fine-tuning stage faster, the selection of samples for the pre-training stage is usually made by two criteria. The first criterion is to get as close as possible to the final target sample; The second criterion is to make it as easy as possible to obtain. Usually, the pre-trained dataset is an open-access dataset with labels (Zhang et al. 2017; DeVries & Taylor, 2017; Yun et al., 2019; Cubuk et al., 2020). This method can obtain a large number of labeled pre-training samples quickly. Transfer learning and fine-tuning have been widely used in agricultural applications including plant species classification, plant disease detection, and weed detection (Dyrmann et al., 2016; Ferreira et al., 2017; Barbedo, 2018; Picon et al., 2019). For example, Abdalla et al. (2019) studied weed density semantic segmentation method based on a fine-tuning convolutional neural network with transfer learning. The highest semantic segmentation accuracy was 96%. From the above studies, it can be seen that transfer learning can effectively reduce the number of samples used in weed segmentation network training. However, an open-source weeds dataset suitable for this study did not find. Therefore, it is necessary to investigate a training method for this research.
This paper hypothesized that semantic segmentation neural networks can achieve an accurate assessment of weed density than the traditional method and transfer learning can effectively improve the training efficiency of the network. The main objective of this article is to investigate a weed species and density assessment method. The specific research objectives are: (1) design a semantic segmentation neural network for segmenting different species of weeds; (2) investigate a convenient and fast neural network training method to train this neural network.
Materials and methods
Imaging system
In this study, a spray vehicle based image acquisition device was used to acquire weed images in the field (Zou et al., 2021). A spray vehicle (FJ 3WP 500A) was used as a mobile platform. The camera, the positioning module, and the control unit were mounted on this vehicle. Three cameras (RMONCAM HD908) were mounted in the front of the vehicle to capture images. A POS sensor (WIT-IMU, WIT-MOTION Technology Co, China) was mounted on the vehicle to record the ground coordinates. The camera was installed horizontally with its lens facing down vertically, 140 cm from the ground. The size of the imaging area was 90 × 120 cm. Vehicle movement speed did not exceed 0.1 m/s during image acquisition. The images of the acquisition device and camera are shown in Fig. 1.
Weeds image acquisition
The images for this study were taken on 4th June 2021 at 15:30–17:30, 9th June 2021 at 8:00–12:00, and 15th June 2021 at 13:00–15:00 in a maize field and a soybean field, located at Qixing Farm, Jiansanjiang, Jiamusi, Heilongjiang Province, China (132° 35′ 32″ E, 47° 10′ 58″ N), shown in Fig. 2. The original image resolutions were 3000 × 4000 pixels and 2250 × 4000 pixels. It was too large for processing, especially for the deep-learning-based method, so a series of small-size images of 256 × 256 pixels were randomly cut out from the original images. There was no interaction or overlap between sub-images.
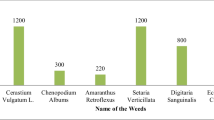
There were mainly five species of weeds in this area, namely Green bristlegrass (EPPO: SETVI), Thistle (EPPO: CIRJA), Acalypha copperleaf (EPPO: ACCAU), Equisetum arvense (EPPO: EQUAR), Goosefoots (EPPO: CHEAL) (shown in Fig. 3). One thousand images were collected for each weed species. There were no green plants other than weeds in these images, shown in Fig. 3. There are also 1000 images containing crops, maize (EPPO: ZEAMX), and soybeans (EPPO: GLXMA). In total 6000 images were used in the pre-training stage, called the "pre-training set". One thousand images contained crops, and different species of weeds were manually labeled. Some of the images and labels are shown in Fig. 4. Eight hundred of the images were used in the fine-tuning stage of training, called the "fine-tuning set". The remaining 200 images were used in the test stage, called the "testing set".
General steps of weed density evaluation
The weed density evaluation method described in this paper relies on a semantic segmentation neural network. The main process consists of the following steps: (1) Original image cutting: original images were cut into sub-images of a specified size; (2) Pre-training set auto labeling: sub-images containing only one species of weed were selected as pre-training images. Labels were automatically added to the images by an algorithm; (3) Fine-tuning set labeling: sub-images containing different species of weeds and crops were selected. And labels were added to the images manually; (4) Semantic segmentation network construction: based on the complexity of the images and the requirements for segmentation accuracy, the neural network was constructed based on the encoding and decoding structure; (5) Network training: the neural network was trained in two stages, pre-training and fine-tuning; (6) Image segmentation: the sub-images were segmented by the network. After segmentation, the results were stitched into one map again; (7) Weed density calculation: the ratio of the area occupied by each species of weed to the total area was calculated to obtain the density of each weed. The weed contamination type and density could be obtained by the above steps from the field image. The results could be used to guide accurate spraying. The flow chart was shown in Fig. 5.
Pre-training set automatic labeling
Transfer learning is divided into two phases: pre-training and fine-tuning (Abdalla et al., 2019). The pre-training phase can use synthetic samples to train the model. This can reduce the amount of real labeled samples required (Zou et al., 2021). In this paper, to solve the problem of difficult labeling of semantic segmentation neural networks, a two-stage training method combining pre-training and fine-tuning was used. The pre-training sample set consists of 6000 images. One thousand of them were crop images. All pixels, including bare land, maize and soybeans, on these images were labeled as 0. And there were 1000 images for each species of weed. In these images, bare land was labeled as 0. Green bristlegrass, Thistle, Acalypha copperleaf, Equisetum arvense, and Goosefoots were labeled as 1, 2, 3, 4, 5, respectively. These images contained only one species of weed on each image. The rest of the areas were bare land. Therefore, the automatic labeling of the weed samples was to segment the weeds and bare land and then label the weed areas according to the weed types. The weed species on each image were manually labeled.
There is a significant difference in color between weeds and bare land. Therefore, weed segmentation on the pre-training set can be performed by a color index with threshold segmentation. Excess Green (ExG) was a common color index (Meyer & Neto, 2008). It could enlarge the difference between green areas and other areas. It was often used in green plant image segmentation. The calculation method was shown in Eq. 1.
The minimum error method was a segmentation threshold finding method based on the normal distribution (Hsu et al., 2019). In this paper, it was used to find the optimal segmentation threshold in the Excess Green. In the natural environment, the data usually distribution obeys the normal (Gaussian) distribution. The normal test can be used to detect the extent to which the sample points conformed to a normal distribution (Ghasemi & Zahediasl, 2012; Virtanen et al., 2020). The images in this study were collected from the natural environment, so this method could be used. The distribution curve of one sample obeys the normal distribution curve (a) (shown in Fig. 6a). The other sample distribution curve obeys the normal distribution curve (b) (shown in Fig. 6b). When the centers of these two curves do not overlap. The value of the horizontal coordinate corresponding to the intersection point "o" ( shown in Fig. 6o) of these two curves was the segmentation threshold with the minimum error.
The error could be defined as Eq. 2. In order to obtain the minimum value of E(t), let the partial derivative of E(t) with respect to t be 0, and the conditions for minimizing the error fraction were as Eq. 3. When both weeds and bare land obey normal distribution, the equation to solve for o could be expressed as Eq. 4. The t value of Eq. 4 could be solved as Eqs. 5 and 6:
where w represents the proportion of one kind of samples in the whole image, while \(1-w\) represents the proportion of the other kind of samples in the whole image.
where \(\mu _a\) and \(\mu _b\) are the mean value of two kind of smple pixels, \(\sigma _a\) and \(\sigma _b\) are the root means square error of two kind of smple pixels.
In this paper, 30 images were randomly selected and 3000 pixels were randomly selected from these 30 images. Each pixel was manually labeled bare land pixes(the value set as 0) or green plants (the value set as 1). In these pixels, 2000 pixels were used to calculate the segmentation threshold, and 1000 pixels were used to test the accuracy of segmentation. Part of the sample points and labels are shown in Fig. 7.
Determination of sub-image size.
Due to the limitation of computational resources and the difficulty of manual labeling, the original images were cut into sub-images by a Python script. Semantic segmentation was used to segment the sub-images. The segment results were then stitched together into a complete result map. Since the size of the segmented sub-images had an impact on the segmentation accuracy (Strudel et al., 2021). So an experiment was used to find the optimal sub-image size. The images were cut into \(256\times 256\), \(128\times 128\), \(64\times 64\), \(32\times 32\), respectively (shown in Fig. 8). FCN, Segnet, and U-net neural networks were used for semantic segmentation of the sub-images (Huang et al., 2018; Badrinarayanan et al., 2017; Zou et al., 2021). A suitable sub-image size was selected based on the segmentation accuracy.
Neural network structure
A semantic segmentation neural network commonly consists of two parts, the encoding part, and the decoding part (Huang et al., 2020). The encoding part is the backbone part of the neural network. It consists of convolutional layers and pooling layers. Usually, the encoding part consists of several abstract feature extraction units. These units are composed of two consecutive convolutional layers and a pooling layer. Multiple such units are successively lined up and extracted to the abstract features. In complex classification tasks, such as the ImageNet challenge, where there are thousands of categories to be classified, the feature extraction units are usually relatively more (Deng et al., 2009). The classification in this paper was relatively simple, with only 7 species of plants and bare ground. Therefore, there were only two feature extraction units in the encoding part. However, the task described in this paper required more information in the shallow layers to improve the accuracy of the neural network concerning details. Therefore, the number of convolutional kernels in the convolutional layer of the encoding part was doubled. The decoding part usually consists of upsampling layers, concatenation layers, and convolutional layers. In the research, this part was inspired by U-net(shown in Fig. 9a). In the U-Net, the decoding part is usually connected with two consecutive convolutional layers after the concatenate layer. The concatenate layer connects the upsampling layer with the corresponding convolutional layer of the encoding part. It is used to restore the abstract features extracted by the encoding part to the segmentation result. In this research, the number of convolutional layers was reduced to one. Although the image class in this paper was relatively few, the detailed accuracy of segmentation was required to be high. For example, very thin weed leaves should also be accurately segmented. Therefore, the input layer is connected with the last convolutional layer. A convolutional layer was added after the concatenate layer. This was used to improve the processing ability of details. The structure of the proposed semantic segmentation network is shown in Fig. 9b. The parameters of the network are shown in Table 1.
Training of neural network training
The hardware environment was Intel Core i7-9700 K CPU, 16 GB memory, NVIDIA GeForce RTX 2080 Super. The software environment was Windows 10, CUDA 10.1, Python 3.6, and Tensorflow 2.3.
In this paper, the training of neural networks consists of two stages: pre-training and fine-tuning (Zou et al., 2021). In the pre-training stage, automatically labeled, single-species weed pre-training set images were used in this stage. The "Adam" optimizer was used to optimize the network. The learning rate was set to \(1 \times 10^{-4}\). The loss function was "sparse categorical cross-entropy". The batch size of pre-training was set to 10 and epochs were set to 80. During the training process, the loss and pixel accuracy of the pre-training set and validation set were recorded. Since there is a gap between the data in the pre-training set and the data in the real field. Most of the time, real field images are of multiple weeds existing simultaneously. The error of using the pre-trained model directly is relatively large. Therefore fine-tuning is needed. In the fine-tuning stage, due to the network being pre-trained before, the weights of the parameters of the neural network were close to the optimal solution. At this time, if a larger learning rate was set, the weight combination would deviate from the optimal solution, so it was necessary to reduce the learning rate. In this study, the learning rate of the fine-tuning stage was set to \(1 \times 10^{-5}\). The batch size of fine-tuning was set to 10 and epochs were set to 40. And other parameters were consistent with the pre-training stage.
Crop segment network performance evaluation
In this study, 6 quantitative criteria were used to evaluate the performance of the segmentation network. The accuracy (Acc), precision (Pr), recall (Re), F1-Score(F1) and mean average precision (mAP) were used to assess the network (Eqs. 7–10). The Acc, Pr, and Re were averaged over all images in the testing dataset. The mAP was the average value of Pr for all species. The segmentation time of different methods were also compared. The segment time (ST) was the time needed to segment a single image. It was recorded by the program during segmenting the images.
where: TP is true positive; TN is true negative; FP is false positive; FN is false negative.
Weed coverage rate is a common indicator to assess weed density. It can be calculated by Eq. 11. In this paper, this metric was used to assess the weed density in images. The original image was cut into sub-images and segmented by the proposed semantic segmentation network. Then the coverage rate for each species of weed was calculated. The relationship between the Rw calculated from the image (predicted Rw) and the Rw by expertise visual observation (observed Rw) was evaluated by regression analysis (Virtanen et al., 2020).Two criteria, coefficient of determination and root mean square error, were calculated and used to assess errors. The coefficient of determination (\(R^2\)) was computed by Eq. 12. The root means square error (\(\sigma \)) was determined by Eq. 13.
where Rw is the ratio of weed area, \(S_{weed}\) is the area of weeds and \(S_{total}\) is the area of whole field.
where, \(Y_{oi}\) and \(Y_{pi}\) are respectively the i-th observed and predicted Rw from N total data.
Results and disscussion
Automatic labeling of results for pre-training sets
After the normal test, the P-value of the weed samples on the ExG color index was 0.88; The P-value of the bare land samples on the ExG color index was 0.02. All the P-values of the normal test results were more than 0.01, indicating that they all obeyed the normal distribution significantly. The \(\mu _a\) of the weed sample was calculated to be 123.88, the \(\sigma _a\) was 39.87. The \(\mu _b\) of the bare land sample was 7.38, the \(\sigma _b\) was 13.34; The split point t was 42. Histograms and fitted probability density function curves of weeds and bare land on ExG are shown in Fig. 10. After testing on 1000 pixels, the accuracy of weed and bare land segmentation using this method was 98.57%. Some sample images of the pre-training set after partial segmentation and labeled by kind are shown in Fig. 11.
The results show that the ExG can effectively expand the color difference between weed samples and bare land, making the threshold segmentation easier. The segmentation thresholds obtained using the minimum error method could effectively segment weed samples and bare land samples. The automatically labeling method would theoretically make the pre-trained samples have little error. However, a manual labeling sample was used in the fine-tuning stage, which can correct the error generated in the automatic labeling process.
Sub-image size evaluation result
The segmentation results of the sub-image in different sizes are shown in Table 2. It could be seen that the segmentation accuracy of all neural networks was higher when the sub-image size was \(256\times 256\). The accuracy of U-net was 0.97, Segnet was 0.95, and FCN was 0.92. As the sub-image size decreases, the segmentation accuracy of the neural networks gradually decreases. When the sub-image size was \(128\times 128\), the segmentation accuracy of U-net was 0.96; Segnet was 0.92, and FCN was 0.89. When the sub-image size was \(64\times 64\), the segmentation accuracy of U-net was 0.92, Segnet was 0.91, and FCN was 0.89. When the sub-image size was \(32\times 32\), the segmentation accuracy of the segmented U-net was 0.89, Segnet was 0.89, and FCN was 0.88. From the table, it could also be seen that the precision of bare land was relatively high, and was above 90%. But the precision of weeds was changing with the change of sub-image size. Especially, Thistle and Acalypha copperleaf decreased precision with the decrease of sub-image size.
By observing Fig. 8, it could be found that when the image was relatively large, the structure of the weed could be retained in the image relatively intact. Such an image could be recognized and accurately segmented by the neural network. But when the image was cut smaller, some of the weed structures were incomplete, and the neural network would be wrongly segmented because it did not get the complete image information. Since Acalypha copperleaf, Equisetum arvense, and Goosefoots were relatively small, they were more likely to have incomplete images after being cut. So their accuracy was lower. Bare land was more uniform, and the color and texture features remain unchanged, so they could be better segmented. Therefore, when the computational resources were insufficient and the image needed to be cut, the larger the cut sub-image size was, the better segment performance was. In this paper, due to the limitation of computational resources, the final sub-image size was determined as \(256\times 256\).
Neural network training results
The changes in accuracy and loss during pre-training and fine-tuning of the neural network as the training proceeds are shown in Figs. 12 and 13. As could be seen from Fig. 12, the loss value of the neural network decreases steadily and the accuracy value increases steadily as the pre-training proceeds. Although there were some fluctuations, the overall trend tends to be stable. The loss and accuracy tend to stabilize in the later stage of training. There was no significant difference between the accuracy and loss values of the training and validation sets. Therefore, the pre-training was adequate and no overfitting. From Fig. 13, it could be seen that the accuracy of the training and validation sets steadily decrease during fine-tuning. It stabilizes when it rose to about 0.97. The loss value decreases and then stabilizes. There was no significant difference between the training set loss and the validation set loss, so no overfitting occurs. Figure 14 shows that when the neural network was trained directly with the fine-tuning set, the accuracy went through a growth phase and then stagnated at around 0.87. The loss stagnates after a decline phase. This indicated that the training process reached a local optimal solution and then stalled.
The results show that the combination of pre-training and fine-tuning used in this paper was effective. This method could effectively reduce the need for manually labeled samples. At the same time, it could avoid the neural network falling into a locally optimal solution. Direct training with the fine-tuning set without pre-training was much less effective than the combination of pre-training and fine-tuning.
Image segmentation results
The original images, labels, and segmentation results of different neural networks for some of the test sets are shown in Fig. 15. By observing the images, it was clear that the segmentation results of the proposed semantic segmentation network were better. Its segmentation accuracy was the highest, reaching 97%. However, there were still errors at the edges. For example, some pixels were segmented as Green bristlegrass at the edges of some Thistle. U-net also had good segmentation results, but some details were segmented incorrectly, so the segmentation accuracy was slightly reduced to 96%. Segnet network was better for the segmentation of large areas. But the detail processing ability was poor. Its segmentation accuracy was 0.95. In the segmentation of Equisetum arvense, the detail processing ability was weak, so the bare land in the weed gap was segmented as weeds. FCN had the weakest segmentation ability. This network could only segment large and independent weeds accurately, such as Acalypha copperleaf. It was less effective in the case of overlapping crops and weeds, or when the weeds themselves were complex. There was also some weed segmented wrongly. The accuracy, precision, recall, F1-score and mean average precision of the proposed semantic segmentation network segmentation results are shown in Table 3. The confusion matrix is shown in Fig. 16. The average segmentation time (ST) of one image of the proposed semantic segmentation network, U-net, Segnet, and FCN was 56.28 ms, 74.35 ms, 68.23 ms, and 63.51 ms respectively. The proposed semantic segmentation network was less complex and could run faster because the backbone network was simplified.
Weed density assessment results
Weed assessment results of a sample are shown in Fig. 17. The results of the regression analysis between the algorithm-assessed weed density results and the manually-assessed weed density results are shown in Fig. 18. Firstly, it could be seen from Fig. 18a, regression analysis showed that the relationship between the algorithm assessed weed density and the manually observed weed density was \(y = 0.95x+0.02 \). The coefficient of determination \(R^2\) was 0.90, and the root means square error (\(\sigma \)) 0.05. This indicated that the correlation between the two was good. It was effective to evaluate weed density by the algorithm. The slope of the relationship was 0.95, and the intercept was 0.02. This indicates that the weed density assessed by the algorithm was slightly higher than the weed density assessed manually. From the images, it could be seen that some of the weeds were incorrectly segmented as non-weed. Therefore, the weed density assessed by the algorithm was slightly less than that assessed manually. The regression analysis of Green bristlegrass is shown in Figure 18b. Where the slope was 1.02, which indicates that the results of the algorithm assessment were higher than those of the manual assessment. Some of the other types of weeds were classified as Green bristlegrass by the algorithm. It could be found from images that some of the pixels at the edges of the other types of weeds were segmented as Green bristlegrass, which led to a higher density of Green bristlegrass evaluated by the algorithm. The same situation occurs in Figure 18d for Acalypha copperleaf. The situation for other types of weeds was consistent with the overall situation.
Discussion
In this paper, a method to assess the density of each species of weed by images was investigated. This method could accurately segment weed images and assessment of weed density in complex environments, where containing multiple weeds and different crops.
In previous studies, Mainstream methods usually employ object detection algorithms to detect crop locations (Jin et al., 2021). After getting the crop, all other locations were sprayed with herbicides. This method was effective in avoiding spraying to the crop. But it failed to accurately exclude bare ground. It also causes some herbicide waste and pollution. This study presents the first identification scheme for weeds. The subjects identified were five common weeds. This method can accurately identify the location and density of each weed. It can weed more precisely.
To solve the problem of difficult manual labeling samples, the training method in this research consisted of pre-training and fine-tuning. In the previous pre-training, the open-access datasets with labels are often used for training (Abdalla et al., 2019; Bosilj et al., 2020). However, there was no suitable labeled weed dataset available for this study. Therefore, a training method using automatically labeled samples was proposed for the first time. And these samples were automatically labeled by an algorithm. Meanwhile, this paper adopted single-class samples pre-training method for the first time. There were multiple types of weeds in the real samples. But due to the difficulty of labeling, in the pre-trained samples, there was only one type of weed on each sample. The experimental results proved that such samples can also be effective for pre-training the neural network. The training method in this paper reduced the number of manually labeled samples and made sample labeling easier. Labor and time were saved.
Due to the limitation of computational resources, the whole images could not be fed into the neural network for semantic segmentation at one time. This paper was the first to propose semantic segmentation after cutting the image into sub-images. For this purpose, the relationship between the size of the cut sub-images and the accuracy of the model was investigated. After an experiment, it was found that the error was smaller when the sub-images were larger. This was because the weed images were cut incomplete when the sub-images were too smaller. The neural network could not make an accurate classification based on the limited image.
Based on the characteristics of weeds, a new neural network structure was proposed. This neural network had less number of network layers in the encoding part. The number of convolution kernels became more. In the decoding part, the number of convolutional layers was reduced. The connections of the input layers were added. Such a neural network could operate faster and have better accuracy for detail segmentation than the classical neural network used in previous studies (Su et al., 2021; Huang et al., 2018; Khan et al., 2020).
Conclusions
This paper introduces a method for assessing multiple weed densities and species by images. A more efficient neural network structure was proposed. This network was trained by pre-training and fine-tuning. And a method to automatically label pre-training samples was proposed. To solve the limitation of computer resources, a method of cutting images into serval sub-images for processing was proposed and studied. By analyzing the experimental results, the following conclusions were obtained:
-
(1)
The image segmentation method by a combination of ExG and minimum error threshold could be used to automatically label the pre-training samples. The accuracy of these labels reaches 98.75%.
-
(2)
In the task of multiple weed segmentation, using images with only one type of weed on each image for pre-training was effective in improving model performance.
-
(3)
The larger the sub-image, the higher the accuracy obtained, as far as the computational conditions allow. The sub-image size of \(256\times 256\) or more was acceptable.
-
(4)
The proposed semantic segmentation network could achieve better weed segmentation results. The accuracy rate reached 97%. The average segmentation time of each sum-image was 56.28 ms. The coefficient of determination (\(R^2\)) between algorithm-assessed weed density and the manually-assessed weed density was 0.90, and the root means square error (\(\sigma \)) was 0.05.
-
(5)
The results show that the algorithm can effectively evaluate the density of various weeds. It provides a reference for accurate weed control for weed species.
References
Abdalla, A., Cen, H., Wan, L., Rashid, R., Weng, H., Zhou, W., & He, Y. (2019). Fine-tuning convolutional neural network with transfer learning for semantic segmentation of ground-level oilseed rape images in a field with high weed pressure. Computers and Electronics in Agriculture, 167, 105091.
Alenya, G., Dellen, B., Foix, S., & Torras, C. (2013). Robotized plant probing: Leaf segmentation utilizing time-of-flight data. IEEE Robotics & Automation Magazine, 20(3), 50–59.
Aversano, L., Bernardi, M. L., Cimitile, M., Iammarino, M., & Rondinella, S. (2020). Tomato diseases classification based on vgg and transfer learning. In IEEE international workshop on metrology for agriculture and forestry (MetroAgriFor) (pp. 129–133). IEEE.
Badrinarayanan, V., Kendall, A., & Cipolla, R. (2017). Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Transactions on Pattern Analysis and Machine Intelligence, 39(12), 2481–2495.
Bakhshipour, A., & Jafari, A. (2018). Evaluation of support vector machine and artificial neural networks in weed detection using shape features. Computers and Electronics in Agriculture, 145, 153–160.
Barbedo, J. G. A. (2018). Impact of dataset size and variety on the effectiveness of deep learning and transfer learning for plant disease classification. Computers and Electronics in Agriculture, 153, 46–53.
Berge, T., Aastveit, A., & Fykse, H. (2008). Evaluation of an algorithm for automatic detection of broad-leaved weeds in spring cereals. Precision Agriculture, 9(6), 391–405.
Bosilj, P., Aptoula, E., Duckett, T., & Cielniak, G. (2020). Transfer learning between crop types for semantic segmentation of crops versus weeds in precision agriculture. Journal of Field Robotics, 37(1), 7–19.
Champ, J., Mora-Fallas, A., Goëau, H., Mata-Montero, E., Bonnet, P., & Joly, A. (2020). Instance segmentation for the fine detection of crop and weed plants by precision agricultural robots. Applications in Plant Sciences, 8(7), e11373.
Chen, Y., Wu, Z., Zhao, B., Fan, C., & Shi, S. (2021). Weed and corn seedling detection in field based on multi feature fusion and support vector machine. Sensors, 21(1), 212.
Cubuk, E. D., Zoph, B., Shlens, J., Le, Q. V.(2020). Randaugment: Practical automated data augmentation with a reduced search space. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition workshops (pp. 702–703).
Deng, J., Dong, W., Socher, R., Li, L.-J., Li, K., & Fei-Fei, L. (2009). Imagenet: A large-scale hierarchical image database. In IEEE conference on computer vision and pattern recognition (pp. 248–255). IEEE.
Deng, Z., Sun, H., Zhou, S., Zhao, J., Lei, L., & Zou, H. (2018). Multi-scale object detection in remote sensing imagery with convolutional neural networks. ISPRS Journal of Photogrammetry and Remote Sensing, 145, 3–22.
DeVries, T., & Taylor, G. W. Improved regularization of convolutional neural networks with cutout. arXiv:1708.04552.
Dyrmann, M., Karstoft, H., & Midtiby, H. S. (2016). Plant species classification using deep convolutional neural network. Biosystems Engineering, 151, 72–80.
Ferreira, A. D. S., Freitas, D. M., Silva, G. G. D., Pistori, H., & Folhes, M. T. (2017). Weed detection in soybean crops using convnets. Computers and Electronics in Agriculture, 143, 314–324.
Fu, L., Gao, F., Wu, J., Li, R., Karkee, M., & Zhang, Q. (2020). Application of consumer rgb-d cameras for fruit detection and localization in field: A critical review. Computers and Electronics in Agriculture, 177, 105687.
Ge, L., Yang, Z., Sun, Z., Zhang, G., Zhang, M., Zhang, K., et al. (2019). A method for broccoli seedling recognition in natural environment based on binocular stereo vision and gaussian mixture model. Sensors, 19(5), 1132.
Ghasemi, A., & Zahediasl, S. (2012). Normality tests for statistical analysis: A guide for non-statisticians. International journal of endocrinology and metabolism, 10(2), 486.
Gongal, A., Amatya, S., Karkee, M., Zhang, Q., & Lewis, K. (2015). Sensors and systems for fruit detection and localization: A review. Computers and Electronics in Agriculture, 116, 8–19.
Hamuda, E., Glavin, M., & Jones, E. (2016). A survey of image processing techniques for plant extraction and segmentation in the field. Computers and Electronics in Agriculture, 125, 184–199.
Hsu, C.-Y., Shao, L.-J., Tseng, K.-K., & Huang, W.-T. (2019). Moon image segmentation with a new mixture histogram model. Enterprise Information Systems, 1–24.
Huang, H., Deng, J., Lan, Y., Yang, A., Deng, X., Wen, S., et al. (2018). Accurate weed mapping and prescription map generation based on fully convolutional networks using uav imagery. Sensors, 18(10), 3299.
Huang, H., Lin, L., Tong, R., Hu, H., Zhang, Q., Iwamoto, Y., Han, X., Chen, Y.-W., & Wu, J. (2020). Unet 3+: A full-scale connected unet for medical image segmentation. In ICASSP 2020-2020 IEEE international conference on acoustics, speech and signal processing (ICASSP) (pp. 1055–1059). IEEE.
Jin, X., Che, J., & Chen, Y. (2021). Weed identification using deep learning and image processing in vegetable plantation. IEEE Access, 9, 10940–10950.
Kalin, U., Lang, N., Hug, C., Gessler, A., & Wegner, J. D. (2019). Defoliation estimation of forest trees from ground-level images. Remote Sensing of Environment, 223, 143–153.
Kamilaris, A., & Prenafetaboldu, F. X. (2018). Deep learning in agriculture: A survey. Computers and Electronics in Agriculture, 147, 70–90.
Kazmi, W., Foix, S., Alenya, G., & Andersen, H. J. (2014). Indoor and outdoor depth imaging of leaves with time-of-flight and stereo vision sensors: Analysis and comparison. ISPRS Journal of Photogrammetry and Remote Sensing, 88, 128–146.
Kazmi, W., Garcia-Ruiz, F., Nielsen, J., Rasmussen, J., & Andersen, H. J. (2015). Exploiting affine invariant regions and leaf edge shapes for weed detection. Computers and Electronics in Agriculture, 118, 290–299.
Kemker, R., Salvaggio, C., & Kanan, C. (2018). Algorithms for semantic segmentation of multispectral remote sensing imagery using deep learning. ISPRS Journal of Photogrammetry and Remote Sensing, 145, 60–77.
Khan, A., Ilyas, T., Umraiz, M., Mannan, Z. I., & Kim, H. (2020). CED-Net: Crops and weeds segmentation for smart farming using a small cascaded encoder-decoder architecture. Electronics, 9(10), 1602.
Khan, M. J., Khan, H. S., Yousaf, A., Khurshid, K., & Abbas, A. (2018). Modern trends in hyperspectral image analysis: A review. IEEE Access, 6, 14118–14129.
Kusumam, K., Krajník, T., Pearson, S., Duckett, T., & Cielniak, G. (2017). 3D‐vision based detection, localization, and sizing of broccoli heads in the field. Journal of Field Robotics, 34(8), 1505−1518.
Lammie, C., Olsen, A., Carrick, T., & Azghadi, M. R. (2019). Low-power and high-speed deep fpga inference engines for weed classification at the edge. IEEE Access, 7, 51171–51184.
Le, V. N. T.,Truong, G., & Alameh, K. (2021). Detecting weeds from crops under complex field environments based on faster RCNN. In 2020 IEEE eighth international conference on communications and electronics (ICCE) (pp. 350–355). IEEE.
Lee, M.-K., Golzarian, M. R., & Kim, I. (2021). A new color index for vegetation segmentation and classification. Precision Agriculture, 22(1), 179–204.
Li, J., & Tang, L. (2018). Crop recognition under weedy conditions based on 3d imaging for robotic weed control. Journal of Field Robotics, 35(4), 596–611.
Lottes, P., Behley, J., Milioto, A., & Stachniss, C. (2018). Fully convolutional networks with sequential information for robust crop and weed detection in precision farming. IEEE Robotics and Automation Letters, 3(4), 2870–2877.
Meyer, G. E., & Neto, J. C. (2008). Verification of color vegetation indices for automated crop imaging applications. Computers and Electronics in Agriculture, 63(2), 282–293.
Pan, B., Shi, Z., & Xu, X. (2017). Mugnet: Deep learning for hyperspectral image classification using limited samples. ISPRS Journal of Photogrammetry and Remote Sensing, 145, 108–119.
Picon, A., Alvarezgila, A., Seitz, M., Ortizbarredo, A., Echazarra, J., & Johannes, A. (2019). Deep convolutional neural networks for mobile capture device-based crop disease classification in the wild. Computers and Electronics in Agriculture, 161, 280–290.
Rico-Fernández, M., Rios-Cabrera, R., Castelan, M., Guerrero-Reyes, H.-I., & Juarez-Maldonado, A. (2019). A contextualized approach for segmentation of foliage in different crop species. Computers and Electronics in Agriculture, 156, 378–386.
Rodrigo, M., Oturan, N., & Oturan, M. A. (2014). Electrochemically assisted remediation of pesticides in soils and water: A review. Chemical Reviews, 114(17), 8720–8745.
Sabzi, S., Abbaspour-Gilandeh, Y., & García-Mateos, G. (2018). A fast and accurate expert system for weed identification in potato crops using metaheuristic algorithms. Computers in Industry, 98, 80–89.
Stroppiana, D., Villa, P., Sona, G., Ronchetti, G., Candiani, G., Pepe, M., et al. (2018). Early season weed mapping in rice crops using multi-spectral uav data. International Journal of Remote Sensing, 39(15–16), 5432–5452.
Strudel, R., Garcia, R., Laptev, I., & Schmid, C. (2021). Segmenter: Transformer for semantic segmentation. In Proceedings of the IEEE/CVF international conference on computer vision (pp. 7262–7272).
Su, D., Qiao, Y., Kong, H., & Sukkarieh, S. (2021). Real time detection of inter-row ryegrass in wheat farms using deep learning. Biosystems Engineering, 204, 198–211.
Tiwari, O., Goyal, V., Kumar, P., Vij, S. (2019). An experimental set up for utilizing convolutional neural network in automated weed detection. In 2019 4th international conference on internet of things: Smart innovation and usages (IoT-SIU) (pp. 1–6). IEEE.
Too, E. C., Yujian, L., Njuki, S., & Yingchun, L. (2019). A comparative study of fine-tuning deep learning models for plant disease identification. Computers and Electronics in Agriculture, 161, 272–279.
Virtanen, P., Gommers, R., Oliphant, T. E., Haberland, M., Reddy, T., Cournapeau, D., Burovski, E., Peterson, P., Weckesser, W., Bright, J., van der Walt, S. J., Brett, M., Wilson, J., Millman, K. J., Mayorov, N., Nelson, A. R. J., Jones, E., Kern, R., Larson, E., Carey, C. J., Polat, İ., Feng, Y., Moore, E. W., VanderPlas, J., Laxalde, D., Perktold, J., Cimrman, R., Henriksen, I., Quintero, E. A., Harris, C. R., Archibald, A. M., Ribeiro, A. H., Pedregosa, F., & van Mulbregt, P. (2020). SciPy 1.0 contributors, SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nature Methods, 17, 261–272. https://doi.org/10.1038/s41592-019-0686-2.
Wang, A., Zhang, W., & Wei, X. (2019). A review on weed detection using ground-based machine vision and image processing techniques. Computers and Electronics in Agriculture, 158, 226–240.
Yun, S., Han, D., Oh, S. J., Chun, S., Choe, J., & Yoo, Y. (2019). Cutmix: Regularization strategy to train strong classifiers with localizable features. In Proceedings of the IEEE/CVF international conference on computer vision (pp. 6023–6032).
Zhang, C., Zou, K., & Pan, Y. (2020). A method of apple image segmentation based on color-texture fusion feature and machine learning. Agronomy, 10(7), 972.
Zhang, H., Cisse, M., Dauphin, Y. N., & Lopez-Paz, D. mixup: Beyond empirical risk minimization. arXiv:1710.09412.
Zhang, S., Huang, W., & Wang, Z. (2021). Combing modified grabcut, k-means clustering and sparse representation classification for weed recognition in wheat field. Neurocomputing, 452, 665–674.
Zou, K., Chen, X., Wang, Y., Zhang, C., & Zhang, F. (2021). A modified u-net with a specific data argumentation method for semantic segmentation of weed images in the field. Computers and Electronics in Agriculture, 187, 106242.
Zou, K., Ge, L., Zhang, C., Yuan, T., & Li, W. (2019). Broccoli seedling segmentation based on support vector machine combined with color texture features. IEEE Access, 7, 168565–168574.
Acknowledgements
This research was funded by National Key Research and Development Project (2019YFB1312303).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zou, K., Wang, H., Yuan, T. et al. Multi-species weed density assessment based on semantic segmentation neural network. Precision Agric 24, 458–481 (2023). https://doi.org/10.1007/s11119-022-09953-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11119-022-09953-9